Automated Segmentation of Microvessels in Intravascular OCT Images Using Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Manual Labeling
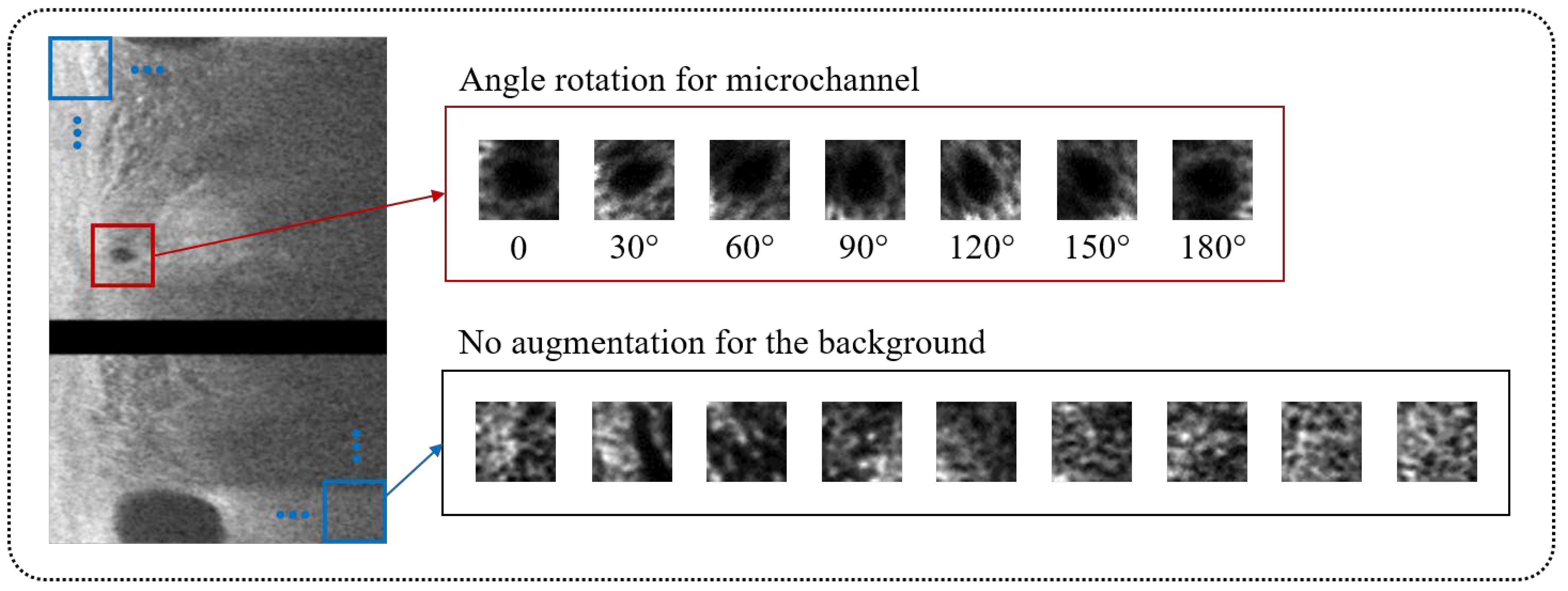
2.2. Data Augmentation
2.3. Pre-Processing
2.4. Segmentation of Microvessel Candidates Using DeepLab-v3+
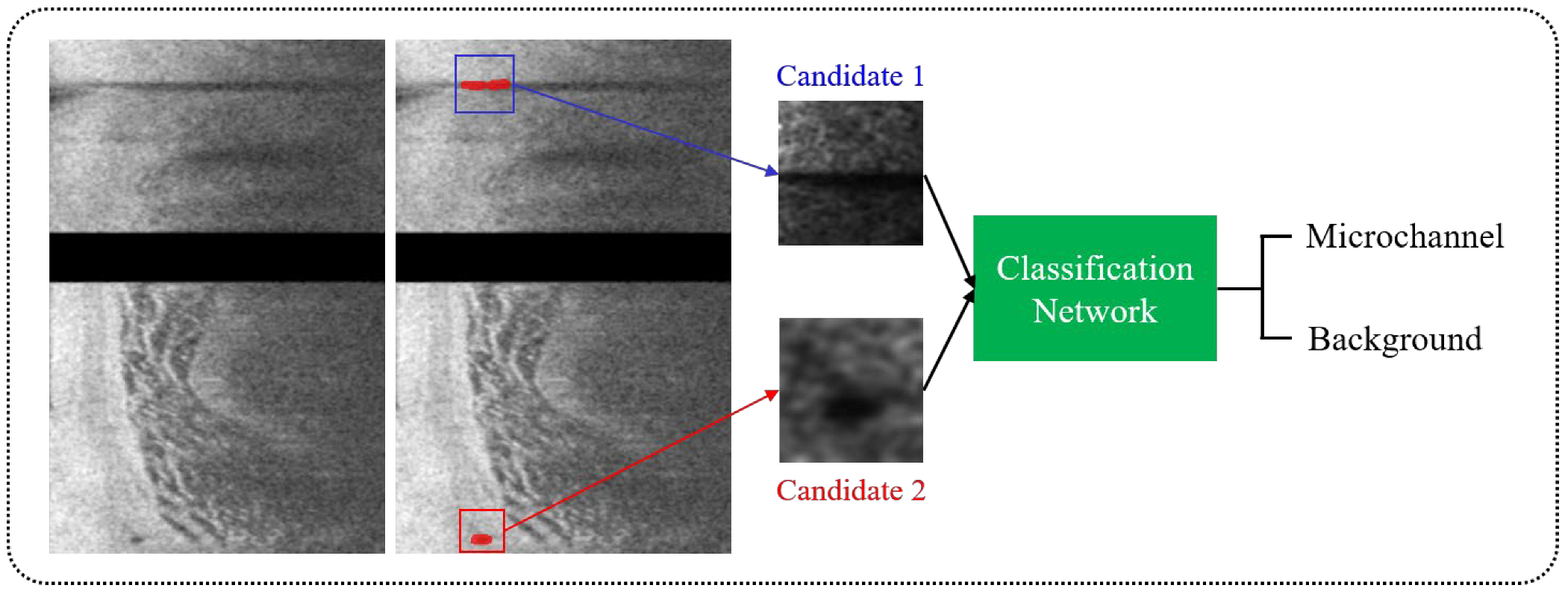
2.5. Classification of Microvessel Candidates Using a Shallow CNN
2.6. Network Training
2.7. Performance Evaluation
2.8. Statistical Analysis
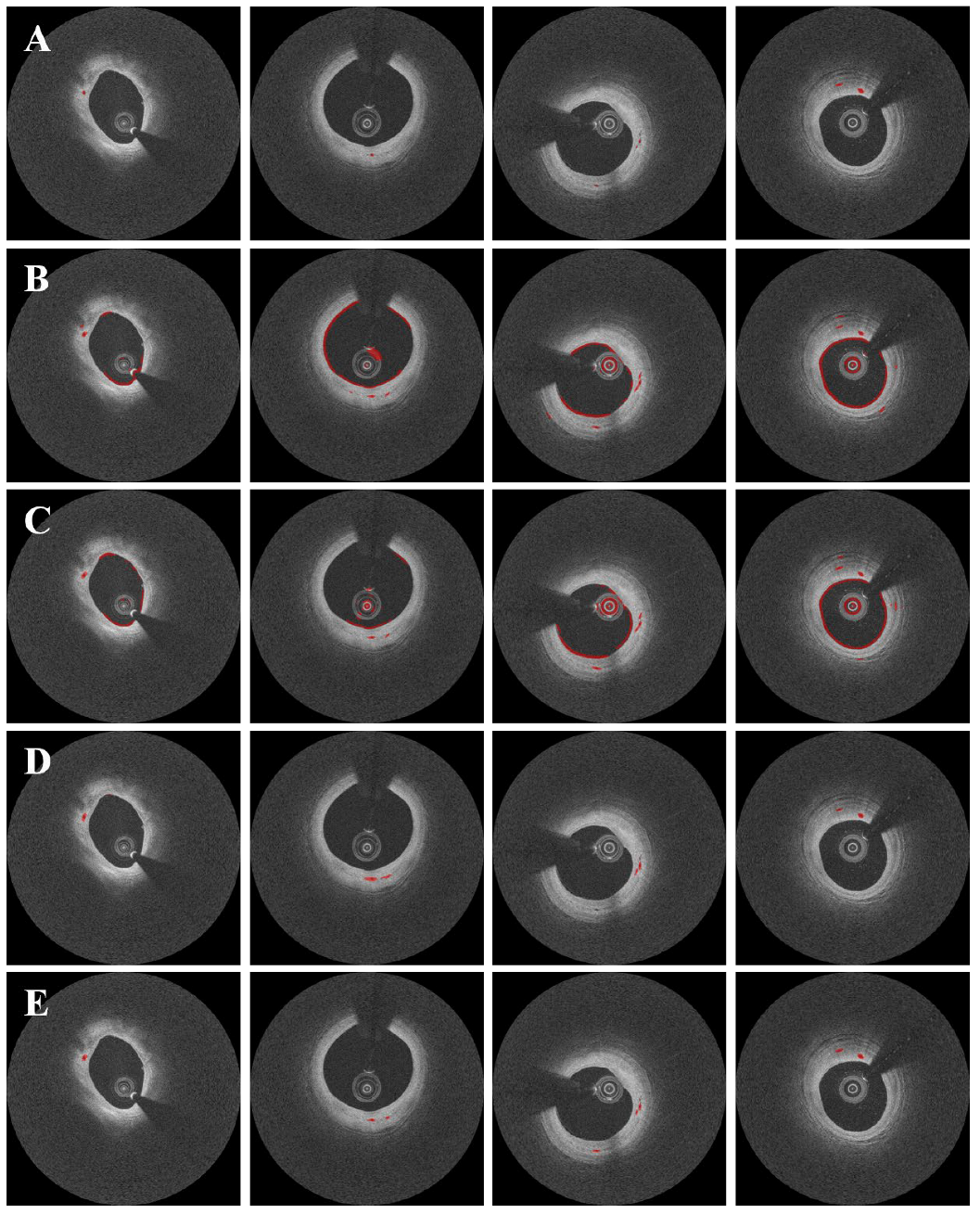
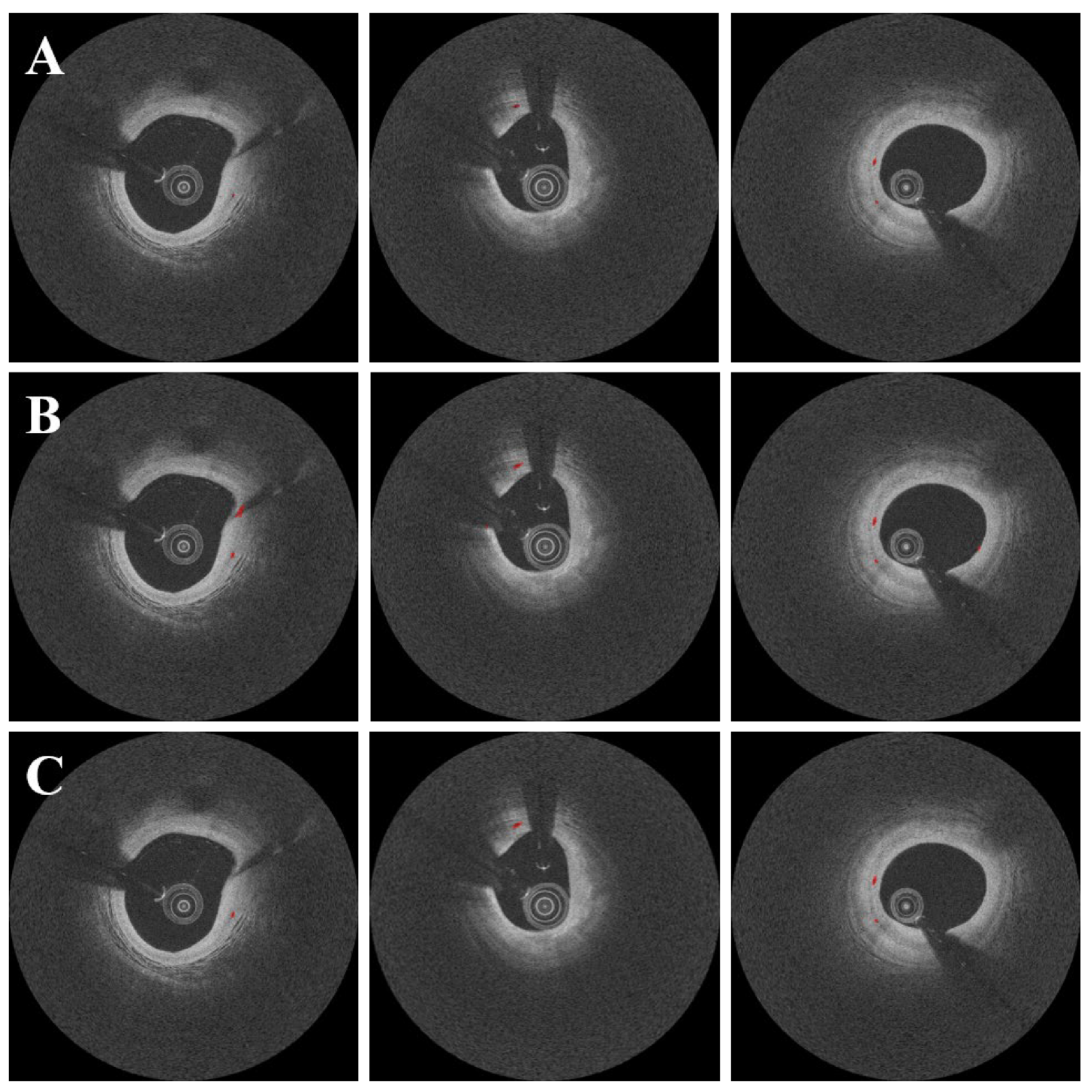
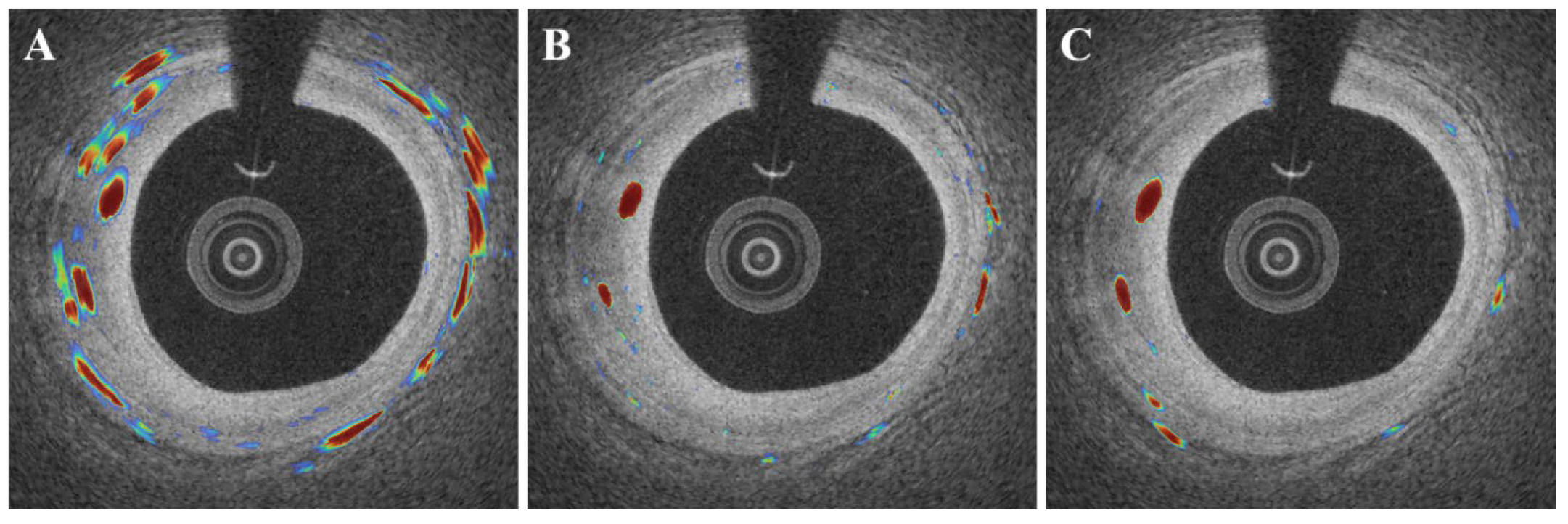
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Finn, A.V.; Nakano, M.; Narula, J.; Kolodgie, F.D.; Virmani, R. Concept of Vulnerable/Unstable Plaque. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, J.C.; Kolodgie, F.D.; Bijnens, A.P.J.J.; Maxfield, K.; Pacheco, E.; Kutys, B.; Duimel, H.; Frederik, P.M.; van Hinsbergh, V.W.M.; Virmani, R.; et al. Thin-Walled Microvessels in Human Coronary Atherosclerotic Plaques Show Incomplete Endothelial Junctions: Relevance of Compromised Structural Integrity for Intraplaque Microvascular Leakage. J. Am. Coll. Cardiol. 2009, 53, 1517–1527. [Google Scholar] [CrossRef]
- Kitabata, H.; Tanaka, A.; Kubo, T.; Takarada, S.; Kashiwagi, M.; Tsujioka, H.; Ikejima, H.; Kuroi, A.; Kataiwa, H.; Ishibashi, K.; et al. Relation of Microchannel Structure Identified by Optical Coherence Tomography to Plaque Vulnerability in Patients With Coronary Artery Disease. Am. J. Cardiol. 2010, 105, 1673–1678. [Google Scholar] [CrossRef]
