Meta-Analysis: A Convenient Tool for the Choice of Nose-to-Brain Nanocarriers
Abstract
1. Introduction
2. Methods
2.1. Data Mining
2.2. Inclusion Data and Criteria
2.3. Meta Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giunchedi, P.; Gavini, E.; Bonferoni, M.C. Nose-to-Brain Delivery. Pharmaceutics 2020, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.D.; Vanjari, Y.H.; Sancheti, K.H.; Belgamwar, V.S.; Surana, S.J.; Pardeshi, C.V. Nanotechnology-Mediated Nose to Brain Drug Delivery for Parkinson’s Disease: A Mini Review. J. Drug Target. 2015, 23, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Phukan, K.; Nandy, M.; Sharma, R.B.; Sharma, H.K. Nanosized Drug Delivery Systems for Direct Nose to Brain Targeting: A Review. Recent Patents Drug Deliv. Formul. 2016, 10, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Abd-Algaleel, S.A.; Metwally, A.A.; Abdel-Bar, H.M.; Kassem, D.H.; Hathout, R.M. Synchronizing In Silico, In Vitro, and In Vivo Studies for the Successful Nose to Brain Delivery of an Anticancer Molecule. Mol. Pharm. 2021, 18, 3763–3776. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; El-Ahmady, S.H.; Metwally, A.A. Curcumin or Bisdemethoxycurcumin for Nose-to-Brain Treatment of Alzheimer Disease? A Bio/Chemo-Informatics Case study. Nat. Prod. Res. 2018, 32, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; Abdelhamid, S.G.; El-Housseiny, G.S.; Metwally, A.A. Comparing Cefotaxime and Ceftriaxone in Combating Meningitis through Nose-to-Brain Delivery Using Bio/Chemoinformatics Tools. Sci. Rep. 2020, 10, 21250. [Google Scholar] [CrossRef]
- Shah, S.A.A.; Firlak, M.; Berrow, S.R.; Halcovitch, N.R.; Baldock, S.J.; Yousafzai, B.M.; Hathout, R.M.; Hardy, J.G. Electrochemically Enhanced Drug Delivery Using Polypyrrole Films. Materials 2018, 11, 1123. [Google Scholar] [CrossRef]
- ElMasry, S.R.; Hathout, R.M.; Abdel-Halim, M.; Mansour, S. In Vitro Transdermal Delivery of Sesamol Using Oleic Acid Chemically-Modified Gelatin Nanoparticles as a Potential Breast Cancer Medication. J. Drug Deliv. Sci. Technol. 2018, 48, 30–39. [Google Scholar] [CrossRef]
- Fagir, W.; Hathout, R.M.; Sammour, O.A.; ElShafeey, A.H. Self-Microemulsifying Systems of Finasteride with Enhanced Oral Bioavailability: Multivariate Statistical Evaluation, Characterization, Spray-Drying and In Vivo Studies in Human Volunteers. Nanomedicine 2015, 10, 3373–3389. [Google Scholar] [CrossRef]
