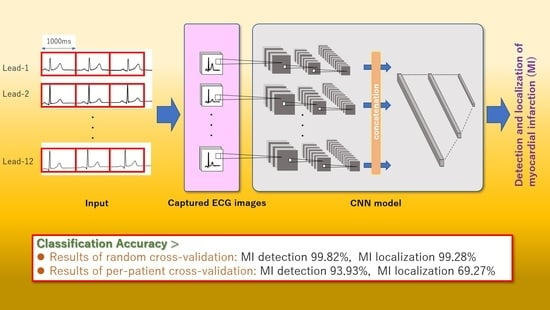
End-to-End Convolutional Neural Network Model to Detect and Localize Myocardial Infarction Using 12-Lead ECG Images without Preprocessing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Methods
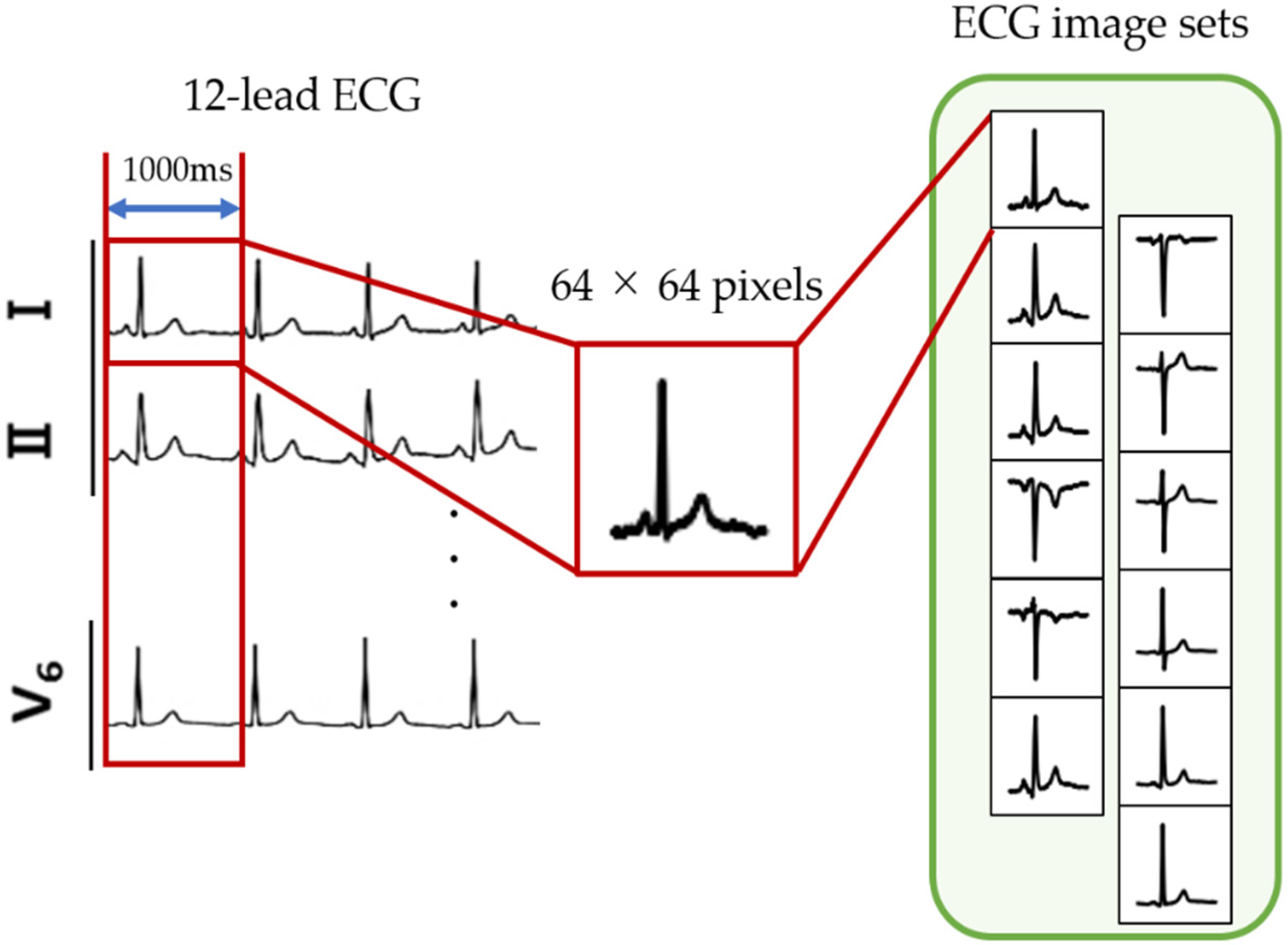
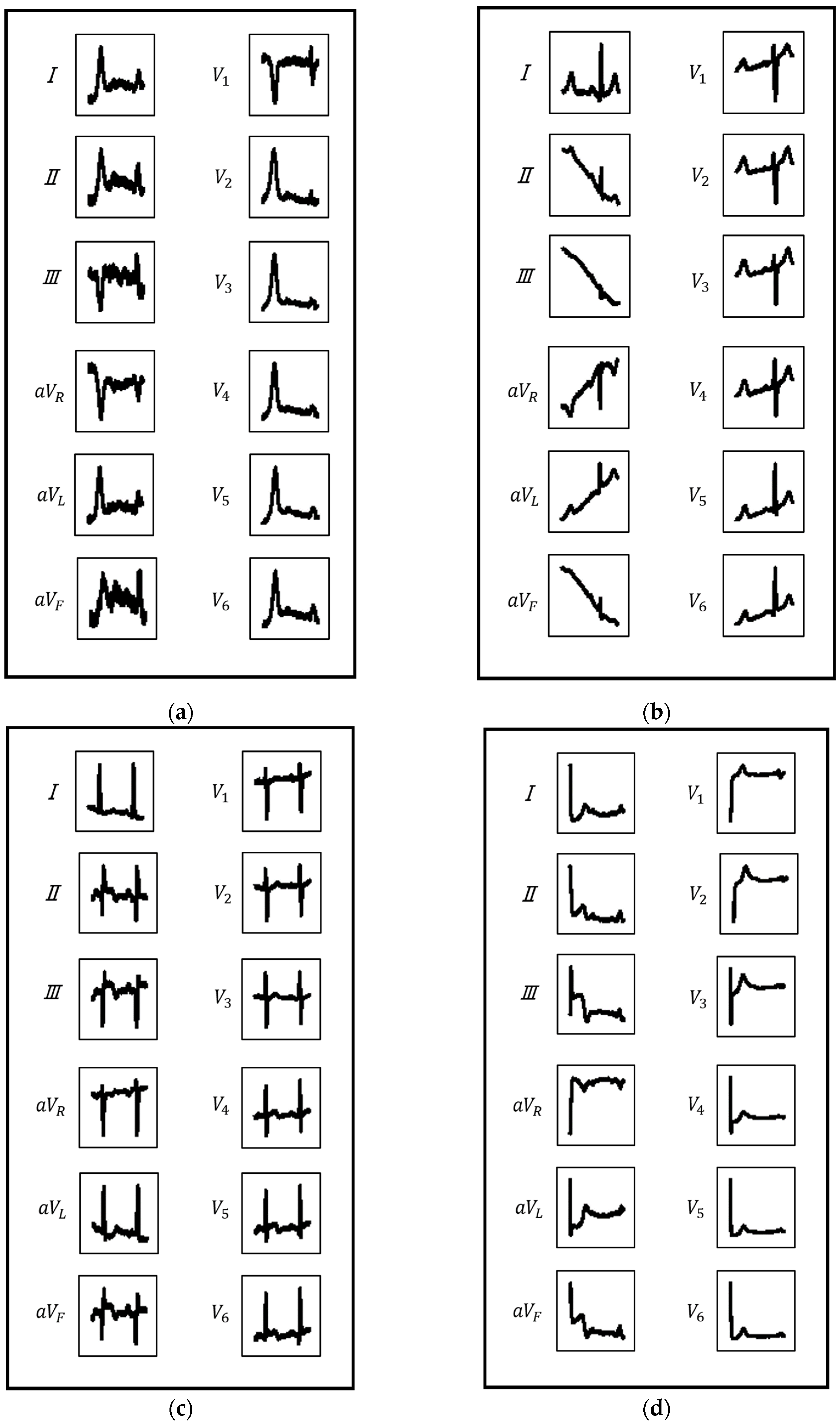
2.2.1. ECG Image Generation
2.2.2. CNN Model Construction and Classification
3. Experiments
3.1. Experimental Setup
- Setting 1:
- Setting 2:
3.2. Evaluation Indices
4. Experimental Results
4.1. Classification Results for Setting 1
4.1.1. MI Detection Results
4.1.2. MI Localization Results
4.2. Classification Results for Setting 2
4.2.1. MI Detection Results
4.2.2. MI Localization Results
5. Discussion
5.1. Results of Setting 1
5.2. Results of Setting 2
5.3. Study Advantages and Limitations
- The proposed model does not require complicated preprocessing, e.g., noise reduction, trend removal, beat segmentation, and feature selection.
