1. Introduction
Chronic kidney disease is among the leading causes of death globally. A recent medical report states that approximately 324 million people suffer from CKD globally [
1]. The glomerular filtration rate (GFR) is a widely used CKD screening test [
2]. Though CKD affects people worldwide, it is more prevalent in developing countries [
3]. Meanwhile, early detection is vital in reducing the progression of CKD. However, people from developing countries have not benefitted from early-stage CKD screening due to the cost of diagnosing the disease and limited healthcare infrastructure. While the global prevalence of CKD is reported to be 13.4% [
4], it is said to have a 13.9% prevalence in Sub-Saharan Africa [
5,
6]. Another study reported a 16% pooled prevalence of CKD in West Africa [
7], the highest in Africa. Numerous research works have specified that CKD is more prevalent in developing countries [
8]. Notably, it is reported that 1 out of every 10 persons suffers from CKD in South Asia, including Pakistan, India, Bhutan, Bangladesh, and Nepal [
3].
Therefore, several researchers have proposed machine learning (ML)-based methods for the early detection of CKD. These ML methods could provide effective, convenient, and low-cost computer-aided CKD diagnosis systems to enable early detection and intervention, especially in developing countries. Researchers have proposed different methods to detect CKD effectively using the CKD dataset [
9] available at the University of California, Irvine (UCI) machine learning repository. For example, Qin et al. [
9] proposed an ML approach for the early detection of CKD. The approach involved using the k-Nearest Neighbor (KNN) imputation to handle the missing values in the dataset. After filling the missing data, six ML classifiers were trained and tested with the preprocessed data. The classifiers include logistic regression, SVM, random forest, KNN, naïve Bayes, and a feed-forward neural network. Due to the misclassification of these classifiers, the authors developed an integrated classifier that uses a perceptron to combine the random forest and logistic regression classifiers, which produced an enhanced accuracy of 99.83%.
Meanwhile, Ebiaredoh-Mienye et al. [
10] proposed a method for improved medical diagnosis using an improved sparse autoencoder (SAE) network with a softmax layer. The neural network achieved sparsity through weight penalty, unlike the traditional sparse autoencoders that penalize the hidden layer activations. When used for CKD prediction, the proposed SAE achieved an accuracy of 98%. Chittora et al. [
11] studied how to effectively diagnose CKD using ML methods. The research employed seven algorithms, including the C5.0 decision tree, logistic regression, linear support vector machine (LSVM) with 𝐿1 and 𝐿2 norms, artificial neural network, etc. The authors studied the performance of the selected classifiers when they were trained under different experimental conditions. These conditions include instances where the complete and reduced feature sets were used to train the classifiers. The experimental results showed that the LSVM with
norm trained with the reduced feature set obtained an accuracy of 98.46%, which outperformed the other classifiers.
Furthermore, Silveira et al. [
12] developed a CKD prediction approach using a variety of resampling techniques and ML algorithms. The resampling techniques include the synthetic minority oversampling technique (SMOTE) and Borderline-SMOTE, while the classifiers include random forest, decision tree, and AdaBoost. The experimental results showed that the decision tree with the SMOTE technique achieved the best performance with 98.99%. Generally, these ML research works utilize many attributes such as albumin, hemoglobin level, white blood cell count, red blood cell count, packed cell volume, blood pressure, specific gravity, etc., to flag patients at risk of CKD, thereby allowing clinicians to provide early and cost-efficient medical intervention. Despite the attention given to CKD prediction using machine learning, only a few research works have focused on identifying the most relevant features needed to improve CKD detection [
13,
14,
15]. If identified correctly in suspected CKD patients, these features could be utilized for efficient computer-aided CKD diagnosis.
In machine learning tasks, the algorithms employ discriminative abilities of features in classifying the samples. The ML models’ performance relies not only on the specific training algorithm but also on the input data characteristics, such as the number of features and the correlation between the features [
16]. Moreover, in most ML applications, especially in medical diagnosis, all the input features may not have equal importance. The goal of feature selection is to remove redundant attributes from the input data, ensuring the training algorithm learns the data more effectively. By removing non-informative variables, the computational cost of building the model is reduced, leading to faster and more efficient learning with enhanced classification performance.
