Anti-Coagulant and Antimicrobial Recombinant Heparin-Binding Major Ampullate Spidroin 2 (MaSp2) Silk Protein
Abstract
:1. Introduction
2. Materials and Methods
Protein Expression and Purification
3. Protein Characterization
3.1. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
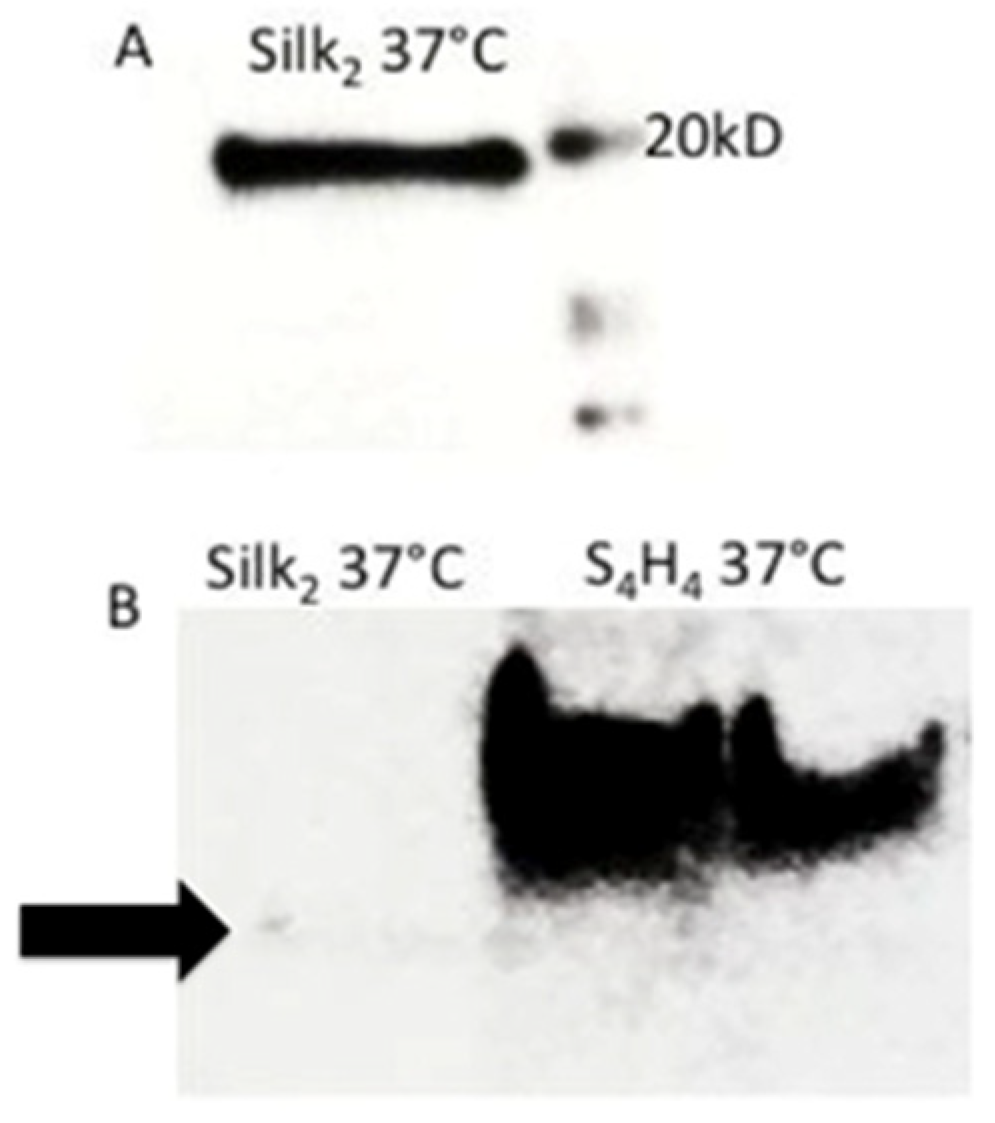
3.2. Western Blot
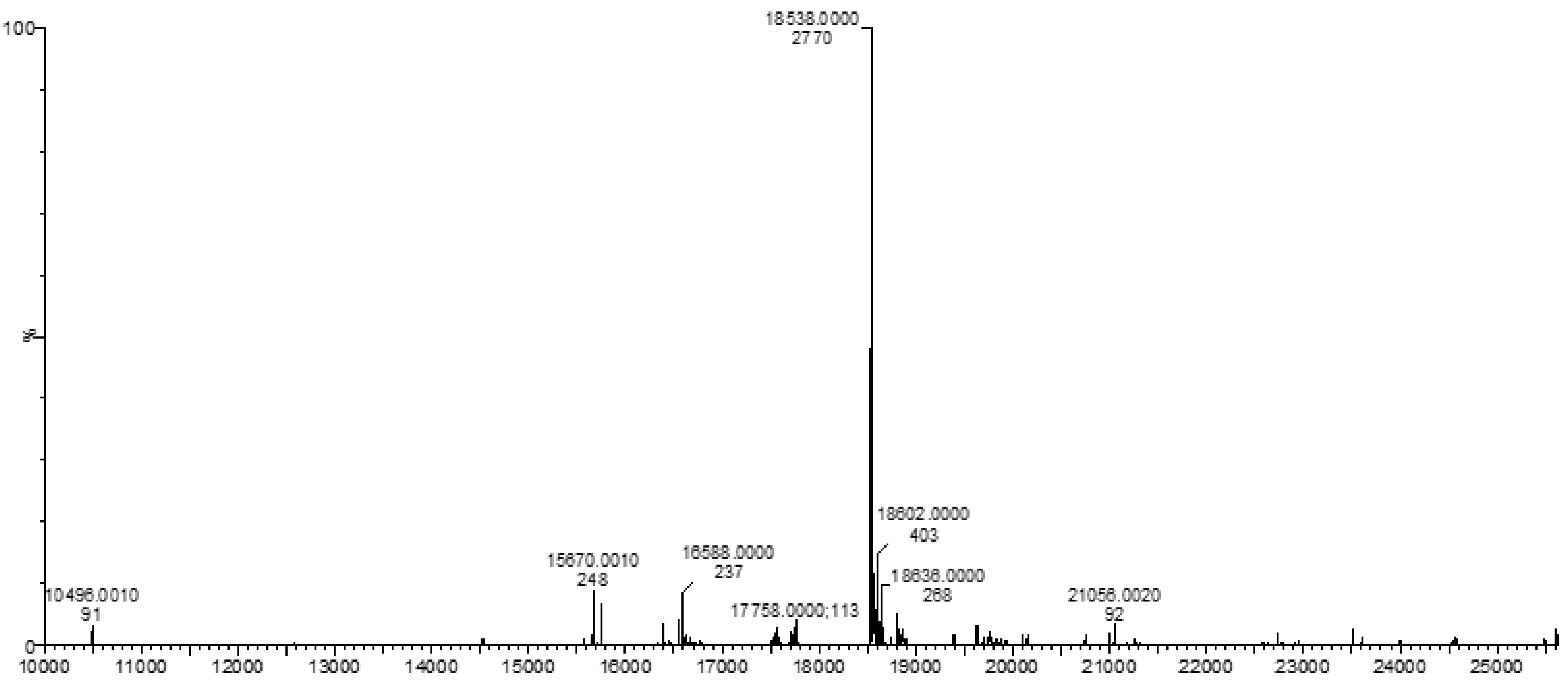
3.3. Mass Spectrometry (MS)
4. Heparin-Binding Characterization
4.1. Heparin Affinity Dot Blot
4.2. Heparin Affinity ELISA
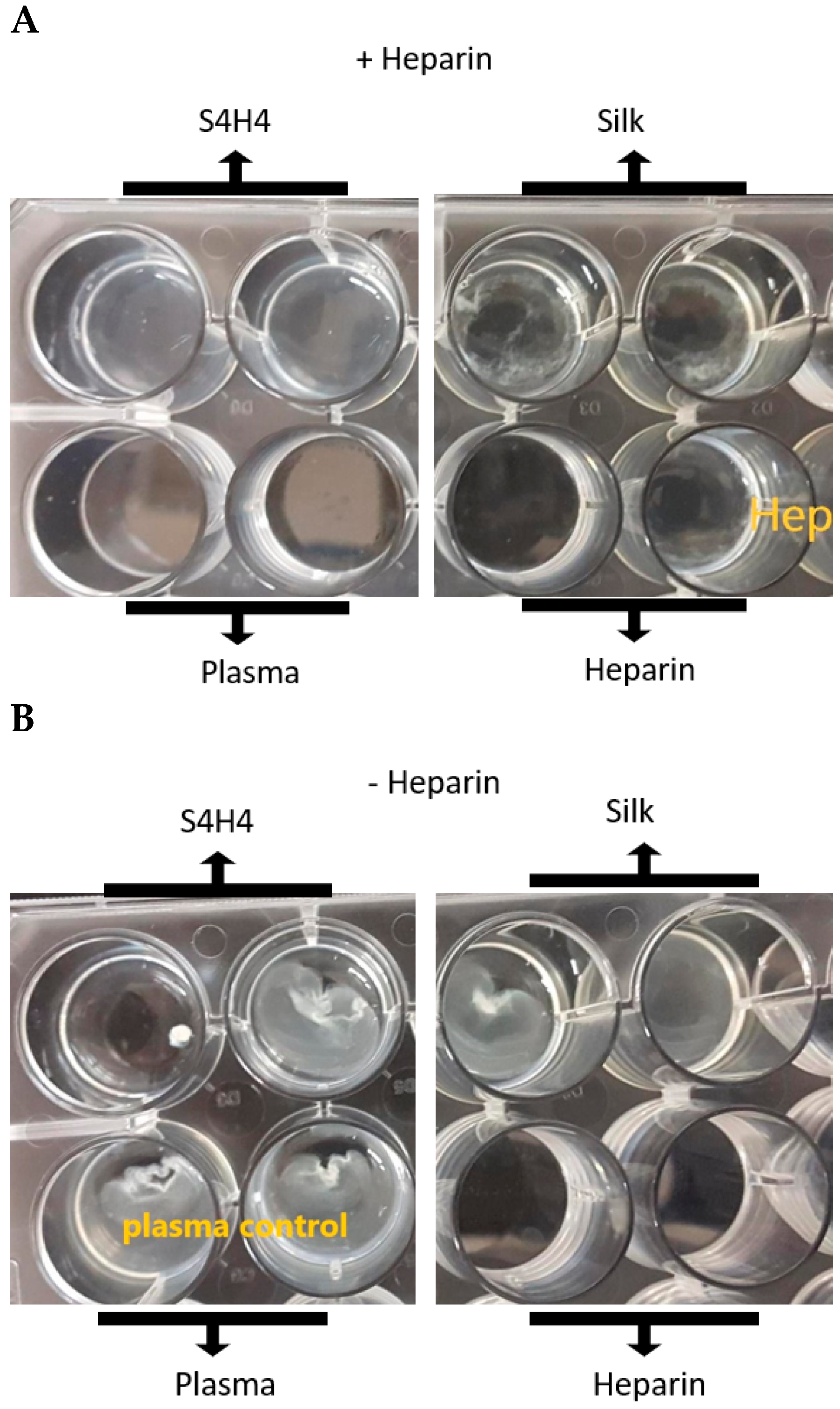
4.3. APTT Coagulation Assay
5. Antibacterial Activity
5.1. Kirby Bauer Zone of Inhibition Assay
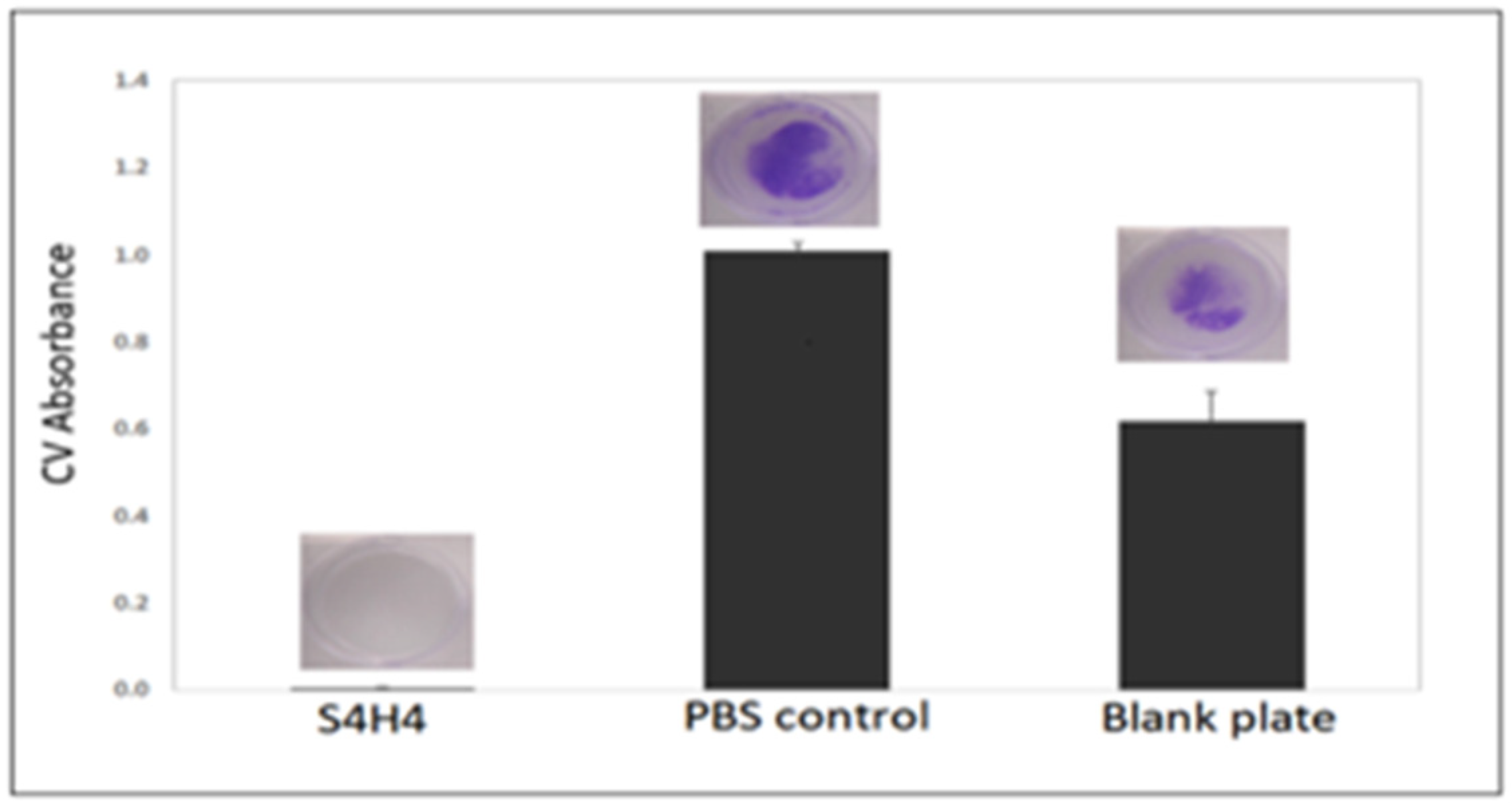
5.2. Biofilm Formation
6. Mechanical Testing of Silk Fibers
7. Statistical Analysis
8. Results
9. Discussion
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| S4H4 | fusion protein containing four repetitive units of silk and four repetitive units of heparin binding peptide |
| SX2 | two repetitive units of silk |
| HBM | heparin binding motif |
| AMP | antimicrobial peptide |
References
- Brooks, A.E.; Nelson, S.R.; Jones, J.A.; Koenig, C.; Hinman, M.; Stricker, S.; Lewis, R.V. Distinct contributions of model MaSp1 and MaSp2 like peptides to the mechanical properties of synthetic major ampullate silk fibers as revealed in silico. Nanotechnol. Sci. Appl. 2008, 1, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Hinman, M.B.; Jones, J.A.; Lewis, R.V. Synthetic spider silk: A modular fiber. Trends Biotechnol. 2000, 18, 374–379. [Google Scholar] [CrossRef]
- Chaw, R.C.; Correa-Garhwal, S.M.; Clarke, T.H.; Ayoub, N.A.; Hayashi, C.Y. Proteomic Evidence for Components of Spider Silk Synthesis from Black Widow Silk Glands and Fibers. J. Proteome Res. 2015, 14, 4223–4231. [Google Scholar] [CrossRef]
- Holland, G.P.; Jenkins, J.E.; Creager, M.S.; Lewis, R.V.; Yarger, J.L. Solid-state NMR investigation of major and minor ampullate spider silk in the native and hydrated states. Biomacromolecules 2008, 9, 651–657. [Google Scholar] [CrossRef]
- Blamires, S.J.; Tseng, Y.-H.; Wu, C.-L.; Toft, S.; Raubenheimer, D.; Tso, I.-M. Spider web and silk performance landscapes across nutrient space. Sci. Rep. 2016, 6, 26383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, C.Y.; Shipley, N.H.; Lewis, R.V. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int. J. Biol. Macromol. 1999, 24, 271–275. [Google Scholar] [CrossRef]