- Safwat, S.; Ishak, R.A.; Hathout, R.M.; Mortada, N.D. Statins Anticancer Targeted Delivery Systems: Re-Purposing an Old Molecule. J. Pharm Pharm. 2017, 69, 613–624. [Google Scholar] [CrossRef]
- Nasser, N.; Hathout, R.M.; Abd-Allah, H.; Sammour, O.A. Enhancement of Oral Bioavailability of Drugs Using Lipid-Based Carriers: A Meta-Analysis Study. Drug Dev. Ind. Pharm. 2020, 46, 2105–2110. [Google Scholar] [CrossRef]
- Hathout, R.M. Do Polymeric Nanoparticles Really Enhance the Bioavailability of Oral Drugs? A Quantitative Answer Using Meta-Analysis. Gels 2022, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- des Rieux, A.; Fievez, V.; Garinot, M.; Schneider, Y.J.; Préat, V. Nanoparticles as Potential Oral Delivery Systems of Proteins and Vaccines: A Mechanistic Approach. J. Control. Release 2006, 116, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; Metwally, A.A. Gelatin Nanoparticles. Methods Mol. Biol 2019, 2000, 71–78. [Google Scholar] [PubMed]
- Hathout, R.M.; Mahmoud, O.A.; Ali, D.S.; Mamdouh, M.; Metwally, A.A. Modeling Drugs-PLGA Nanoparticles Interactions Using Gaussian Processes: Pharmaceutics Informatics Approach. J. Clust. Sci. 2022, 33, 2031–2036. [Google Scholar] [CrossRef]
- Kassem, D.H.; Kamal, M.M. Therapeutic Efficacy of Umbilical Cord-Derived Stem Cells for Diabetes Mellitus: A Meta–Analysis study. Stem Cell Res. Ther. 2020, 11, 484. [Google Scholar] [CrossRef]
- Elmeligy, S.; Hathout, R.M.; Khalifa, S.A.M.; El-Seedi, H.R.; Farag, M.A. Pharmaceutical Manipulation of Citrus Flavonoids Towards Improvement of Its Bioavailability and Stability. A Mini Review and a Meta-Analysis Study. Food Biosci. 2021, 44, 101428. [Google Scholar] [CrossRef]
- Burns, P.B.; Rohrich, R.J.; Chung, K.C. The Levels of Evidence and their Role in Evidence-Based Medicine. Plast. Reconstr. Surg. 2011, 128, 305–310. [Google Scholar] [CrossRef]
- Fong, S.Y.; Brandl, M.; Bauer-Brandl, A. Phospholipid-Based Solid Drug Formulations for Oral Bioavailability Enhancement: A Meta-Analysis. Eur. J. Pharm. Sci 2015, 80, 89–110. [Google Scholar] [CrossRef]
- Hathout, R.M.; Metwally, A.A. Towards Better Modeling of Drug-Loading in Solid Lipid Nanoparticles: Molecular Dynamics, Docking Experiments and Gaussian Processes Machine Learning. Eur. J. Pharm. Biopharm. 2016, 108, 262–268. [Google Scholar] [CrossRef]