Filter- and wrapper-based methods are the two widely used feature selection mechanisms [
17]. Wrapper-based feature selection techniques use a classifier to build ML models with different predictor variables and select the variable subset that leads to the best model. In contrast, filter-based methods are statistical techniques independent of a learning algorithm used to compute the correlation between the predictor and independent variables [
18]. The predictor variables are scored according to their relevance to the target variable. The variables with higher scores are then used to build the ML model. Therefore, this research aims to use information gain (IG), a filter-based feature selection method, to identify the most relevant features for improved CKD detection. IG is a robust algorithm for evaluating the gain of the various features with respect to the target variable [
19]. The attributes with the least IG values are removed, and those whose IG values are above a particular threshold are used to train the classifiers.
Meanwhile, a significant challenge in applying machine learning algorithms for medical diagnosis is the imbalanced class problem [
20,
21]. Most ML classifiers underperform when trained with imbalanced datasets. Class imbalance implies there is an uneven distribution of samples in each class. The class with the most samples is the majority class, while the class with the lesser samples is the minority class. Imbalance learning can be divided into data and algorithm-level approaches [
22]. Data level methods are based on resampling techniques. Several studies have employed resampling techniques such as undersampling and oversampling to solve the class imbalance problem [
23,
24,
25]. In order to create a balanced dataset, undersampling methods remove samples from the majority class, while oversampling techniques artificially create and add more data in the minority class. However, there are limitations to using these resampling techniques. For example, the samples discarded from the majority class could be vital in efficiently training the classifiers [
20]. Therefore, several studies have resorted to using algorithm-level methods such as ensemble learning and cost-sensitive learning to effectively handle the imbalanced data instead of data-level techniques [
26,
27,
28,
29].
Ensemble learning is a breakthrough in ML research and application, which is used to obtain a very accurate classifier by combining two or more classifiers. Boosting [
30] and Bagging [
31] are widely used ensemble learning techniques. Adaptive boosting [
