The Landscape of Nucleic-Acid-Based Aptamers for Treatment of Hematologic Malignancies: Challenges and Future Directions
Abstract
1. Introduction
2. CD Markers Are Great Therapeutic Targets for Hematologic Malignancy
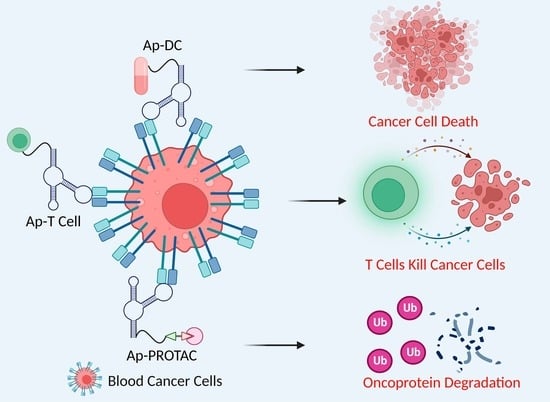
3. Aptamer-Mediated Precision Therapy for Hematologic Malignancy
3.1. Synthesis of Aptamer–Drug Conjugates through Chemical Linkers
3.2. Direct Synthesis of Aptamer–Drug Conjugates
4. Aptamer–T Cell (AP–T) Targeted Therapy for Hematologic Malignancy
5. Aptamer–PROTAC Conjugates (ApPCs)
6. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodson, D.J.; Screen, M.; Turner, M. RNA-binding proteins in hematopoiesis and hematological malignancy. Blood 2019, 133, 2365–2373. [Google Scholar] [CrossRef]
- Sheth, A.; de Melo, V.A.; Szydlo, R.; Szydlo, R.; Macdonald, D.H.; Reid, A.G.; Wagner, S.D. Specific patterns of chromosomal gains and losses associate with t(3;14), t(8;14), and t(14;18) in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 2009, 194, 48–52. [Google Scholar] [CrossRef]
- Ozery-Flato, M.; Linhart, C.; Trakhtenbrot, L.; Izraeli, S.; Shamir, R. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol. 2011, 12, R61. [Google Scholar] [CrossRef]
- Rustad, E.H.; Yellapantula, V.D.; Glodzik, D.; Maclachlan, K.H.; Diamond, B.; Boyle, E.M.; Ashby, C.; Blaney, P.; Gundem, G.; Hultcrantz, M.; et al. Revealing the Impact of Structural Variants in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 258–273. [Google Scholar] [CrossRef]
- Bergh, J.C. Gene amplification in human lung cancer. The myc family genes and other proto-oncogenes and growth factor genes. Am. Rev. Respir. Dis. 1990, 142, S20–S26. [Google Scholar] [CrossRef]
- Young, D.J.; Nguyen, B.; Li, L.; Higashimoto, T.; Levis, M.J.; Liu, J.O.; Small, D. A Method for Overcoming Plasma Protein Inhibition of Tyrosine Kinase Inhibitors. Blood Cancer Discov. 2021, 2, 532–547. [Google Scholar] [CrossRef]
- Soverini, S.; Martelli, M.; Bavaro, L.; Benedittis, C.D.; Iurlo, A.; Galimberti, S.; Pregno, P.; Bonifacio, M.; Lunghi, F.; Castagnetti, F.; et al. Detection of Actionable BCR-ABL1 Kinase Domain (KD) Mutations in Chronic Myeloid Leukemia (CML) Patients with Failure and Warning Response to Tyrosine Kinase Inhibitors (TKIs): Potential Impact of Next-Generation Sequencing (NGS) and Droplet Digital PCR (ddPCR) on Clinical Decision Making. Blood 2019, 134 (Suppl. 1), 661. [Google Scholar]
- Shastri, A.; Gonzalez-Lugo, J.; Verma, A. Understanding FLT3 Inhibitor Resistance to Rationalize Combinatorial AML Therapies. Blood Cancer Discov. 2020, 2, 113–115. [Google Scholar] [CrossRef]
- Zhang, X.H.; Chen, J.; Han, M.Z.; Huang, H.; Jiang, E.L.; Jiang, M.; Lai, Y.R.; Liu, D.H.; Liu, Q.F.; Liu, T.; et al. The consensus from The Chinese Society of Hematology on indications, conditioning regimens and donor selection for allogeneic hematopoietic stem cell transplantation: 2021 update. J. Hematol. Oncol. 2021, 14, 1–20. [Google Scholar] [CrossRef]