- Creager, M.S.; Jenkins, J.E.; Thagard-Yeaman, L.A.; Brooks, A.E.; Jones, J.A.; Lewis, R.V.; Holland, G.P.; Yarger, J.L. Solid-State NMR Comparison of Various Spiders’ Dragline Silk Fiber. Biomacromolecules 2010, 11, 2039–2043. [Google Scholar] [CrossRef] [Green Version]
- Yazawa, K.; Yamaguchi, E.; Knight, D.; Asakura, T. 13C solid-state NMR study of the 13C-labeled peptide, (E)8GGLGGQGAG(A)6GGAGQGGYGG as a model for the local structure of Nephila clavipes dragline silk (MaSp1) before and after spinning. Biopolymers 2012, 97, 347–354. [Google Scholar] [CrossRef]
- Ohgo, K.; Bagusat, F.; Asakura, T.; Scheler, U. Investigation of structural transition of regenerated silk fibroin aqueous solution by Rheo-NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 4182–4186. [Google Scholar] [CrossRef]
- Xiao, H.; Miller, S.J.; Bang, N.U.; Faulk, W.P. Protein-bound heparin/heparan sulfates in human adult and umbilical cord plasma. Haemostasis 1999, 29, 237–246. [Google Scholar] [CrossRef]
- Krishnaji, S.T.; Kaplan, D.L. Bioengineered chimeric spider silk-uranium binding proteins. Macromol. Biosci. 2013, 13, 256–264. [Google Scholar] [CrossRef]
- Gomes, S.; Gallego-Llamas, J.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Biological responses to spider silk-antibiotic fusion protein. J. Tissue Eng. Regen. Med. 2012, 6, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Canabady-Rochelle, L.L.; Belton, D.J.; Deschaume, O.; Currie, H.A.; Kaplan, D.L.; Perry, C.C. Bioinspired Silicification of Silica-Binding Peptide-Silk Protein Chimeras: Comparison of Chemically and Genetically Produced Proteins. Biomacromolecules 2012, 13, 683–690. [Google Scholar] [CrossRef]
- Wong Po Foo, C.; Patwardhan, S.V.; Belton, D.J.; Kitchel, B.; Anastasiades, D.; Huang, J.; Naik, R.R.; Perry, C.C.; Kaplan, D.L. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 9428–9433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, S.; Numata, K.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. AFM Study of Morphology and Mechanical Properties of a Chimeric Spider Silk and Bone Sialoprotein Protein for Bone Regeneration. Biomacromolecules 2011, 12, 1675–1685. [Google Scholar] [CrossRef] [Green Version]