- With the proposed model, it is possible to detect and localize MI by comprehensively checking the characteristics of the ECG images for each lead (similar to the diagnoses of medical professionals).
- It is possible to misclassify ECG images with extremely strong noise and trends.
- It is possible to misclassify ECG images with multiple beats or ECG images with most of the beats missing.
5.4. Discussion for Practical Application of the Model
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Heart, Lung, and Blood Institute. What Is a Heart Attack? Available online: https://www.nhlbi.nih.gov/health/heart-attack (accessed on 20 May 2022).
- Guyton, A.C.; Hall, J.E. Textbook of Medical Physiology, 11th ed.; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Simoons, M.J.; Chaitman, B.R.; White, H.D.; Bax, J.J.; Baumgartner, H.; Ceconi, C.; Dean, V.; et al. Third universal definition of myocardial infarction. Circulation 2012, 126, 2020–2035. [Google Scholar] [CrossRef] [PubMed]
- Timmis, A.D. Will serum enzymes and other proteins find a clinical application in the early diagnosis of myocardial infarction? Br. Heart J. 1994, 71, 309–310. [Google Scholar] [CrossRef]
- Lewis, K.M.; Handal, K.A. Sensible Analysis of the 12 Lead ECG, 1st ed.; Delmar Pub: Delmar, NY, USA, 2000. [Google Scholar]
- Timmis, A.D. Early diagnosis of acute myocardial infarction. Br. Med. J. 1990, 301, 941–942. [Google Scholar] [CrossRef] [PubMed]
- Meloni, L.; Marchetti, M.F.; Cacace, C.; Congia, M.; Scotto, R.; Caddeo, P.; Montisci, R. Prognosis and first diagnostic ECG in STEMI patients referred to the emergency medical system for primary PCI. J. Electrocardiol. 2018, 51, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Hayıroğlu, M.İ.; Türkkan, C.; Tekkeşin, A.İ. Ideal admission electrocardiographic parameters in STEMI: What else do we need to learn? J. Electrocardiol. 2020, 63, 187. [Google Scholar] [CrossRef]
- Hayıroğlu, M.İ.; Lakhani, I.; Tse, G.; Çınar, T.; Çinier, G.; Tekkeşin, A.İ. In-hospital prognostic value of electrocardiographic parameters other than ST-segment changes in acute myocardial infarction: Literature review and future perspectives. Heart. Lung. Circ. 2020, 29, 1603–1612. [Google Scholar] [CrossRef]
- Arif, M.; Malagore, I.A.; Afsar, F.A. Detection and localization of myocardial infarction using K-nearest neighbor classifier. J. Med. Syst. 2012, 36, 279–289. [Google Scholar] [CrossRef]
- Safdarian, N.; Dabanloo, N.J.; Attarodi, G. A new pattern recognition method for detection and localization of myocardial infarction using T-wave integral and total integral as extracted feature from one cycle of ECG signal. J. Biomed. Sci. Eng. 2014, 7, 818–824. [Google Scholar] [CrossRef]
- Sharma, L.N.; Tripathy, R.K.; Dandapat, S. Multiscale Energy and eigenspace approach to detection and localization of myocardial infarction. IEEE Trans. Biomed. Eng. 2015, 62, 1827–1837. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated Detection and localization of myocardial infarction using electrocardiogram: A comparative study of different leads. Knowl. Based. Syst. 2016, 99, 146–156. [Google Scholar] [CrossRef]
- Baloglu, U.B.; Talo, M.; Yildirim, O.; Tan, R.S.; Acharya, U.R. Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern. Recognit. Lett. 2019, 122, 23–30. [Google Scholar] [CrossRef]