- Metwally, A.A.; Hathout, R.M. Computer-Assisted Drug Formulation Design: Novel Approach in Drug Delivery. Mol. Pharm. 2015, 12, 2800–2810. [Google Scholar] [CrossRef] [PubMed]
- Metwally, A.A.; El-Ahmady, S.H.; Hathout, R.M. Selecting Optimum Protein Nano-Carriers for Natural Polyphenols Using Chemoinformatics Tools. Phytomedicine 2016, 23, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.J.; Bansback, N.; Ghement, I.; Thorlund, K.; Kelly, S.; Puhan, M.A.; Wright, J. Multiple Treatment Comparison Meta-Analyses: A Step Forward into Complexity. Clin. Epidemiol. 2011, 3, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M. Particulate Systems in the Enhancement of the Antiglaucomatous Drug Pharmacodynamics: A Meta-Analysis Study. ACS Omega 2019, 4, 21909–21913. [Google Scholar] [CrossRef]
- Hathout, R.M.; Metwally, A.A. Pharmaceutics Informatics: Bio/Chemoinformatics in Drug Delivery. In Computer Aided Pharmaceutics and Drug Delivery: An Application Guide for Students and Researchers of Pharmaceutical Sciences; Saharan, V.A., Ed.; Springer Nature Singapore: Singapore, 2022; pp. 705–724. [Google Scholar]
- Muntimadugu, E.; Dhommati, R.; Jain, A.; Challa, V.G.S.; Shaheen, M.; Khan, W. Intranasal Delivery of Nanoparticle Encapsulated Tarenflurbil: A Potential Brain Targeting Strategy for Alzheimer's Disease. Eur. J. Pharm. Sci. 2016, 92, 224–234. [Google Scholar] [CrossRef]
- Junior, E.R.D.O.; Nascimento, T.L.; Salomão, M.A.; da Silva, A.C.G.; Valadares, M.C.; Lima, E.M. Increased Nose-to-Brain Delivery of Melatonin Mediated by Polycaprolactone Nanoparticles for the Treatment of Glioblastoma. Pharm. Res. 2019, 36, 131. [Google Scholar] [CrossRef]
- Shamarekh, K.S.; Gad, H.A.; Soliman, M.E.; Sammour, O.A. Development and Evaluation of Protamine-Coated PLGA Nanoparticles for Nose-To-Brain Delivery of Tacrine: In-Vitro and In-Vivo Assessment. J. Drug Deliv. Sci. Technol. 2020, 57, 101724. [Google Scholar] [CrossRef]
- Ahmad, N.; Ahmad, R.; Al Qatifi, S.; Alessa, M.; Al Hajji, H.; Sarafroz, M. A Bioanalytical UHPLC Based Method Used for the Quantification of Thymoquinone-Loaded-PLGA-Nanoparticles in the Treatment of Epilepsy. BMC Chem. 2020, 14, 10. [Google Scholar] [CrossRef]