- Walker, B.A. The Chromosome 13 Conundrum in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 16–17. [Google Scholar] [CrossRef]
- Zarbo, A.; Axelson, A. Common Cutaneous Side Effects of Anti-cancer Agents. In Practical Guide to Dermatology: The Henry Ford Manual, 2nd ed.; Lim, H.W., Kohen, L.L., Schneider, S.F., Yeager, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 289–306. [Google Scholar]
- Stoddart, A.; Wang, J.; Fernald, A.A.; Davis, E.M.; Johnson, C.R.; Hu, C.; Cheng, J.X.; McNerney, M.E.; Le Beau, M.M. Cytotoxic Therapy-Induced Effects on Both Hematopoietic and Marrow Stromal Cells Promotes Therapy-Related Myeloid Neoplasms. Blood Cancer Discov. 2020, 1, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Padma, V.V. An overview of targeted cancer therapy. Biomedicine (Taipei) 2015, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, J.; Vaikari, V.P.; Beckford, J.S.; Wu, S.; Akhtari, M.; Alachkar, H. Apolipoprotein C2—CD36 Promotes Leukemia Growth and Presents a Targetable Axis in Acute Myeloid Leukemia. Blood Cancer Discov. 2020, 1, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Panchagnula, R.; Dey, C.S. Monoclonal antibodies in drug targeting. J. Clin. Pharm. Ther. 1997, 22, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, U.; Parakh, S.; Gan, H.K.; Scott, A.M. Antibody-Drug Conjugates for Cancer Therapy. Molecules 2020, 25, 4764. [Google Scholar] [CrossRef]
- Kantarjian, H.; Short, N.J.; DiNardo, C.; Stein, E.M.; Daver, N.; Perl, A.E.; Wang, E.S.; Wei, A.; Tallman, M. Harnessing the benefits of available targeted therapies in acute myeloid leukaemia. Lancet Haematol. 2021, 8, E922–E933. [Google Scholar] [CrossRef]
- Pittaluga, S.; Nicolae, A.; Wright, G.W.; Melani, C.; Roschewski, M.; Steinberg, S.; Huang, D.; Staudt, L.M.; Jaffe, E.S. Wilson, W.H. Gene Expression Profiling of Mediastinal Gray Zone Lymphoma and Its Relationship to Primary Mediastinal B-cell Lymphoma and Classical Hodgkin Lymphoma. Blood Cancer Discov. 2020, 1, 155–161. [Google Scholar] [CrossRef]
- Estey, E.H. Acute myeloid leukemia: 2021 update on risk-stratification and management. Am. J. Hematol. 2020, 95, 1368–1398. [Google Scholar] [CrossRef]
- Schurch, C.M. Therapeutic Antibodies for Myeloid Neoplasms-Current Developments and Future Directions. Front. Oncol. 2018, 8, 152. [Google Scholar] [CrossRef]
- Chesi, M.; Stein, C.K.; Garbitt, V.M.; Sharik, M.E.; Asmann, Y.W.; Bergsagel, M.; Riggs, D.L.; Welsh, S.J.; Meermeier, E.W.; Kumar, S.K. Monosomic loss of MIR15A/MIR16-1 is a driver of multiple myeloma proliferation and disease progression. Blood Cancer Discov. 2020, 1, 68–81. [Google Scholar] [CrossRef]