- Bezerra, H.G.; Costa, M.A.; Guagliumi, G.; Rollins, A.M.; Simon, D.I. Intracoronary Optical Coherence Tomography: A Comprehensive Review Clinical and Research Applications. JACC Cardiovasc. Interv. 2009, 2, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Guagliumi, G.; Shimamura, K.; Sirbu, V.; Garbo, R.; Boccuzzi, G.; Vassileva, A.; Valsecchi, O.; Fiocca, L.; Canova, P.; Colombo, F.; et al. Temporal Course of Vascular Healing and Neoatherosclerosis after Implantation of Durable- or Biodegradable-Polymer Drug-Eluting Stents. Eur. Heart J. 2018, 39, 2448–2456. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, J.N.; Gharaibeh, Y.; Zimin, V.N.; Dallan, L.A.P.; Pereira, G.T.R.; Vergara-Martel, A.; Kolluru, C.; Hoori, A.; Bezerra, H.G.; et al. OCTOPUS—Optical Coherence Tomography Plaque and Stent Analysis Software. arXiv 2022. [Google Scholar] [CrossRef]
- Gharaibeh, Y.; Prabhu, D.S.; Kolluru, C.; Lee, J.; Zimin, V.; Bezerra, H.G.; Wilson, D.L. Coronary Calcification Segmentation in Intravascular OCT Images Using Deep Learning: Application to Calcification Scoring. J. Med. Imaging 2019, 6, 045002. [Google Scholar] [CrossRef]
- Kolluru, C.; Prabhu, D.; Gharaibeh, Y.; Bezerra, H.; Guagliumi, G.; Wilson, D. Deep Neural Networks for A-Line-Based Plaque Classification in Coronary Intravascular Optical Coherence Tomography Images. J. Med. Imaging 2018, 5, 044504. [Google Scholar] [CrossRef]