30] is a type of boosting technique that creates many classifiers by assigning weights to the training data and adjusting these weights after every boosting cycle. The wrongly classified training instances are given higher weights in the next iteration, whereas the weight of correctly predicted examples is decreased. However, the AdaBoost algorithm does not treat the minority class and majority class weight updates differently when faced with imbalanced data. Therefore, in this study, we develop an AdaBoost classifier that gives higher weight to examples in the minority class, thereby enhancing the prediction of the minority class samples and the overall classification performance. A cost-sensitive classifier is obtained by biasing the weighting technique to focus more on the minority class. Recent findings have demonstrated that cost-sensitive learning is an efficient technique suitable for imbalanced classification problems [
32,
33,
34]. The contribution of this study is to obtain the most important CKD attributes needed to improve the performance of CKD detection and develop a cost-sensitive AdaBoost classifier that gives more attention to samples in the minority class.
The rest of this paper is structured as follows:
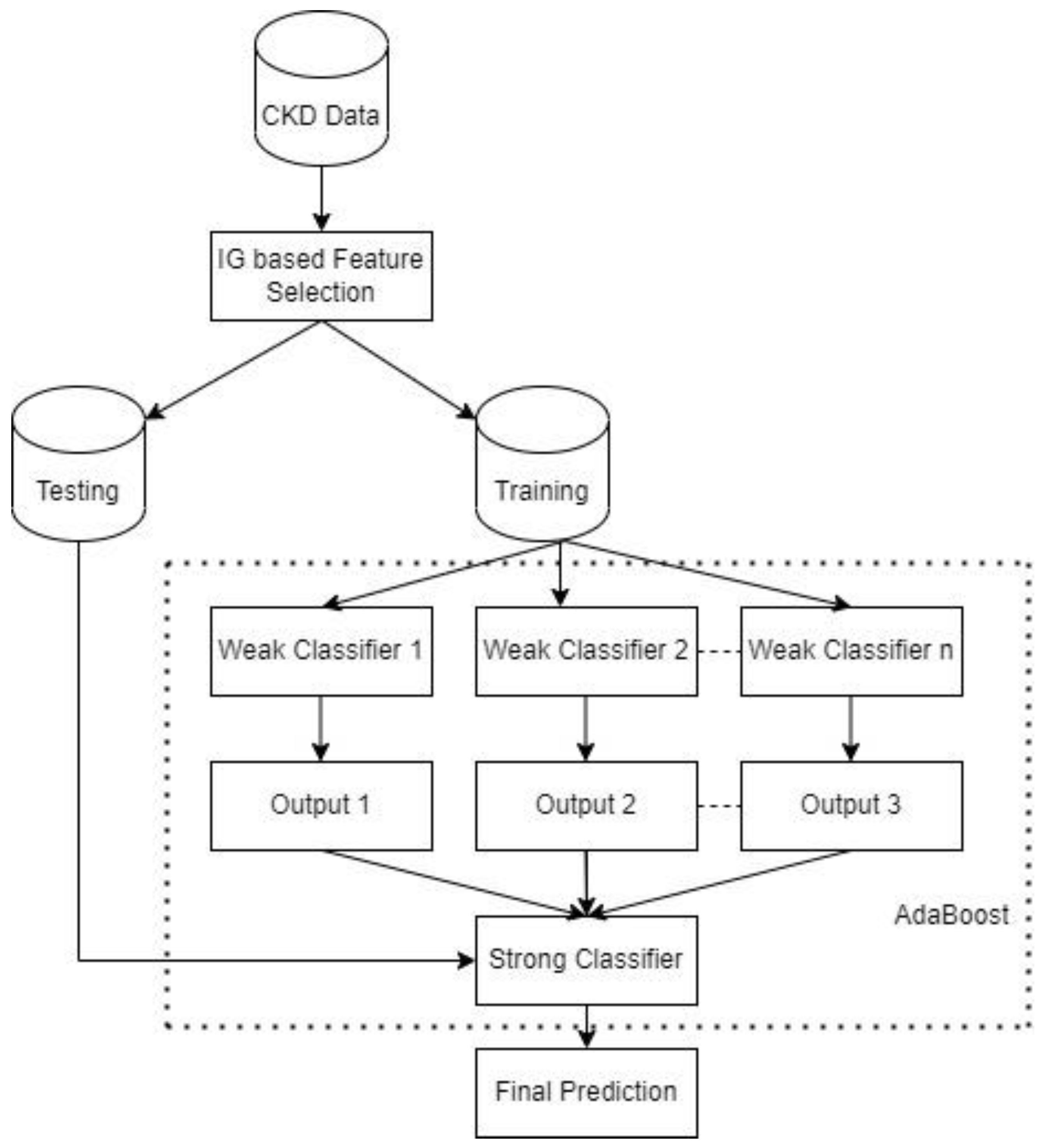
Section 2 presents the materials and methods, including an overview of the CKD dataset, the information gain technique, the traditional AdaBoost method, the proposed cost-sensitive AdaBoost, and the performance evaluation metrics.
Section 3 presents the experimental results and discussion, while
Section 4 concludes the paper.
3. Results
In this section, the experimental results are presented and discussed. All experiments used the preprocessed data, as discussed in
Section 2.1, and the ML models were developed using scikit-learn [
58], a machine learning library for Python programming. The experiments were conducted using a computer with the following specifications: Intel(R) Core(TM) i7-113H @ 3.30 GHz, 4 Core(s), and 16 GB RAM. Furthermore, to have a baseline for comparing the proposed cost-sensitive AdaBoost (CS AdaBoost), this research implements the traditional AdaBoost [
30] presented in Algorithm 1 and other well-known classifiers, including logistic regression [
59], decision tree [
60], XGBoost [
61], random forest [
62], and SVM [
63]. The classifiers are trained with the complete feature set and the reduced feature set to demonstrate the impact of the feature selection. Meanwhile, the 10-fold cross-validation method is employed to evaluate the performance of the various models. The decision tree algorithm is the base learner for the AdaBoost and CS AdaBoost implementations.