- Maimaitiyiming, Y.; Ye, L.; Yang, T.; Yu, W.; Naranmandura, H. Linear and Circular Long Non-Coding RNAs in Acute Lymphoblastic Leukemia: From Pathogenesis to Classification and Treatment. Int. J. Mol. Sci. 2022, 23, 4442. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Ward, A.C.; De, A.; Yang, C.J.; Wei, M.; Duan, W. Clinical applications of aptamers and nucleic acid therapeutics in haematological malignancies. Br. J. Haematol. 2011, 155, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Sicco, E.; Baez, J.; Fernández, M.; Fernández, M.; Cabral, P.; Moreno, M.; Cerecetto, H.; Calzada, V. Sgc8-c Aptamer as a Potential Theranostic Agent for Hemato-Oncological Malignancies. Cancer Biother. Radiopharm. 2020, 35, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Seo, J.M.; Shin, K.J.; Yang, S.G. Design and clinical developments of aptamer-drug conjugates for targeted cancer therapy. Biomater Res. 2021, 25, 42. [Google Scholar] [CrossRef] [PubMed]
- Eladl, E.; Tremblay-LeMay, R.; Rastgoo, N.; Musani, R.; Chen, W.; Liu, A.; Chang, H. Role of CD47 in Hematological Malignancies. J. Hematol. Oncol. 2020, 13, 96. [Google Scholar] [CrossRef]
- Louvet, C.; Nadeem, O.; Smith, E.L. Finding the optimal partner to pair with bispecific antibody therapy for multiple myeloma. Blood Cancer Discov. 2021, 2, 297–299. [Google Scholar] [CrossRef]
- Rezaeeyan, H.; Shahrabi, S.; McKee, T.D.; Saki, N. The expression of CD markers in solid tumors: Significance in metastasis and prognostic value. Histol. Histopathol. 2018, 33, 1005–1012. [Google Scholar]
- Russ, A.; Hua, A.B.; Montfort, W.R.; Rahman, B.; Riaz, I.B.; Khalid, M.U.; Carew, J.S.; Nawrocki, S.T.; Persky, D.; Anwer, F. Blocking “don’t eat me” signal of CD47-SIRPalpha in hematological malignancies, an in-depth review. Blood Rev. 2018, 32, 480–489. [Google Scholar] [CrossRef]
- Walter, R.B. The role of CD33 as therapeutic target in acute myeloid leukemia. Expert Opin. Ther. Targets 2014, 18, 715–718. [Google Scholar] [CrossRef]
- Dillon, L.W.; Ghannam, J.; Nosiri, C.; Gui, G.; Goswami, M.; Calvo, K.R.; Lindblad, K.E.; Oetjen, K.A.; Wilkerson, M.D.; Soltis, A.R.; et al. Personalized Single-Cell Proteogenomics to Distinguish Acute Myeloid Leukemia from Nonmalignant Clonal Hematopoiesis. Blood Cancer Discov. 2021, 2, 319–325. [Google Scholar] [CrossRef]
- Rubnitz, J.E.; Kaspers, G.J.L. How I treat pediatric acute myeloid leukemia. Blood 2021, 138, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Norsworthy, K.J.; Ko, C.-W.; Lee, J.E.; Liu, J.; John, C.S.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Mylotarg for Treatment of Patients with Relapsed or Refractory CD33-Positive Acute Myeloid Leukemia. Oncologist 2018, 23, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Abou-El-Enein, M.; Elsallab, M.; Feldman, S.A.; Fesnak, A.D.; Heslop, H.E.; Marks, P.; Till, B.G.; Bauer, G.; Savoldo, B. Scalable Manufacturing of CAR T cells for Cancer Immunotherapy. Blood Cancer Discov. 2021, 2, 408–422. [Google Scholar] [CrossRef] [PubMed]
- Raman, T.; Mohanraj, S.; Muthu, A.; Prabhakar, V.; Ramakrishnan, B.; Vaidhyanathan, L.; Easow, J.; Raja, T. Independent diagnostic utility of CD20, CD200, CD43 and CD45 in chronic lymphocytic leukaemia. Leuk. Lymphoma 2021, 63, 1–8. [Google Scholar]