- Esim, O.; Savaser, A.; Ozkan, C.K.; Oztuna, A.; Goksel, B.A.; Ozler, M.; Tas, C.; Ozkan, Y. Nose to Brain Delivery of Eletriptan Hydrobromide Nanoparticles: Preparation, In Vitro/In Vivo Evaluation and Effect on Trigeminal Activation. J. Drug Deliv. Sci. Technol. 2020, 59, 101919. [Google Scholar] [CrossRef]
- Yasir, M.; Sara, U.V.S. Solid Lipid Nanoparticles for Nose to Brain Delivery of Haloperidol: In Vitro Drug Release and Pharmacokinetics Evaluation. Acta Pharm. Sin. B 2014, 4, 454–463. [Google Scholar] [CrossRef]
- Yasir, M.; Sara, U.V.S.; Chauhan, I.; Gaur, P.K.; Singh, A.P.; Puri, D.; Ameeduzzafar, A. Solid Lipid Nanoparticles for Nose to Brain Delivery of Donepezil: Formulation, Optimization by Box–Behnken Design, In Vitro and In Vivo Evaluation. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1–14. [Google Scholar] [CrossRef]
- Patil, N.L.; Mahajan, H.S. Quercetin Loaded Nanostructured Lipid Carriers for Nose to Brain Delivery: In Vitro and In Vivo Studies. Am. J. Adv. Drug Deliv. 2018, 6. [Google Scholar] [CrossRef]
- Yasir, M.; Chauhan, I.; Zafar, A.; Verma, M.; Noorulla, K.M.; Tura, A.J.; Alruwaili, N.K.; Haji, M.J.; Puri, D.; Gobena, W.G.; et al. Buspirone Loaded Solid Lipid Nanoparticles for Amplification of Nose to Brain Efficacy: Formulation Development, Optimization by Box-Behnken Design, In-Vitro Characterization and In-Vivo Biological Evaluation. J. Drug Deliv. Sci. Technol. 2021, 61, 102164. [Google Scholar] [CrossRef]
- Saka, R.; Chella, N.; Khan, W. Development of Imatinib Mesylate-Loaded Liposomes for Nose to Brain Delivery: In Vitro and In Vivo Evaluation. AAPS PharmSciTech 2021, 22, 192. [Google Scholar] [CrossRef]
- Ahmad, N. Rasagiline-Encapsulated Chitosan-Coated PLGA Nanoparticles Targeted to the Brain in the Treatment of Parkinson's Disease. J. Liq. Chromatogr. Relat. Technol. 2017, 40, 677–690. [Google Scholar] [CrossRef]
- Singh, S.K.; Dadhania, P.; Vuddanda, P.R.; Jain, A.; Velaga, S.; Singh, S. Intranasal Delivery of Asenapine Loaded Nanostructured Lipid Carriers: Formulation, Characterization, Pharmacokinetic and Behavioural Assessment. RSC Adv. 2016, 6, 2032–2045. [Google Scholar] [CrossRef]
- Haque, S.; Md, S.; Sahni, J.K.; Ali, J.; Baboota, S. Development and Evaluation of Brain Targeted Intranasal Alginate Nanoparticles for Treatment of Depression. J. Psychiatr. Res. 2014, 48, 1–12. [Google Scholar] [CrossRef]