- Lee, J.; Prabhu, D.; Kolluru, C.; Gharaibeh, Y.; Zimin, V.N.; Bezerra, H.G.; Wilson, D.L. Automated Plaque Characterization Using Deep Learning on Coronary Intravascular Optical Coherence Tomographic Images. Biomed. Opt. Express 2019, 10, 6497–6515. [Google Scholar] [CrossRef]
- Wang, Z.; Kyono, H.; Bezerra, H.G.; Wilson, D.L.; Costa, M.A.; Rollins, A.M. Automatic Segmentation of Intravascular Optical Coherence Tomography Images for Facilitating Quantitative Diagnosis of Atherosclerosis. In Proceedings of the Optical Coherence Tomography and Coherence Domain Optical Methods in Biomedicine XV, San Francisco, CA, USA, 22–27 January 2011; Fujimoto, J.G., Izatt, J.A., Tuchin, V.V., Eds.; SPIE: Bellingham, WA, USA, 2011; p. 78890N. [Google Scholar]
- Cormen, T.H.; Leiserson, C.E.; Rivest, R.L.; Stein, C. Introduction to Algorithms, 3rd ed.; MIT Press: Cambridge, MA, USA, 2009; ISBN 978-0-262-03384-8. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Lee, J.; Kim, J.N.; Pereira, G.T.R.; Gharaibeh, Y.; Kolluru, C.; Zimin, V.N.; Dallan, L.A.P.; Motairek, I.K.; Hoori, A.; Guagliumi, G.; et al. Automatic Microchannel Detection Using Deep Learning in Intravascular Optical Coherence Tomography Images. In Proceedings of the Medical Imaging 2022: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, CA, USA, 20 February–28 March 2022; Volume 12034, pp. 166–173. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 3–6 December 2012; Curran Associates Inc.: Red Hook, NY, USA, 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Wang, Z.; Chamié, D.; Bezerra, H.G.; Yamamoto, H.; Kanovsky, J.; Wilson, D.L.; Costa, M.A.; Rollins, A.M. Volumetric