3.1. Performance of the Classifiers without Feature Selection
This subsection presents the experimental results obtained when the complete feature set was used to train the various classifiers. These results are tabulated in
Table 2. Additionally,
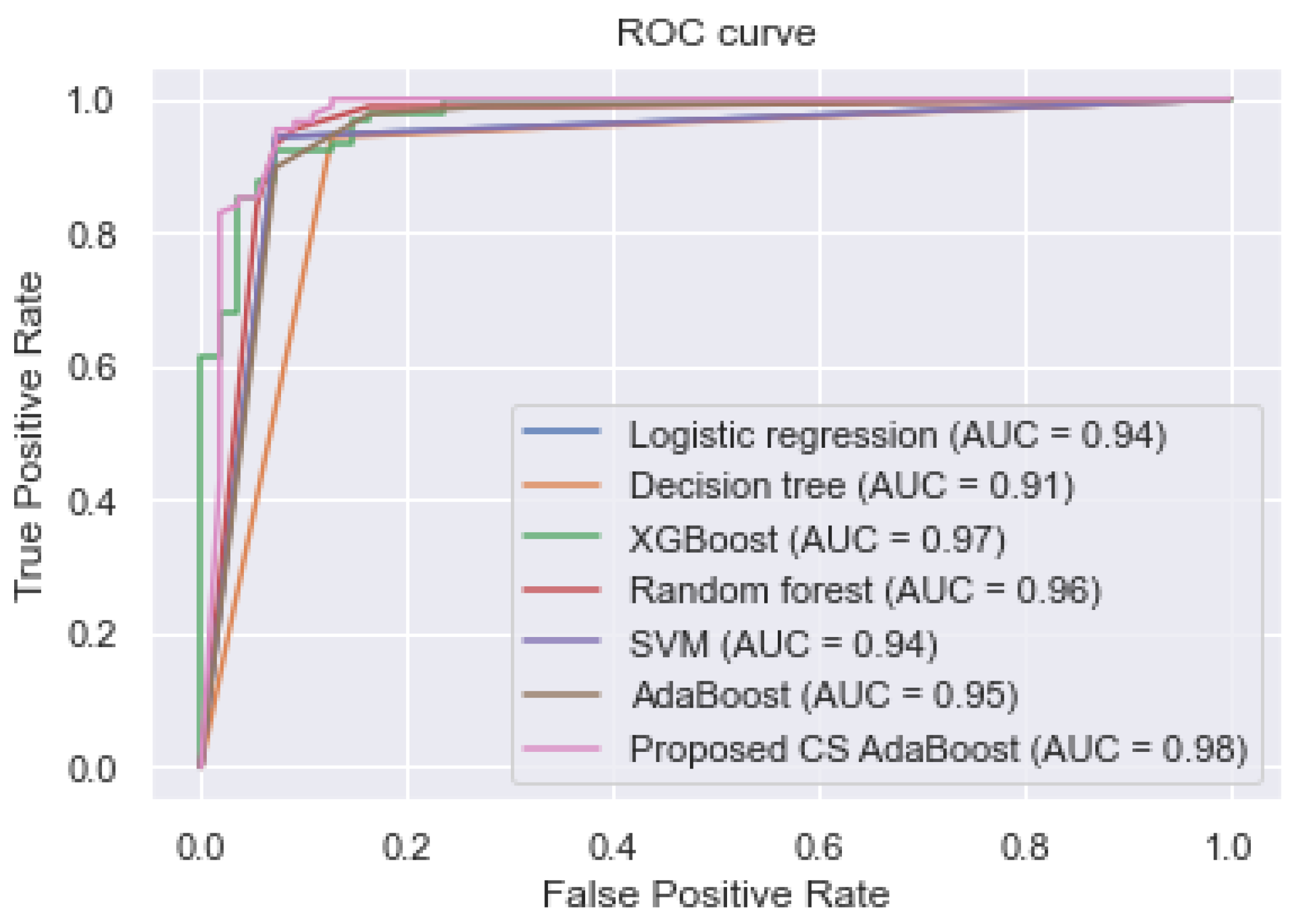
Figure 2 shows the AUC values and the ROC curves of the different classifiers.
Table 2 and
Figure 2 show that the proposed cost-sensitive AdaBoost obtained excellent performance by outperforming the traditional AdaBoost and the other classifiers, having obtained an AUC, accuracy, sensitivity, and specificity of 0.980, 0.967, 0.975, and 0.960, respectively.
3.2. Performance of the Classifiers after Feature Selection
The information-gain-based feature selection ranked the chronic kidney disease attributes. This step aims to select the features with the highest information gain with respect to the target variable. The ranked features and their IG values are shown in
Table 3. After obtaining the IG values of the various features, the standard deviation [
19] of the values is computed, which serves as the threshold value for the feature selection. The standard deviation measure has been used in recent research to obtain a reasonable threshold for feature selection [
19,
64,
65]. The threshold value obtained is 0.156. Therefore, the IG values equal to or greater than 0.156 are selected as the informative features and used for building the models. In contrast, the attributes with IG values lower than the threshold are discarded. Hence, from
Table 3, the top 18 features are selected as the optimal feature set since their IG values (rounded to three decimal values) are greater than 0.156, and the following features are discarded: f22, f5, f24, f8, f9, and f21.