- Bari, N.K.; Fazil, M.; Hassan, M.Q.; Haider, M.R.; Gaba, B.; Narang, J.K.; Baboota, S.; Ali, J. Brain Delivery of Buspirone Hydrochloride Chitosan Nanoparticles for the Treatment of General Anxiety Disorder. Int. J. Biol. Macromol. 2015, 81, 49–59. [Google Scholar] [CrossRef]
- Shah, B.; Khunt, D.; Misra, M.; Padh, H. Application of Box-Behnken design for optimization and development of quetiapine fumarate loaded chitosan nanoparticles for brain delivery via intranasal route. Int. J. Biol. Macromol. 2016, 89, 206–218. [Google Scholar] [CrossRef]
- Liu, S.; Ho, P.C. Intranasal Administration of Brain-Targeted HP-ß-CD/Chitosan Nanoparticles for Delivery of Scutellarin, A Compound with Protective Effect in Cerebral Ischaemia. J. Pharm. Pharmacol. 2017, 69, 1495–1501. [Google Scholar] [CrossRef]
- Sridhar, V.; Gaud, R.; Bajaj, A.; Wairkar, S. Pharmacokinetics and Pharmacodynamics of Intranasally Administered Selegiline Nanoparticles with Improved Brain Delivery in Parkinson's Disease. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2609–2618. [Google Scholar] [CrossRef]
- Liu, S.; Yang, S.; Ho, P.C. Intranasal Administration of Carbamazepine-Loaded Carboxymethyl Chitosan Nanoparticles for Drug Delivery to the Brain. Asian J. Pharm. Sci. 2018, 13, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zou, J.; Mu, C.; Yang, W.; Gu, M.; Lin, J.; Song, X.; Zheng, H.; Li, F. Intranasal Administration of Pullulan-Based NanoParticles for Enhanced Delivery of Adriamycin into the Brain: In vitro and in vivo Evaluation. Die Pharm. Int. J. Pharm. Sci. 2019, 74, 39–46. [Google Scholar]
- Rao, G.; Lopez-Jimenez, F.; Boyd, J.; D’amico, F.; Durant, N.H.; Hlatky, M.; Howard, G.; Kirley, K.; Masi, C.; Powell-Wiley, T.M.; et al. Methodological Standards for Meta-Analyses and Qualitative Systematic Reviews of Cardiac Prevention and Treatment Studies: A Scientific Statement from the American Heart Association. Circulation 2017, 136, e172–e194. [Google Scholar] [CrossRef] [PubMed]
- Greenland, S.; Senn, S.J.; Rothman, K.J.; Carlin, J.B.; Poole, C.; Goodman, S.N.; Altman, D.G. Statistical Tests, P Values, Confidence Intervals, and Power: A Guide to Misinterpretations. Eur. J. Epidemiol. 2016, 31, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Zlowodzki, M.; Poolman, R.W.; Kerkhoffs, G.M.; Tornetta, P.; Bhandari, M.; On Behalf of the International Evidence-Based Orthopedic Surgery Working Group. How to Interpret a Meta-Analysis and Judge its Value as a Guide for Clinical Practice. Acta Orthop. 2007, 78, 598–609. [Google Scholar] [CrossRef]