- Scheller, J.; Henggeler, D.; Viviani, A.; Conrad, U. Purification of Spider Silk-elastin from Transgenic Plants and Application for Human Chondrocyte Proliferation. Transgenic Res. 2004, 13, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, W.; Liu, Q.; Zhang, S. Dual-functional composite with anticoagulant and antibacterial properties based on heparinized silk fibroin and chitosan. Colloids Surf. B Biointerfaces 2011, 85, 241–247. [Google Scholar] [CrossRef]
- Yu, H.; Muñoz, E.M.; Edens, R.E.; Linhardt, R.J. Heparin Regulation of the Complement System. In Chemistry and Biology of Heparin and Heparan Sulfate; Elsevier: Amsterdam, The Netherlands, 2005; pp. 313–343. ISBN 978-0-08-044859-6. [Google Scholar]
- Diekjürgen, D.; Grainger, D.W. Polysaccharide matrices used in 3D in vitro cell culture systems. Biomaterials 2017, 141, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Edward Conrad, H. Chapter 1—Heparin vs. Heparan Sulfate. In Heparin-Binding Proteins; Edward Conrad, H., Ed.; Academic Press: San Diego, CA, USA, 1998; pp. 1–5. ISBN 978-0-12-186060-8. [Google Scholar]
- Edward Conrad, H. Chapter 8—Heparin-Binding Proteins in Hemostasis. In Heparin-Binding Proteins; Edward Conrad, H., Ed.; Academic Press: San Diego, CA, USA, 1998; pp. 239–300. ISBN 978-0-12-186060-8. [Google Scholar]
- Edward Conrad, H. Chapter 4—Structural Modification of Heparinoids. In Heparin-Binding Proteins; Edward Conrad, H., Ed.; Academic Press: San Diego, CA, USA, 1998; pp. 115–136. ISBN 978-0-12-186060-8. [Google Scholar]
- Edward Conrad, H. Chapter 6—Heparinoid/Protein Interactions. In Heparin-Binding Proteins; Edward Conrad, H., Ed.; Academic Press: San Diego, CA, USA, 1998; pp. 183–202. ISBN 978-0-12-186060-8. [Google Scholar]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler. Off. J. Am. Heart Assoc. Inc. 1989, 9, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [Green Version]
- Martins, M.C.L.; Curtin, S.A.; Freitas, S.C.; Salgueiro, P.; Ratner, B.D.; Barbosa, M.A. Molecularly designed surfaces for blood deheparinization using an immobilized heparin-binding peptide. J. Biomed. Mater. Res. Part A 2009, 88, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Verrecchio, A.; Germann, M.W.; Schick, B.P.; Kung, B.; Twardowski, T.; Antonio, J.D.S. Design of Peptides with High Affinities for Heparin and Endothelial Cell Proteoglycans. J. Biol. Chem. 2000, 275, 7701–7707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmsten, M.; Kasetty, G.; Pasupuleti, M.; Alenfall, J.; Schmidtchen, A. Highly Selective End-Tagged Antimicrobial Peptides Derived from PRELP. PLoS ONE 2011, 6, e16400. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Choo, J.-E.; Choi, Y.-S.; Lee, K.-Y.; Min, D.-S.; Pi, S.-H.; Seol, Y.-J.; Lee, S.-J.; Jo, I.-H.; Chung, C.-P.; et al. Characterization of the surface immobilized synthetic heparin binding domain derived from human fibroblast growth factor-2 and its effect on osteoblast differentiation. J. Biomed. Mater. Res. Part A 2007, 83, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Vepari, C.; Matheson, D.; Drummy, L.; Naik, R.; Kaplan, D.L. Surface modification of silk fibroin with poly(ethylene glycol) for antiadhesion and antithrombotic applications. J. Biomed. Mater. Res. Part A 2010, 93, 595–606. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Castellot, J.; Herman, I.; Iafrati, M.; Kaplan, D.L. Controlled release from multilayer silk biomaterial coatings to modulate vascular cell responses. Biomaterials 2008, 29, 894–903. [Google Scholar] [CrossRef] [Green Version]
- Panilaitis, B.; Altman, G.H.; Chen, J.; Jin, H.J.; Karageorgiou, V.; Kaplan, D.L. Macrophage responses to silk. Biomaterials 2003, 24, 3079–3085. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, H.-J.; Vunjak-Novakovic, G.; Kaplan, D.L. Stem cell-based tissue engineering with silk biomaterials. Biomaterials 2006, 27, 6064–6082. [Google Scholar] [CrossRef]
- Nagaoka, M.; Jiang, H.-L.; Hoshiba, T.; Akaike, T.; Cho, C.-S. Application of recombinant fusion proteins for tissue engineering. Ann. Biomed. Eng. 2010, 38, 683–693. [Google Scholar] [CrossRef]
- Gomes, S.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Antimicrobial functionalized genetically engineered spider silk. Biomaterials 2011, 32, 4255–4266. [Google Scholar] [CrossRef] [Green Version]
- Belton, D.J.; Mieszawska, A.J.; Currie, H.A.; Kaplan, D.L.; Perry, C.C. Silk-silica composites from genetically engineered chimeric proteins: Materials properties correlate with silica condensation rate and colloidal stability of the proteins in aqueous solution. Langmuir ACS J. Surf. Colloids 2012, 28, 4373–4381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bini, E.; Foo, C.W.P.; Huang, J.; Karageorgiou, V.; Kitchel, B.; Kaplan, D.L. RGD-Functionalized Bioengineered Spider Dragline Silk Biomaterial. Biomacromolecules 2006, 7, 3139–3145. [Google Scholar] [CrossRef] [PubMed]