To demonstrate the effectiveness of the feature selection, the reduced feature set is used to train the proposed CS AdaBoost and the other classifiers. The experimental results are shown in
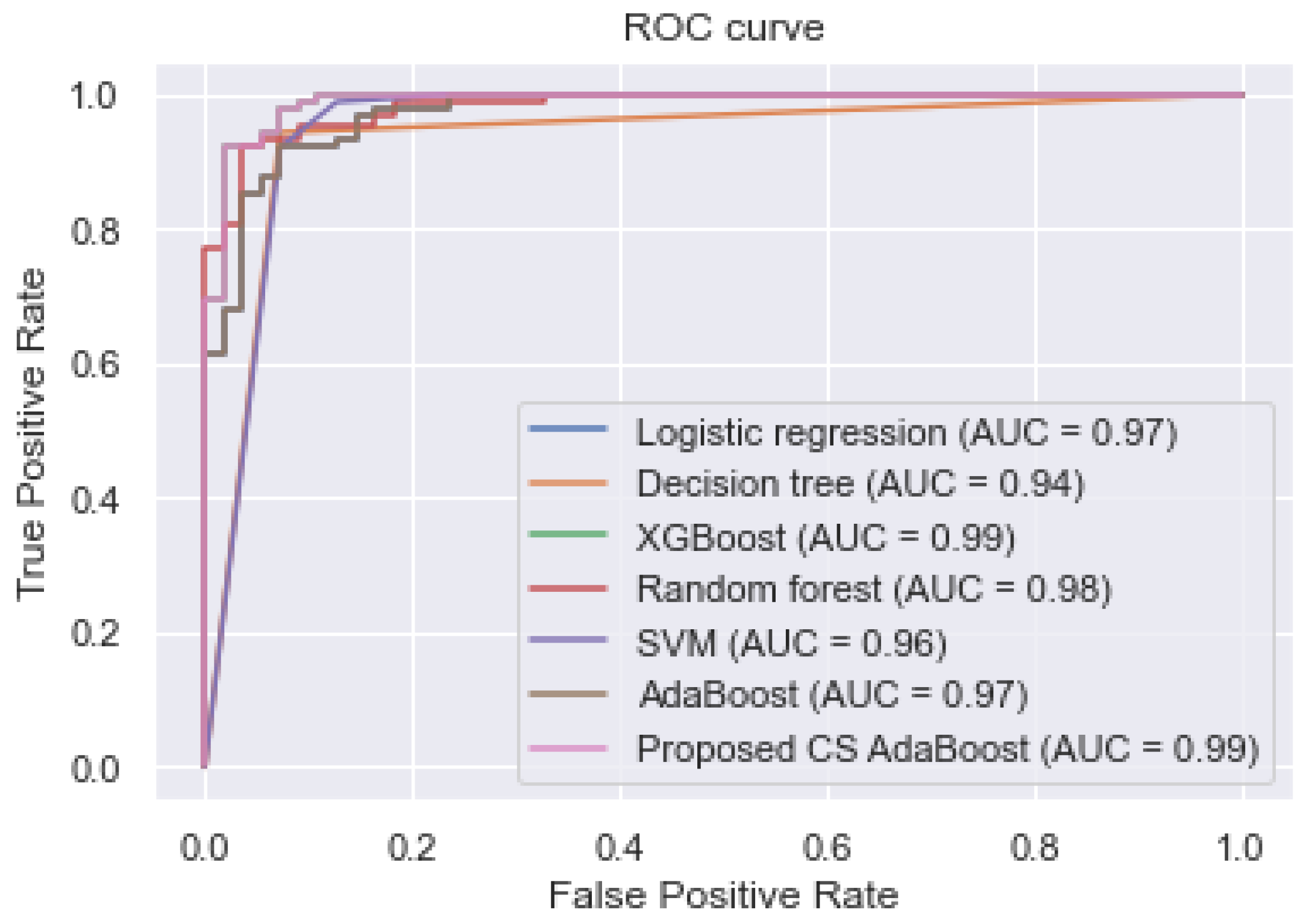
Table 4. Additionally, the ROC curve and the various AUC values are shown in
Figure 3. The experimental results in
Table 4 and
Figure 3 show that the proposed CS AdaBoost obtained an AUC, accuracy, sensitivity, and specificity of 0.990, 0.998, 1.000, and 0.998, respectively, which outperformed the logistic regression, decision tree, XGBoost, random forest, SVM, and conventional AdaBoost. Secondly, it is observed that the performance of the various classifiers in
Table 4 is better than their corresponding performance in
Table 2. This improvement demonstrates the effectiveness of the feature selection step. Therefore, the combination of feature selection and cost-sensitive AdaBoost is an effective method for predicting CKD.