- Patsopoulos, N.A.; Evangelou, E.; Ioannidis, J.P. Sensitivity of Between-Study Heterogeneity in Meta-Analysis: Proposed Metrics and Empirical Evaluation. Int. J. Epidemiol. 2008, 37, 1148–1157. [Google Scholar] [CrossRef]
- Sabir, F.; Ismail, R.; Csoka, I. Nose-To-Brain Delivery of Antiglioblastoma Drugs Embedded into Lipid Nanocarrier Systems: Status Quo And Outlook. Drug Discov. Today 2020, 25, 185–194. [Google Scholar] [CrossRef]
- Von Hippel, P.T. The heterogeneity statistic I2 can be biased in small meta-analyses. BMC Med. Res. Methodol. 2015, 15, 35. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies that Evaluate Health Care Interventions: Explanation and Elaboration. PLoS Med. 2009, 62, e1000100. [Google Scholar]
- Schäfer, T.; Schwarz, M.A. The Meaningfulness of Effect Sizes in Psychological Research: Differences Between Sub-Disciplines and the Impact of Potential Biases. Front. Psychol. 2019, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Akel, H.; Csóka, I. Formulation and In Vitro Comparison Study between Lipid-Based and Polymeric-Based Nanoparticles for Nose-to-Brain Delivery of a Model Drug for Alzheimer′s Disease. Proceedings 2021, 78, 51. [Google Scholar]
- Hill, M.; Cunningham, R.N.; Hathout, R.M.; Johnston, C.; Hardy, J.G.; Migaud, M.E. Formulation of Antimicrobial Tobramycin Loaded PLGA Nanoparticles via Complexation with AOT. J. Funct. Biomater. 2019, 10, 26. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Harbord, R.M. Funnel Plots in Meta-analysis. Stata J. 2004, 4, 127–141. [Google Scholar] [CrossRef]
- Akel, H.; Csoka, I.; Ambrus, R.; Bocsik, A.; Grof, I.; Meszaros, M.; Szecsko, A.; Kozma, G.; Veszelka, S.; Deli, M.A.; et al. In Vitro Comparative Study of Solid Lipid and PLGA Nanoparticles Designed to Facilitate Nose-to-Brain Delivery of Insulin. Int. J. Mol. Sci. 2021, 22, 13258. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.S.; Kairuz, T.; Thiruchelvam, K.; Babar, Z.U.-D. Systematic Reviews and Meta-Analysis in Pharmacy Practice. In Pharmacy Practice Research Methods; Babar, Z.U.-D., Ed.; Springer Singapore: Singapore, 2020; pp. 237–250. [Google Scholar]