- Appelgren, P.; Ransjö, U.; Bindslev, L.; Espersen, F.; Larm, O. Surface heparinization of central venous catheters reduces microbial colonization in vitro and in vivo: Results from a prospective, randomized trial. Crit. Care Med. 1996, 24, 1482–1489. [Google Scholar] [CrossRef] [PubMed]
- Maxwell 16 Polyhistidine Protein Purification Kit Automated Protein Purification with Maximum Performance and Convenience. Available online: https://www.promega.com/resources/pubhub/promega-notes-2007/maxwell-16-polyhistidine-purification-kit-automated-protein-purification-with-maximum-performance/ (accessed on 7 December 2019).
- Teulé, F.; Cooper, A.R.; Furin, W.A.; Bittencourt, D.; Rech, E.L.; Brooks, A.; Lewis, R.V. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat. Protoc. 2009, 4, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, A.E.; Stricker, S.M.; Joshi, S.B.; Kamerzell, T.J.; Middaugh, C.R.; Lewis, R.V. Properties of Synthetic Spider Silk Fibers Based on Argiope aurantia MaSp2. Biomacromolecules 2008, 9, 1506–1510. [Google Scholar] [CrossRef]
- Najjam, S.; Mulloy, B.; Theze, J.; Gordon, M.; Gibbs, R.; Rider, C.C. Further characterization of the binding of human recombinant interleukin 2 to heparin and identification of putative binding sites. Glycobiology 1998, 8, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavan, S.A.V.; Dikshit, M. Recent advances in the status and targets of antithrombotic agents. Drugs Future 2002, 27, 669–688. [Google Scholar] [CrossRef]
- Hattori, T.; Kimura, K.; Seyrek, E.; Dubin, P.L. Binding of bovine serum albumin to heparin determined by turbidimetric titration and frontal analysis continuous capillary electrophoresis. Anal. Biochem. 2001, 295, 158–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kugel, A.J.; Jarabek, L.E.; Daniels, J.W.; Vander Wal, L.J.; Ebert, S.M.; Jepperson, M.J.; Stafslien, S.J.; Pieper, R.J.; Webster, D.C.; Bahr, J.; et al. Combinatorial materials research applied to the development of new surface coatings XII: Novel, environmentally friendly antimicrobial coatings derived from biocide-functional acrylic polyols and isocyanates. J. Coat. Technol. Res. 2009, 6, 107–121. [Google Scholar] [CrossRef]