- Sugimoto, K.; Kon, Y.; Lee, S.; Okada, Y. Detection and localization of myocardial infarction based on a convolutional autoencoder. Knowl. Based Syst. 2019, 178, 123–131. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, W.; Zhang, S.; Xu, L.; Zhu, B.; Cui, H.; Geng, N.; Greenwald, S.E. Detection and localization of myocardial infarction based on multi-scale resnet and attention mechanism. Front. Physiol. 2022, 13, 783184. [Google Scholar] [CrossRef]
- Jun, T.J.; Nguyen, H.M.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. ECG arrhythmia classification using a 2-D convolutional neural network. arXiv 2018, arXiv:1804.06812. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Stanley, H.E.; Mietus, J.E.; Moody, G.B.; Peng, C.K. PhysioBank, PhysioToolkit, and Physionet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, 215–220. [Google Scholar] [CrossRef]
- National Institute of General Medical Sciences and National Institute of Biomedical Imaging and Bioengineering, PhysioBank. Available online: https://physionet.org/physiobank/ (accessed on 20 May 2022).
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Chen, T.; Liu, T.; Wang, G.; Wang, X.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Boureau, Y.L.; Ponce, J.; LeCun, Y. A theoretical analysis of feature pooling in visual recognition. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 111–118. [Google Scholar]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. Lect. Notes Comput. Sci. 1999, 1681, 319–345. [Google Scholar]
- Dong, S.; Wang, P.; Abbas, K. A survey on deep learning and its applications. Comput. Sci. Rev. 2021, 40, 100379. [Google Scholar] [CrossRef]
- Liu, W.; Huang, Q.; Chang, S.; Wang, H.; He, J. Multiple-feature-branch convolutional neural network for myocardial infarction diagnosis using electrocardiogram. Biomed. Signal Process. Control 2018, 45, 22–32. [Google Scholar] [CrossRef]
- Remez, T.; Litany, O.; Giryes, R.; Bronstein, A.M. Class-aware fully convolutional Gaussian and Poisson denoising. IEEE Trans. Image Process. 2018, 27, 5707–5722. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual attention network for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 26 July 2017; pp. 3156–3164. [Google Scholar]
- Pires, R.G.; Santos, D.F.; Santos, C.F.; Santana, M.C.; Papa, J.P. Image denoising using attention-residual convolutional neural networks. arXiv 2021, arXiv:2101.07713. [Google Scholar]
- Fu, L.; Lu, B.; Nie, B.; Peng, Z.; Liu, H.; Pi, X. Hybrid network with attention mechanism for detection and location of myocardial infarction based on 12-lead Electrocardiogram signals. Sensors 2020, 20, 1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Shi, L. MLResNet: A novel network to detect and locate myocardial infarction using 12 leads ECG. Comput. Methods Programs Biomed. 2019, 185, 105–138. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Hayıroğlu, M.İ.; Çınar, T.; Çiçek, V.; Asal, S.; Kılıç, Ş.; Keser, N.; Orhan, A.L.; Uzun, M. A simple formula to predict echocardiographic diastolic dysfunction—electrocardiographic diastolic index. Herz 2021, 46, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; You, M.; Xu, W.; Li, J. Fully integer-based quantization for mobile convolutional neural network inference. Neurocomputing 2021, 432, 194–205. [Google Scholar] [CrossRef]