- Safwat, S.; Hathout, R.M.; Ishak, R.A.; Mortada, N.D. Elaborated Survey in the Scope of Nanocarriers Engineering for Boosting Chemotherapy Cytotoxicity: A Meta-Analysis Study. Int. J. Pharm. 2021, 610, 121268. [Google Scholar] [CrossRef] [PubMed]

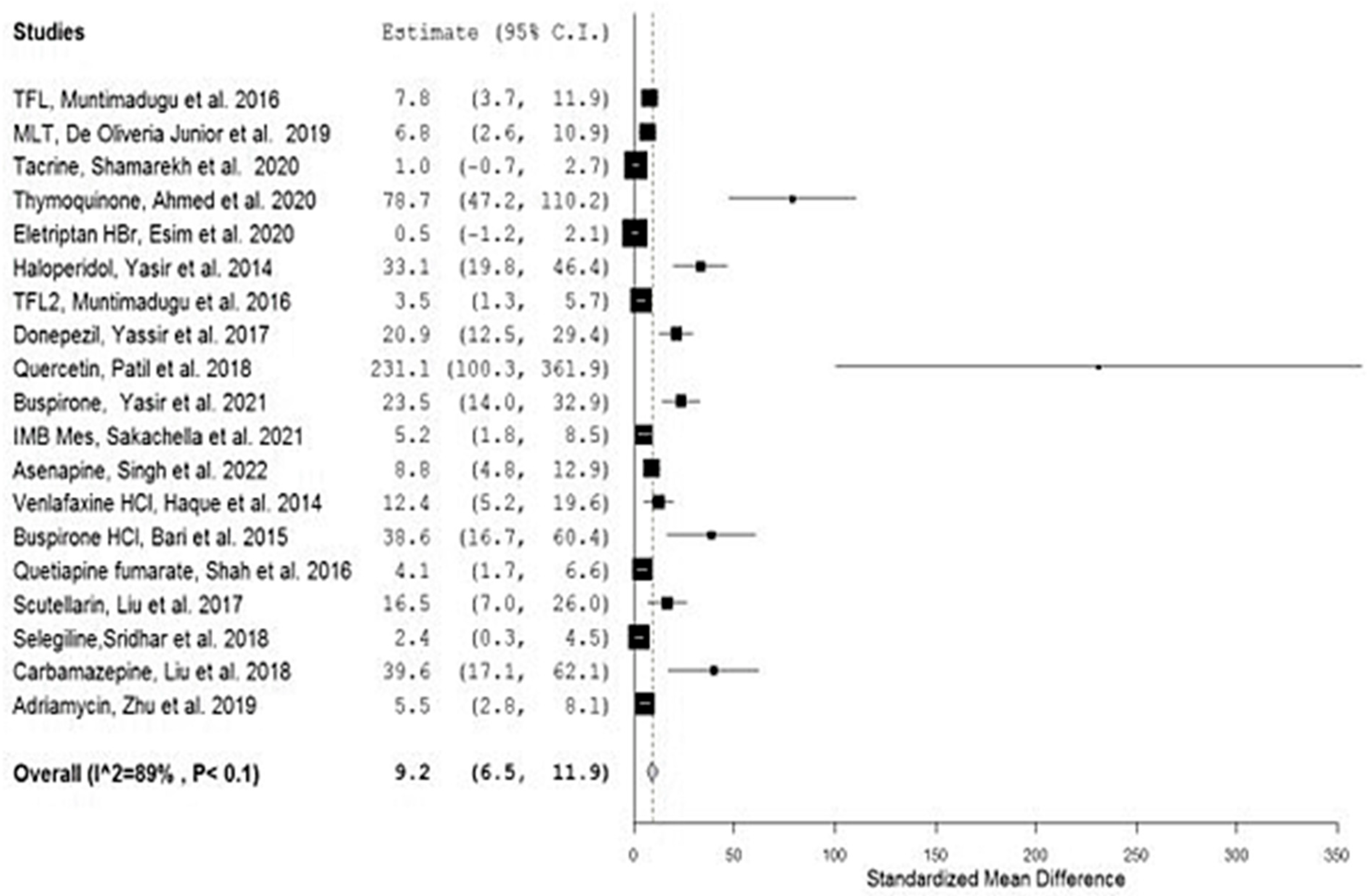



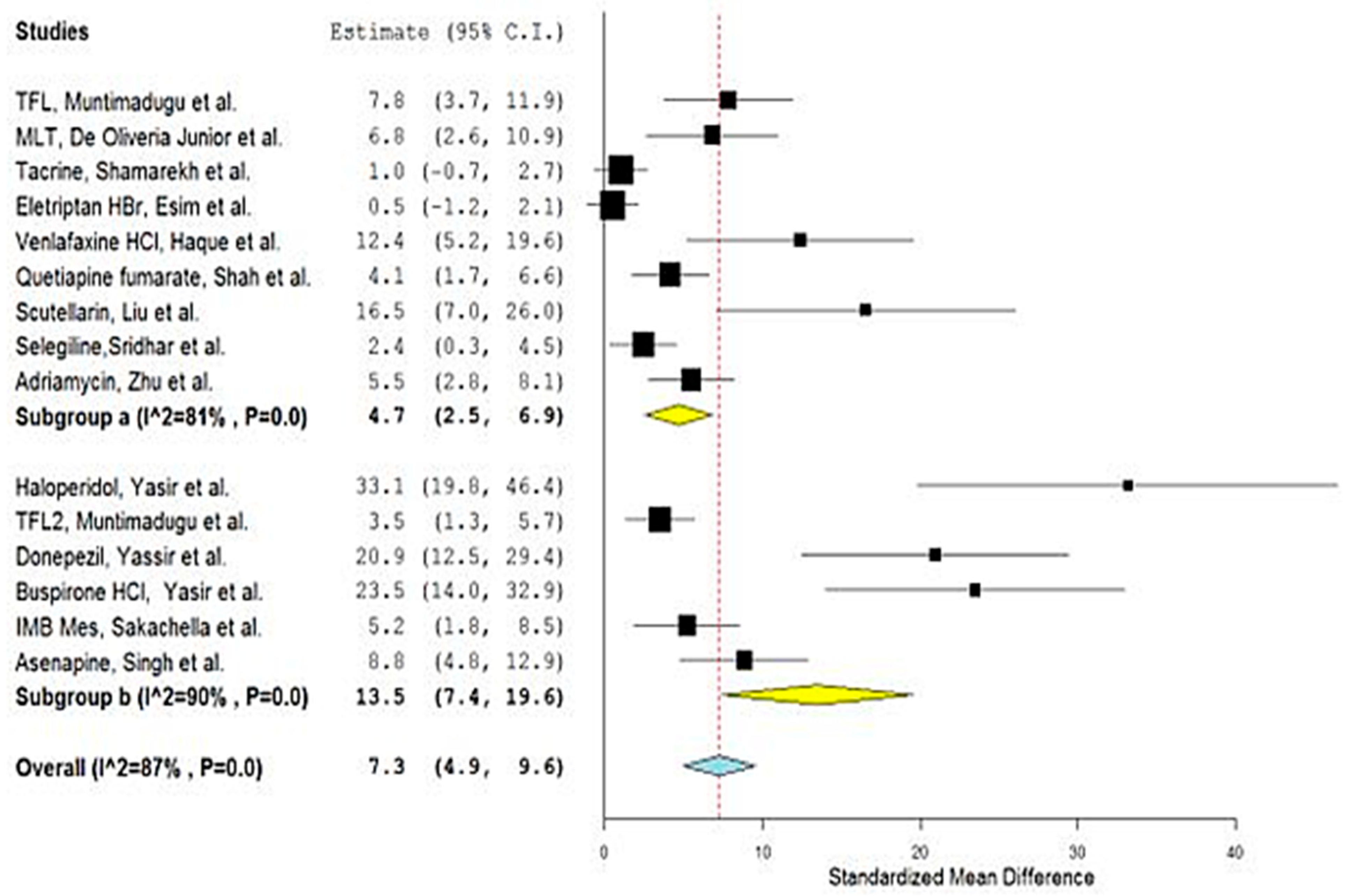
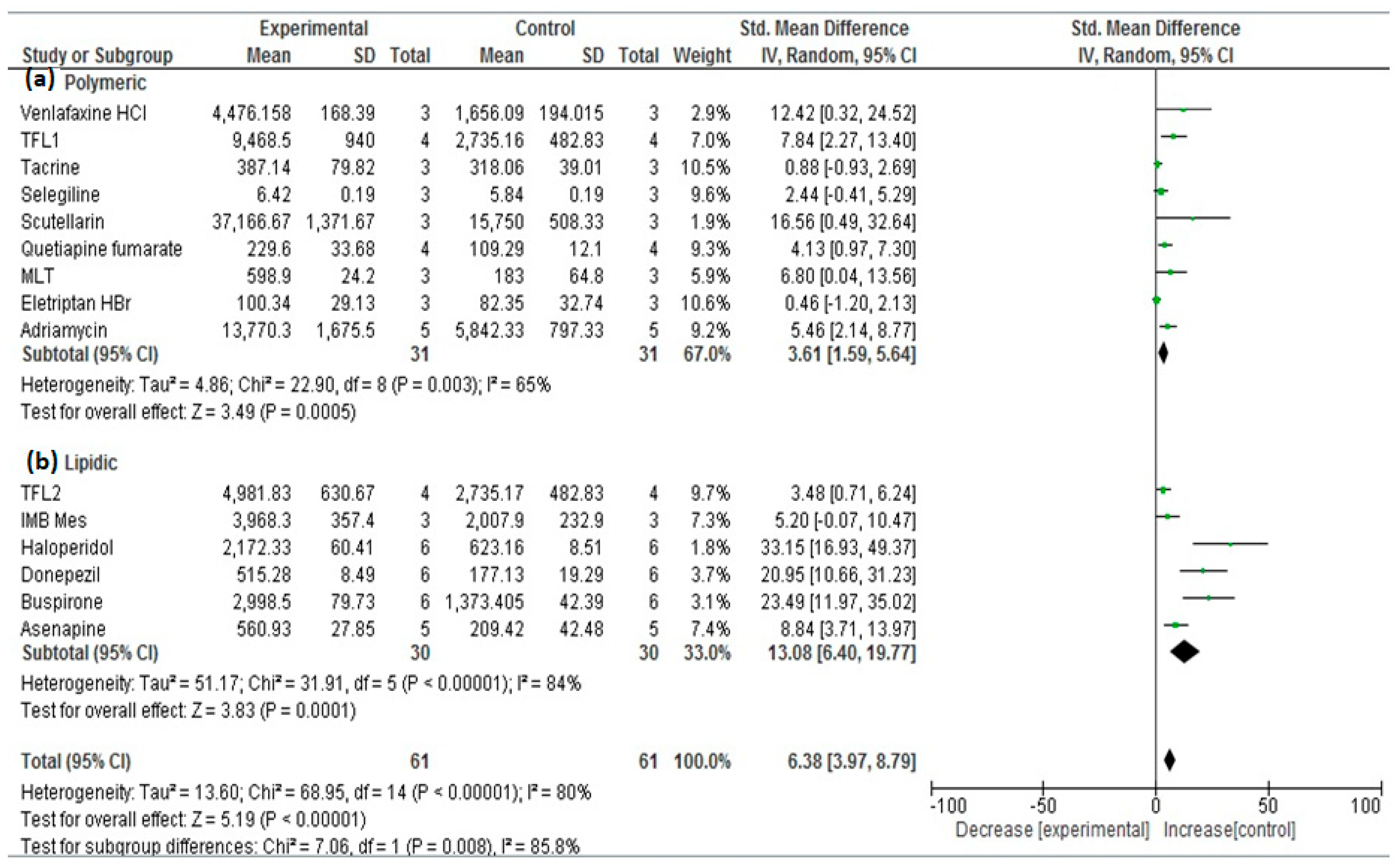
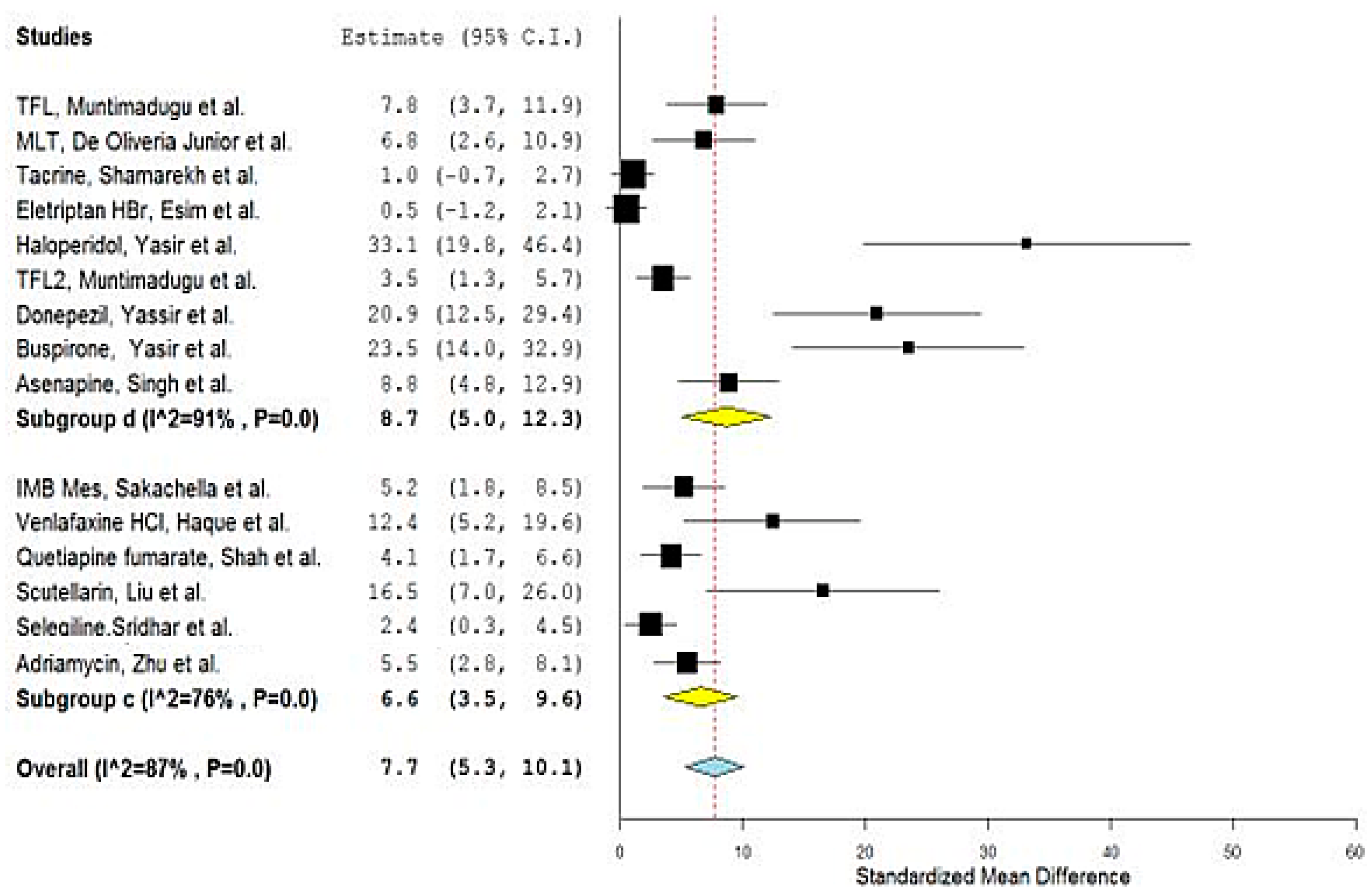
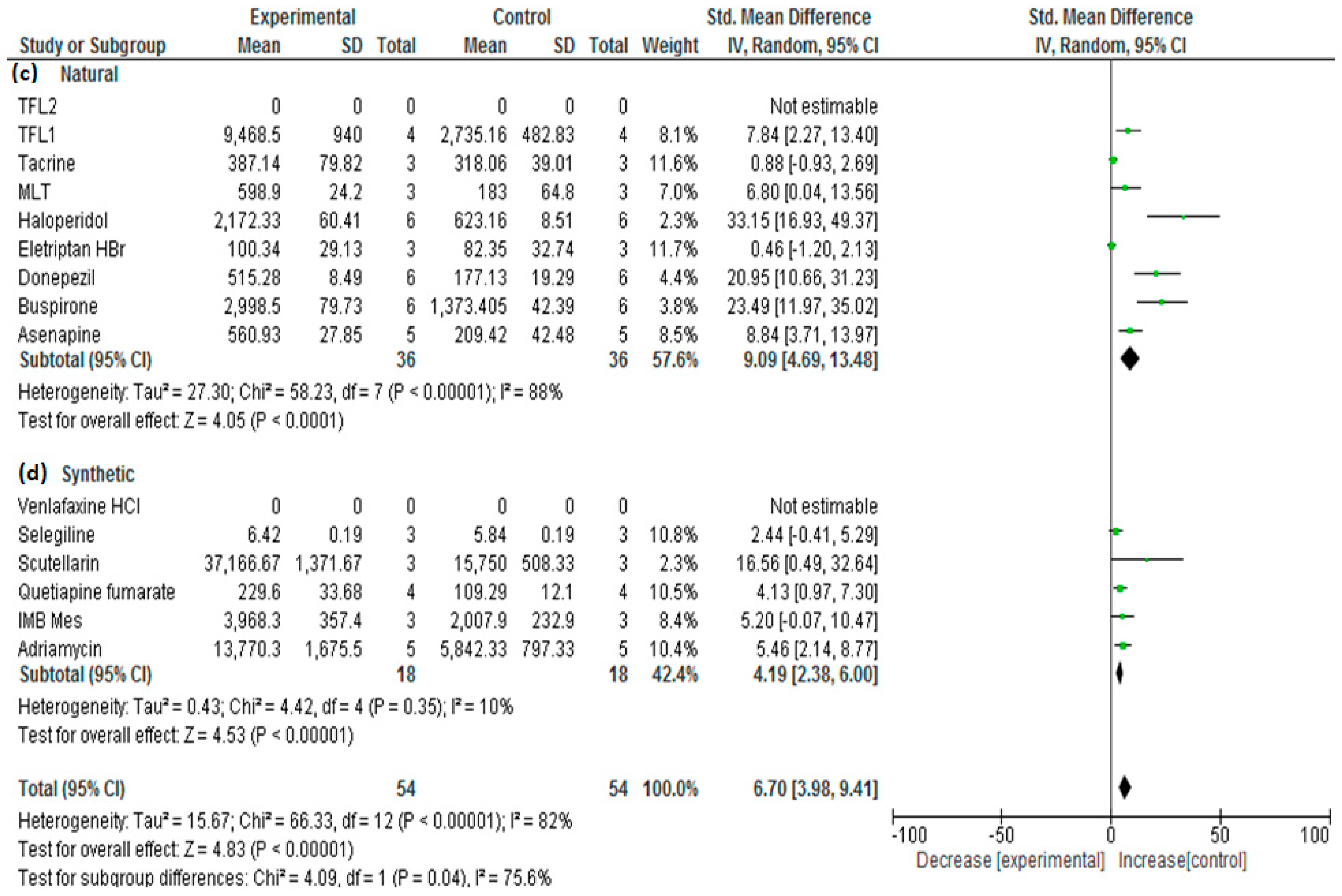

| No. | Drug | Year | No. of Animals 1 | Types | Group A 2 | Nano Carrier Type | Group B 3 | SMD | Lower/Upper Confidence | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NPs Mean AUC 4 | AUC SD | NPs Mean AUC 2 | AUC SD | |||||||||
| 1 | TFL1 | 2016 | 4 | Sprague–Dawley Rats | 9468.50 | 940.00 | PLGA a,d | 2735.16 | 482.83 | 7.827 | 3.749/11.905 | [26] |
| 2 | MLT | 2019 | 3 | Wistar rats | 598.90 | 24.20 | PCL a,d | 183.0 | 64.80 | 6.784 | 2.626/10.943 | [27] |
| 3 | Tacrine | 2020 | 3 | Wister albino rats | 397.14 | 79.82 | PLGA a,d | 318.06 | 39.01 | 1.004 | −0.694/2.703 | [28] |
| 4 | Thymoquinone | 2020 | 6 | Rats | 115.71 | 1.83 | PLGA a,d | 4.18 | 0.266 | 78.704 | 47.196/110.212 | [29] |
| 5 | Eletriptan HBr | 2020 | 3 | Wistar albino rats | 100.34 | 29.13 | PLGA a,d | 82.35 | 32.74 | 0.463 | −1.158/2.085 | [30] |
| 6 | Haloperidol | 2014 | 6 | Wistar albino rats | 2172.33 | 60.41 | SLN b,d | 623.16 | 8.51 | 33.138 | 19.832/46.443 | [31] |
| 7 | TFL2 | 2016 | 4 | Sprague–Dawley Rats | 4981.83 | 630.67 | SLN b,d | 2735.17 | 482.83 | 3.475 | 1.279/5.670 | [26] |
| 8 | Donepezil | 2017 | 6 | Wistar Albino rats | 515.28 | 8.49 | SLN b,d | 177.13 | 19.29 | 20.937 | 12.485/29.390 | [32] |
| 9 | Quercetin | 2018 | 3 | Wistar rats | 537,011 | 1102.33 | NLC b,d | 218,704.667 | 1095.5 | 231.110 | 100.340/361.881 | [33] |
| 10 | Buspirone | 2021 | 6 | Albino Wistar rats | 2998.50 | 79.73 | SLN b,d | 1373.405 | 42.39 | 23.485 | 14.022/32.949 | [34] |
| 11 | IMB Mes | 2021 | 3 | Sprague–Dawley (SD) rats | 3968.3 | 357.4 | Liposomes b,c | 2007.9 | 232.9 | 5.186 | 1.844/8.527 | [35,36] |
| 12 | Asenapine | 2022 | 5 | Charles Foster (CF) rats | 560.93 | 27.85 | NLC b,d | 209.42 | 42.48 | 8.834 | 4.769/12.900 | [37] |
| 13 | Venlafaxine HCl | 2014 | 3 | Rats | 4476.158 | 168.39 | Alginate a,c | 1656.09 | 194.015 | 12.387 | 5.198/19.575 | [38] |