Quantification of Fibrous Caps Using Intravascular Optical Coherence Tomography. Biomed. Opt. Express 2012, 3, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Prabhu, D.; Kolluru, C.; Gharaibeh, Y.; Zimin, V.N.; Dallan, L.A.P.; Bezerra, H.G.; Wilson, D.L. Fully Automated Plaque Characterization in Intravascular OCT Images Using Hybrid Convolutional and Lumen Morphology Features. Sci. Rep. 2020, 10, 2596. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Lee, J.; Jakl, M.; Wang, Z.; Cervinka, P.; Bezerra, H.G.; Wilson, D.L. Application and Evaluation of Highly Automated Software for Comprehensive Stent Analysis in Intravascular Optical Coherence Tomography. Sci. Rep. 2020, 10, 2150. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Lee, J.; Ray, S.; Tanaka, K.; Bezerra, H.G.; Rollins, A.M.; Wilson, D.L. Automated Stent Coverage Analysis in Intravascular OCT (IVOCT) Image Volumes Using a Support Vector Machine and Mesh Growing. Biomed. Opt. Express 2019, 10, 2809–2828. [Google Scholar] [CrossRef]
- Lee, J.; Gharaibeh, Y.; Kolluru, C.; Zimin, V.N.; Dallan, L.A.P.; Kim, J.N.; Bezerra, H.G.; Wilson, D.L. Segmentation of Coronary Calcified Plaque in Intravascular OCT Images Using a Two-Step Deep Learning Approach. IEEE Access 2020, 8, 225581–225593. [Google Scholar] [CrossRef]
- Kolluru, C.; Lee, J.; Gharaibeh, Y.; Bezerra, H.G.; Wilson, D.L. Learning With Fewer Images via Image Clustering: Application to Intravascular OCT Image Segmentation. IEEE Access 2021, 9, 37273–37280. [Google Scholar] [CrossRef]