- Choudhary, T.; Mishra, V.; Goswami, A.; Sarangapani, J. Inference-aware convolutional neural network pruning. Future Gener. Comput. Syst. 2022, 135, 44–56. [Google Scholar] [CrossRef]

| Class (Abbreviation) | Number of Subjects | Number of ECG Data | Number of ECG Image Sets |
|---|---|---|---|
| Normal (N) | 51 | 74 | 4837 |
| Anterior (A) | 17 | 47 | 2812 |
| Anterior–Lateral (AL) | 14 | 39 | 2580 |
| Anterior–Septal (AS) | 27 | 77 | 4620 |
| Inferior (I) | 30 | 87 | 5268 |
| Inferior–Lateral (IL) | 23 | 55 | 3315 |
| Inferior–Posterior (IP) | 1 | 1 | 38 |
| Inferior–Posterior–Lateral (IPL) | 8 | 19 | 1118 |
| Lateral (L) | 1 | 3 | 180 |
| Posterior (P) | 1 | 4 | 240 |
| Posterior–Lateral (PL) | 2 | 5 | 300 |
| Total | 175 | 411 | 25,308 |
| Layer | Number of Input Nodes | Number of ECG Output Nodes | Kernel Size | Batch Normalization | Activation Function |
|---|---|---|---|---|---|
| Convolution 1 | True | ReLU | |||
| Convolution 2 | Ture | ReLU | |||
| Pooling 1 | False | - | |||
| Convolution 3 | True | ReLU | |||
| Convolution 4 | True | ReLU | |||
| Pooling 2 | False | - | |||
| Convolution 5 | True | ReLU | |||
| Convolution 6 | True | ReLU | |||
| Pooling 3 | False | - | |||
| The flattened vectors of the 12 leads are concatenated | |||||
| Fully connected 1 | 3456 | 2048 | - | True | ReLU |
| Fully connected 2 | 2048 | 1024 | - | True | ReLU |
| Fully connected 3 | 1024 | 11 | - | False | SoftMax |
| Loss function | Cross-entropy loss | ||||
| Optimizer | Adam | ||||
| Predicted Class | |||
|---|---|---|---|
| N | MI | ||
| True Class | N | 4822 | 15 |
| MI | 31 | 20,440 | |
| Index | Score |
|---|---|
| Sensitivity | 0.9985 |
| Specificity | 0.9969 |
| Accuracy | 0.9982 |
| Predicted Class | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | A | AL | AS | I | IL | IP | IPL | L | P | PL | Accuracy | ||
| True class | N | 4818 | 1 | 0 | 5 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0.9961 |
| A | 2 | 2782 | 3 | 15 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 0.9893 | |
| AL | 3 | 5 | 2551 | 17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0.9888 | |
| AS | 3 | 11 | 4 | 4590 | 9 | 1 | 0 | 1 | 0 | 1 | 0 | 0.9935 | |
| I | 6 | 5 | 0 | 5 | 5243 | 6 | 0 | 2 | 1 | 0 | 0 | 0.9953 | |
| IL | 1 | 1 | 3 | 1 | 14 | 3286 | 0 | 9 | 0 | 0 | 0 | 0.9913 | |
| IP | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 1.0000 | |
| IPL | 0 | 0 | 1 | 1 | 4 | 6 | 0 | 1106 | 0 | 0 | 0 | 0.9893 | |
| L | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 180 | 0 | 0 | 1.0000 | |
| P | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 236 | 0 | 0.9833 | |
| PL | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 297 | 0.9900 | |
| Total | 0.9928 | ||||||||||||
| Predicted Class | |||
|---|---|---|---|
| N | MI | ||
| True Class | N | 3718 | 1119 |
| MI | 390 | 19,623 | |
| Index | Score |
|---|---|
| Sensitivity | 0.9805 |
| Specificity | 0.7687 |
| Accuracy | 0.9393 |
| Predicted Class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | A | AL | AS | I | IL | IPL | PL | Accuracy | ||
| True class | N | 4012 | 24 | 21 | 85 | 515 | 132 | 26 | 22 | 0.8294 |
| A | 91 | 1524 | 646 | 329 | 177 | 45 | 0 | 0 | 0.5420 | |
| AL | 79 | 311 | 1692 | 329 | 152 | 16 | 1 | 0 | 0.6558 | |
| AS | 156 | 140 | 731 | 3239 | 207 | 145 | 1 | 1 | 0.7011 | |
| I | 471 | 158 | 167 | 75 | 3876 | 429 | 90 | 2 | 0.7358 | |