- Stafslien, S.; Daniels, J.; Chisholm, B.; Christianson, D. Combinatorial materials research applied to the development of new surface coatings III. Utilisation of a high-throughput multiwell plate screening method to rapidly assess bacterial biofilm retention on antifouling surfaces. Biofouling 2007, 23, 37–44. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Adetunji, V.O.; Isola, T.O. Crystal Violet Binding Assay for Assessment of Biofilm Formation by Listeria monocytogenes and Listeria spp. on Wood, Steel and Glass Surfaces. Glob. Vet. 2011, 6, 6–10. [Google Scholar]
- Hoffmann, B.; Gruat-Henry, C.; Mulinti, P.; Jiang, L.; Brooks, B.D.; Brooks, A.E. Using Hydrodynamic Focusing to Predictably Alter the Diameter of Synthetic Silk Fibers. PLoS ONE 2018, 13, e0195522. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0195522 (accessed on 7 December 2019). [CrossRef] [Green Version]
- Kairaitis, L.K.; Gottlieb, T. Outcome and complications of temporary haemodialysis catheters. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc.-Eur. Ren. Assoc. 1999, 14, 1710–1714. [Google Scholar] [CrossRef] [Green Version]
- Stigter, M.; Bezemer, J.; de Groot, K.; Layrolle, P. Incorporation of different antibiotics into carbonated hydroxyapatite coatings on titanium implants, release and antibiotic efficacy. J. Control. Release Off. J. Control. Release Soc. 2004, 99, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Harbers, G.M.; Emoto, K.; Greef, C.; Metzger, S.W.; Woodward, H.N.; Mascali, J.J.; Grainger, D.W.; Lochhead, M.J. A functionalized poly(ethylene glycol)-based bioassay surface chemistry that facilitates bio-immobilization and inhibits non-specific protein, bacterial, and mammalian cell adhesion. Chem. Mater. Publ. Am. Chem. Soc. 2007, 19, 4405–4414. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, S.K.; Kwok, C.; Shen, M.; Horbett, T.A.; Ratner, B.D.; Bryers, J.D. Plasma-deposited membranes for controlled release of antibiotic to prevent bacterial adhesion and biofilm formation. J. Biomed. Mater. Res. 2000, 50, 160–170. [Google Scholar] [CrossRef]
- Rojas, I.A.; Slunt, J.B.; Grainger, D.W. Polyurethane coatings release bioactive antibodies to reduce bacterial adhesion. J. Control. Release Off. J. Control. Release Soc. 2000, 63, 175–189. [Google Scholar] [CrossRef]