| 14 | Buspirone HCl | 2015 | 3 | Albino Wistar rats | 67.47 | 0.472 | Thiolated chitosan a,c | 33.948 | 0.86 | 38.558 | 16.683/60.432 | [39] |
| 15 | Quetiapine fumarate | 2016 | 4 | Sprague–Dawley rats | 229.6 | 33.68 | Chitosan a,c | 109.29 | 12.1 | 4.130 | 1.677/6.582 | [40] |
| 16 | Scutellarin | 2017 | 3 | Mice | 37,166.67 | 1371.67 | HP-ß-CD/chitosan a,c | 15,750 | 508.33 | 16.520 | 7.037/26.003 | [41] |
| 17 | Selegiline | 2018 | 3 | Rats | 6.42 | 0.19 | Chitosan a,c | 5.84 | 0.19 | 2.436 | 0.324/4.548 | [42] |
| 18 | Carbamazepine | 2018 | 3 | Mice | 1551.167 | 39.167 | Carboxymethyl chitosan a,c | 125.167 | 10.83 | 39.596 | 17.136/62.057 | [43] |
| 19 | Adriamycin | 2019 | 5 | Wistar rats | 13,770.3 | 1675.5 | Cholesterol-modified Pullulan a,c | 5842.33 | 797.33 | 5.454 | 2.762/8.147 | [44] |
| Study Name | Weights |
|---|---|
| TFL, Muntimadugu et al. | 7.0% |
| MLT, De Oliveria Junior et al. | 7.0% |
| Tacrine, Shamarekh et al. | 8.0% |
| Thymoquinone, Ahmed et al. | 0.7% |
| Eletriptan HBr, Esim et al. | 8.1% |
| Haloperidol, Yasir et al. | 2.8% |
| TFL2, Muntimadugu et al. | 7.9% |
| Donepezil, Yassir et al. | 4.6% |
| Quercetin, Patil et al. | 0.0% |
| Buspirone, Yasir et al. | 4.2% |
| IMB Mes, Sakachella et al. | 7.4% |
| Asenapine, Singh et al. | 7.0% |
| Venlafaxine HCl, Haque et al. | 5.3% |
| Buspirone HCl, Bari et al. | 1.3% |
| Quetiapine fumarate, Shah et al. | 7.8% |
| Scutellarin, Liu et al. | 4.1% |
| Selegiline, Sridhar et al. | 7.9% |
| Carbamazepine, Liu et al. | 1.3% |
| Adriamycin, Zhu et al. | 7.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hathout, R.M.; El-Marakby, E.M. Meta-Analysis: A Convenient Tool for the Choice of Nose-to-Brain Nanocarriers. Bioengineering 2022, 9, 647. https://doi.org/10.3390/bioengineering9110647
Hathout RM, El-Marakby EM. Meta-Analysis: A Convenient Tool for the Choice of Nose-to-Brain Nanocarriers. Bioengineering. 2022; 9(11):647. https://doi.org/10.3390/bioengineering9110647
Chicago/Turabian StyleHathout, Rania M., and Eman M. El-Marakby. 2022. "Meta-Analysis: A Convenient Tool for the Choice of Nose-to-Brain Nanocarriers" Bioengineering 9, no. 11: 647. https://doi.org/10.3390/bioengineering9110647
APA StyleHathout, R. M., & El-Marakby, E. M. (2022). Meta-Analysis: A Convenient Tool for the Choice of Nose-to-Brain Nanocarriers. Bioengineering, 9(11), 647. https://doi.org/10.3390/bioengineering9110647