- Lee, J.; Pereira, G.T.R.; Gharaibeh, Y.; Kolluru, C.; Zimin, V.N.; Dallan, L.A.P.; Kim, J.N.; Hoori, A.; Al-Kindi, S.G.; Guagliumi, G.; et al. Automated Analysis of Fibrous Cap in Intravascular Optical Coherence Tomography Images of Coronary Arteries. arXiv 2022. [Google Scholar] [CrossRef]
- Gharaibeh, Y.; Lee, J.; Zimin, V.N.; Kolluru, C.; Dallan, L.A.P.; Pereira, G.T.R.; Vergara-Martel, A.; Kim, J.N.; Hoori, A.; Dong, P.; et al. Prediction of Stent Under-Expansion in Calcified Coronary Arteries Using Machine-Learning on Intravascular Optical Coherence Tomography. arXiv 2022. [Google Scholar] [CrossRef]
- Sinclair, H.; Bourantas, C.; Bagnall, A.; Mintz, G.S.; Kunadian, V. OCT for the Identification of Vulnerable Plaque in Acute Coronary Syndrome. JACC Cardiovasc. Imaging 2015, 8, 198–209. [Google Scholar] [CrossRef]
- Nakazato, R.; Otake, H.; Konishi, A.; Iwasaki, M.; Koo, B.-K.; Fukuya, H.; Shinke, T.; Hirata, K.-I.; Leipsic, J.; Berman, D.S.; et al. Atherosclerotic Plaque Characterization by CT Angiography for Identification of High-Risk Coronary Artery Lesions: A Comparison to Optical Coherence Tomography. Eur. Heart J. Cardiovasc. Imaging 2015, 16, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Uemura, S.; Ishigami, K.