- Ghandehari, H. Recombinant biomaterials for pharmaceutical and biomedical applications. Pharm. Res. 2008, 25, 672–673. [Google Scholar] [CrossRef] [PubMed]
- Vepari, C.; Kaplan, D.L. Silk as a Biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef]
- Elahi, M.F.; Guan, G.; Wang, L.; Zhao, X.; Wang, F.; King, M.W. Surface Modification of Silk Fibroin Fabric Using Layer-by-Layer Polyelectrolyte Deposition and Heparin Immobilization for Small-Diameter Vascular Prostheses. Langmuir 2015, 31, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Cestari, M.; Muller, V.; da Rodrigues, J.H.S.; Nakamura, C.V.; Rubira, A.F.; Muniz, E.C. Preparing silk fibroin nanofibers through electrospinning: Further heparin immobilization toward hemocompatibility improvement. Biomacromolecules 2014, 15, 1762–1767. [Google Scholar] [CrossRef]
- Seib, F.P.; Herklotz, M.; Burke, K.A.; Maitz, M.F.; Werner, C.; Kaplan, D.L. Multifunctional silk-heparin biomaterials for vascular tissue engineering applications. Biomaterials 2014, 35, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Li, G.; Zheng, Z.; Li, Y.; Han, Y.; Kaplan, D.L.; Wang, X. Silk fibroin-based woven endovascular prosthesis with heparin surface modification. J. Mater. Sci. Mater. Med. 2018, 29, 1–13. [Google Scholar] [CrossRef]
- Zamani, M.; Khafaji, M.; Naji, M.; Vossoughi, M.; Alemzadeh, I.; Haghighipour, N. A Biomimetic Heparinized Composite Silk-Based Vascular Scaffold with sustained Antithrombogenicity. Sci. Rep. 2017, 7, 4455. [Google Scholar] [CrossRef] [Green Version]
- Li, Y. Recombinant production of antimicrobial peptides in Escherichia coli: A review. Protein Expr. Purif. 2011, 80, 260–267. [Google Scholar] [CrossRef]
- Zorko, M.; Jerala, R. Production of recombinant antimicrobial peptides in bacteria. Methods Mol. Biol. Clifton N.J. 2010, 618, 61–76. [Google Scholar] [CrossRef]
- Muñoz, E.M.; Linhardt, R.J. Heparin-Binding Domains in Vascular Biology. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1549–1557. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osmolarity—An Overview|ScienceDirect Topics. Available online: https://www.sciencedirect.com/topics/immunology-and-microbiology/osmolarity (accessed on 15 March 2019).