| IL | 118 | 64 | 10 | 113 | 790 | 2098 | 92 | 30 | 0.6329 | |
| IPL | 16 | 3 | 8 | 0 | 190 | 256 | 622 | 23 | 0.5564 | |
| PL | 13 | 0 | 9 | 4 | 59 | 59 | 6 | 150 | 0.5000 | |
| Total | 0.6927 | |||||||||
| Author (Year) | Methods | MI Detection Results | MI Localization Results |
|---|---|---|---|
| Arif et al., 2012 [10] | k-NN | Sensitivity = 99.97% | Accuracy = 98.8% |
| Specificity = 99.9% | |||
| Safdarian et al., 2014 [11] | • Probabilistic Neural Network (PNN) | Accuracy = 94% | Accuracy = 76% |
| • k-NN | |||
| • Multilayer Perceptron (MLP) | |||
| • Naive Bayes Classification | |||
| Sharma et al., 2015 [12] | • SVM-Lin | Accuracy = 96% | Accuracy = 99.58% |
| • SVM-RBF | Sensitivity = 93% | ||
| • k-NN | Specificity = 99% | ||
| Acharya et al., 2016 [13] | k-NN | Accuracy = 98.8% | Accuracy = 98.74% |
| Sensitivity = 99.45% | Sensitivity = 99.55% | ||
| Specificity = 96.27% | Specificity = 99.16% | ||
| Baloglu et al., 2019 [14] | Deep CNN | N/A | Accuracy = 99.78% |
| Sugimoto et al., 2019 [15] | • Convolutional autoencoder • k-NN | Accuracy = 99.87% | Accuracy = 99.88% |
| Sensitivity = 99.91% | Sensitivity = 99.12% | ||
| Specificity = 99.59% | Specificity = 99.92% | ||
| Cao et al., 2022 [17] | • SENet • Grad-CAM | Accuracy = 99.98% | Accuracy = 99.79% |
| Sensitivity = 99.94% | Sensitivity = 99.88% | ||
| Specificity = 99.94% | Specificity = 99.98% | ||
| Proposed model | CNN | Accuracy = 99.82% | Accuracy = 99.28% |
| Sensitivity = 99.85% | Sensitivity = 99.21% | ||
| Specificity = 99.69% | Specificity = 99.61% |
| Author (Year) | Methods | MI Detection Results | MI Localization Results |
|---|---|---|---|
| Fu et al., 2020 [30] | MLA-CNN-BiGRU | Accuracy = 96.50% | Accuracy = 62.94% |
| Sensitivity = 97.10% | Sensitivity = 63.97% | ||
| Specificity = 93.34% | Specificity = 63.00% | ||
| Han et al., 2020 [31] | ML-ResNet | Accuracy = 95.49% | Accuracy = 55.74% |
| Sensitivity = 94.85% | Sensitivity = 47.58% | ||
| Specificity = 97.37% | Specificity = 55.37% | ||
| Proposed model | CNN | Accuracy = 93.93% | Accuracy = 69.27% |
| Sensitivity = 98.05% | Sensitivity = 65.96% | ||
| Specificity = 76.87% | Specificity = 82.94% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uchiyama, R.; Okada, Y.; Kakizaki, R.; Tomioka, S. End-to-End Convolutional Neural Network Model to Detect and Localize Myocardial Infarction Using 12-Lead ECG Images without Preprocessing. Bioengineering 2022, 9, 430. https://doi.org/10.3390/bioengineering9090430
Uchiyama R, Okada Y, Kakizaki R, Tomioka S. End-to-End Convolutional Neural Network Model to Detect and Localize Myocardial Infarction Using 12-Lead ECG Images without Preprocessing. Bioengineering. 2022; 9(9):430. https://doi.org/10.3390/bioengineering9090430
Chicago/Turabian StyleUchiyama, Ryunosuke, Yoshifumi Okada, Ryuya Kakizaki, and Sekito Tomioka. 2022. "End-to-End Convolutional Neural Network Model to Detect and Localize Myocardial Infarction Using 12-Lead ECG Images without Preprocessing" Bioengineering 9, no. 9: 430. https://doi.org/10.3390/bioengineering9090430
APA StyleUchiyama, R., Okada, Y., Kakizaki, R., & Tomioka, S. (2022). End-to-End Convolutional Neural Network Model to Detect and Localize Myocardial Infarction Using 12-Lead ECG Images without Preprocessing. Bioengineering, 9(9), 430. https://doi.org/10.3390/bioengineering9090430