-I.; Soeda, T.; Okayama, S.; Sung, J.H.; Nakagawa, H.; Somekawa, S.; Takeda, Y.; Kawata, H.; Horii, M.; et al. Thin-Cap Fibroatheroma and Microchannel Findings in Optical Coherence Tomography Correlate with Subsequent Progression of Coronary Atheromatous Plaques. Eur. Heart J. 2012, 33, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Maehara, A.; Nazif, T.M.; Pollack, A.T.; Saito, S.; Rabbani, L.E.; Apfelbaum, M.A.; Dalton, K.; Moses, J.W.; Jorde, U.P.; et al. Optical Coherence Tomographic Evaluation of Transplant Coronary Artery Vasculopathy With Correlation to Cellular Rejection. Circ. Cardiovasc. Interv. 2014, 7, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Galon, M.Z.; Wang, Z.; Bezerra, H.G.; Lemos, P.A.; Schnell, A.; Wilson, D.L.; Rollins, A.M.; Costa, M.A.; Attizzani, G.F. Differences Determined by Optical Coherence Tomography Volumetric Analysis in Non-Culprit Lesion Morphology and Inflammation in ST-Segment Elevation Myocardial Infarction and Stable Angina Pectoris Patients. Catheter. Cardiovasc. Interv. 2015, 85, E108–E115. [Google Scholar] [CrossRef] [PubMed][Green Version]




| Methods | Dice | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Original image | 0.07 ± 0.01 | 92.1 ± 2.5 | 96.2 ± 1.0 |
| Data augmentation only | 0.07 ± 0.03 | 88.9 ± 9.2 | 96.4 ± 1.8 |
| Pre-processing only | 0.67 ± 0.17 | 83.9 ± 7.7 | 99.7 ± 0.2 |
| Data augmentation and pre-processing | 0.71 ± 0.10 | 87.7 ± 6.6 | 99.8 ± 0.1 |
| After candidate classification | 0.73 ± 0.10 | 85.5 ± 6.9 | 99.8 ± 0.1 |
| Method | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| Candidate classification | 99.5 ± 0.3 | 98.8 ± 1.0 | 99.1 ± 0.5 |
| Methods | Dice | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| U-Net | 0.65 ± 0.19 | 91.1 ± 4.8 | 99.6 ± 0.3 |