- Meneghetti, M.C.Z.; Hughes, A.J.; Rudd, T.R.; Nader, H.B.; Powell, A.K.; Yates, E.A.; Lima, M.A. Heparan sulfate and heparin interactions with proteins. J. R. Soc. Interface 2015, 12, 20150589. [Google Scholar] [CrossRef] [Green Version]
- Andersson, E.; Rydengård, V.; Sonesson, A.; Mörgelin, M.; Björck, L.; Schmidtchen, A. Antimicrobial activities of heparin-binding peptides. Eur. J. Biochem. 2004, 271, 1219–1226. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Rangarajan, N.; Weisshaar, J.C. Lights, Camera, Action! Antimicrobial Peptide Mechanisms Imaged in Space and Time. Trends Microbiol. 2016, 24, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Gorr, S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, M.; Jasensky, J.; Foster, L.; Kuroda, K.; Chen, Z. Monitoring Antimicrobial Mechanisms of Surface-Immobilized Peptides in Situ. Langmuir 2018, 34, 2057–2062. [Google Scholar] [CrossRef]
- Liu, Z.; Brady, A.; Young, A.; Rasimick, B.; Chen, K.; Zhou, C.; Kallenbach, N.R. Length Effects in Antimicrobial Peptides of the (RW)n Series. Antimicrob. Agents Chemother. 2007, 51, 597–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenlund, P.; Lindberg, M.J.; Tibell, L.A.E. Structural Requirements for High-Affinity Heparin Binding: Alanine Scanning Analysis of Charged Residues in the C-Terminal Domain of Human Extracellular Superoxide Dismutase. Biochemistry 2002, 41, 3168–3175. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Fitzpatrick, A.W.; Meehan, S.; Mott, H.R.; Vendruscolo, M.; Dobson, C.M.; Welland, M.E. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 2007, 318, 1900–1903. [Google Scholar] [CrossRef] [PubMed]




| SX2 | GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA|GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA |
| S4H4 | GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA|GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA|GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA|GGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA| ARKKAAKA ARKKAAKA ARKKAAKA ARKKAAKA |
| Sample | Heparin Added | Clotting Time (min) |
|---|---|---|
| Heparin | + | No Clot |
| S4H4 | + | No Clot |
| SX2 | + | 13 ± 5 |
| Plasma only | + | 30 ± 6 |
| S4H4 | - | 9 ± 4 |
| SX2 | - | 9 ± 3 |
| Plasma only | - | 8 ± 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulinti, P.; Diekjürgen, D.; Kurtzeborn, K.; Balasubramanian, N.; Stafslien, S.J.; Grainger, D.W.; Brooks, A.E. Anti-Coagulant and Antimicrobial Recombinant Heparin-Binding Major Ampullate Spidroin 2 (MaSp2) Silk Protein. Bioengineering 2022, 9, 46. https://doi.org/10.3390/bioengineering9020046
Mulinti P, Diekjürgen D, Kurtzeborn K, Balasubramanian N, Stafslien SJ, Grainger DW, Brooks AE. Anti-Coagulant and Antimicrobial Recombinant Heparin-Binding Major Ampullate Spidroin 2 (MaSp2) Silk Protein. Bioengineering. 2022; 9(2):46. https://doi.org/10.3390/bioengineering9020046
Chicago/Turabian StyleMulinti, Pranothi, Dorina Diekjürgen, Kristen Kurtzeborn, Narayanaganesh Balasubramanian, Shane J. Stafslien, David W. Grainger, and Amanda E. Brooks. 2022. "Anti-Coagulant and Antimicrobial Recombinant Heparin-Binding Major Ampullate Spidroin 2 (MaSp2) Silk Protein" Bioengineering 9, no. 2: 46. https://doi.org/10.3390/bioengineering9020046
APA StyleMulinti, P., Diekjürgen, D., Kurtzeborn, K., Balasubramanian, N., Stafslien, S. J., Grainger, D. W., & Brooks, A. E. (2022). Anti-Coagulant and Antimicrobial Recombinant Heparin-Binding Major Ampullate Spidroin 2 (MaSp2) Silk Protein. Bioengineering, 9(2), 46. https://doi.org/10.3390/bioengineering9020046