3.3. Comparison with Other CKD Prediction Studies
Even though the proposed approach showed superior performance to the other algorithms, it is not enough to conclude its robustness. It is, however, necessary to compare it with other state-of-the-art methods in the literature. Hence, the proposed approach is compared with the following methods: a probabilistic neural network (PNN) [
66], an enhanced sparse autoencoder (SAE) neural network [
10], a naïve Bayes (NB) classifier with feature selection [
67], a feature selection method based on cost-sensitive ensemble and random forest [
3], a linear support vector machine (LSVM) and synthetic minority oversampling technique (SMOTE) [
11], a cost-sensitive random forest [
68], a feature selection method based on recursive feature elimination (RFE) and artificial neural network (ANN) [
69], a correlation-based feature selection (CFS) and ANN [
69]. The other methods include optimal subset regression (OSR) and random forest [
9], an approach to identify the essential CKD features using improved linear discriminant analysis (LDA) [
13], a deep belief network (DBN) with Softmax classifier [
70], a random forest (RF) classifier with feature selection (FS) [
71], a model based on decision tree and the SMOTE technique [
12], a logistic regression (LR) classifier with recursive feature elimination (RFE) technique [
14], and an XGBoost model with a feature selection approach combining the extra tree classifier (ETC), univariate selection (US), and RFE [
15].
The proposed approach based on cost-sensitive AdaBoost and feature selection achieved excellent performance compared to several state-of-the-art methods in the literature, as shown in
Table 5.
3.4. Discussion
This study aimed to solve two problems: first, to select the most informative features to enable the effective detection of CKD. The second aim was to develop an effective cost-sensitive AdaBoost classifier that accurately classifies samples in the minority class. The use of more features than necessary sometimes affects ML classifiers’ performance and increases the computational cost of training the models. Hence, this research employed the IG-based feature selection method to obtain the optimal feature set. Furthermore, seven classifiers were used in this study, trained using the complete and the reduced feature sets. From the experimental results, the proposed framework showed improved classification performance with the reduced feature set, i.e., 18 out of 24 input variables. Additionally, the models trained with the reduced feature set performed better than those trained with the complete feature set. Remarkably, the proposed method obtained higher performance than the other classifiers.
Furthermore, the features selected by the IG technique were similar to current medical practices. For example, the IG technique ranked albumin, hemoglobin, packed cell volume, red blood cell count, and serum creatinine as the most informative features, and numerous studies have identified a strong correlation between these variables and chronic kidney disease [
71,
72,
73,
74].
Meanwhile, the class imbalance problem is common in most real-world classification tasks. Another objective of this study was to develop a robust classifier to prevent the misclassification of the minority class that occurs when classifiers are trained using imbalanced data. Hence, this study developed a cost-sensitive AdaBoost classifier, giving more attention to the minority class. The experimental results indicate that the proposed method achieved a higher classification performance than the baseline classifiers and techniques in recent literature. Secondly, the results demonstrate that the combination of the information-gain-based feature selection and the cost-sensitive AdaBoost classifier significantly improved the detection of chronic kidney disease.
4. Conclusions
This paper proposed an approach that combines information-gain-based feature selection and a cost-sensitive AdaBoost classifier to improve the detection of chronic kidney disease. Six other machine learning classifiers were implemented as the baseline for performance comparison. The classifiers include logistic regression, decision tree, random forest, SVM, XGBoost, and the traditional AdaBoost. Firstly, the IG technique was used to compute and rank the importance of the various attributes. Secondly, the classifiers were trained with the reduced and complete feature sets. The experimental results show that selected features enhanced the performance of the classifiers.
Furthermore, the proposed cost-sensitive AdaBoost achieved superior performance to the other classifiers and methods in recent literature. Therefore, combining the IG-based feature selection technique and cost-sensitive AdaBoost is an effective approach for CKD detection and can be potentially applied for early detection of CKD through computer-aided diagnosis. Future research will focus on collecting large amounts of data to train ML models, including datasets that allow for the prediction of the disease severity, duration of the disease, and the age of onset.