| SegNet | 0.71 ± 0.11 | 88.3 ± 1.8 | 99.7 ± 0.1 |
| DeepLab v3+ | 0.73 ± 0.10 | 85.5 ± 6.9 | 99.8 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Kim, J.N.; Gomez-Perez, L.; Gharaibeh, Y.; Motairek, I.; Pereira, G.T.R.; Zimin, V.N.; Dallan, L.A.P.; Hoori, A.; Al-Kindi, S.; et al. Automated Segmentation of Microvessels in Intravascular OCT Images Using Deep Learning. Bioengineering 2022, 9, 648. https://doi.org/10.3390/bioengineering9110648
Lee J, Kim JN, Gomez-Perez L, Gharaibeh Y, Motairek I, Pereira GTR, Zimin VN, Dallan LAP, Hoori A, Al-Kindi S, et al. Automated Segmentation of Microvessels in Intravascular OCT Images Using Deep Learning. Bioengineering. 2022; 9(11):648. https://doi.org/10.3390/bioengineering9110648
Chicago/Turabian StyleLee, Juhwan, Justin N. Kim, Lia Gomez-Perez, Yazan Gharaibeh, Issam Motairek, Gabriel T. R. Pereira, Vladislav N. Zimin, Luis A. P. Dallan, Ammar Hoori, Sadeer Al-Kindi, and et al. 2022. "Automated Segmentation of Microvessels in Intravascular OCT Images Using Deep Learning" Bioengineering 9, no. 11: 648. https://doi.org/10.3390/bioengineering9110648
APA StyleLee, J., Kim, J. N., Gomez-Perez, L., Gharaibeh, Y., Motairek, I., Pereira, G. T. R., Zimin, V. N., Dallan, L. A. P., Hoori, A., Al-Kindi, S., Guagliumi, G., Bezerra, H. G., & Wilson, D. L. (2022). Automated Segmentation of Microvessels in Intravascular OCT Images Using Deep Learning. Bioengineering, 9(11), 648. https://doi.org/10.3390/bioengineering9110648