- Alduailej, H.; Kanfar, S.; Bakhit, K.; Raslan, H.; Alsaber, A.; Bashawri, L.; Aldayel, A.; Alanezi, K. Outcome of CD20-positive Adult B-cell Acute Lymphoblastic Leukemia and the Impact of Rituximab Therapy. Clin. Lymphoma Myeloma Leuk. 2020, 20, e560–e568. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Yasuda, T.; Kojima, S.; Kawazu, M.; Akahane, K.; Inukai, T.; Imaizumi, M.; Morishita, T.; Miyamura, K.; Ueno, T.; et al. Targeting MEF2D-fusion Oncogenic Transcriptional Circuitries in B-cell Precursor Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2020, 1, 82–95. [Google Scholar] [CrossRef]
- Kläsener, K.; Jellusova, J.; Andrieux, G.; Salzer, U.; Böhler, C.; Steiner, S.N.; Albinus, J.B.; Cavallari, M.; Süß, B.; Voll, R.E.; et al. CD20 as a gatekeeper of the resting state of human B cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2021342118. [Google Scholar] [CrossRef]
- Song, Y.; Guo, Y.; Wang, Z.; Wu, M.; Peng, W.; Sun, L.; Sun, J.H.; Li, M.; Zhu, J. A Dose Escalation Phase Ia Study of Anti-CD20 Antibody Drug Conjugate, MRG001 in Relapsed/Refractory Advanced Non-Hodgkin Lymphom. Blood 2021, 138 (Suppl. 1), 2490. [Google Scholar] [CrossRef]
- Katz, B.Z.; Herishanu, Y. Therapeutic targeting of CD19 in hematological malignancies: Past, present, future and beyond. Leuk Lymphoma 2014, 55, 999–1006. [Google Scholar] [CrossRef]
- Haloupek, N. The Landscape of Blood Cancer Research Today-and Where the Field Is Headed. Blood Cancer Discov. 2020, 1, 1–4. [Google Scholar] [CrossRef]
- Simon, S.; Riddell, S.R. Dual Targeting with CAR T Cells to Limit Antigen Escape in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Caimi, P.F.; Ai, W.; Alderuccio, J.P.; Ardeshna, K.M.; Hamadani, M.; Hess, B.; Kahl, B.S.; Radford, J.; Solh, M.; Stathis, A. Loncastuximab tesirine in relapsed or refractory diffuse large B-cell lymphoma (LOTIS-2): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2021, 22, 790–800. [Google Scholar] [CrossRef]
- Abramson, J.S. Anti-CD19 CAR T-Cell Therapy for B-Cell Non-Hodgkin Lymphoma. Transfus. Med. Rev. 2020, 34, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Maciocia, N.C.; Burley, A.; Nannini, F.; Wawrzyniecka, P.A.; Neves, M.P.; Karpanasamy, T.; Ferrari, M.; Marafioti, T.; Onuoha, S.C.; Khwaja, A.I. Anti-CD21 Chimeric Antigen Receptor (CAR)-T Cells for T Cell Acute Lymphoblastic Leukaemia (T-ALL). Blood 2021, 138 (Suppl. 1), 902. [Google Scholar] [CrossRef]
- Lanza, F.; Maffini, E.; Rondoni, M.; Massari, E.; Faini, A.C.; Malavasi, F. CD22 Expression in B-Cell Acute Lymphoblastic Leukemia: Biological Significance and Implications for Inotuzumab Therapy in Adults. Cancers 2020, 12, 303. [Google Scholar] [CrossRef]
- Shaffer, A.L., III; Phelan, J.D.; Wang, J.Q.; Huang, D.; Wright, G.W.; Kasbekar, M.; Choi, J.; Young, R.M.; Webster, D.E.; Yang, Y.; et al. Overcoming Acquired Epigenetic Resistance to BTK Inhibitors. Blood Cancer Discov. 2021, 2, 630–647. [Google Scholar] [CrossRef]
- Kantarjian, H.; Thomas, D.; Jorgensen, J.; Kebriaei, P.; Jabbour, E.; Rytting, M.; York, S.; Ravandi, F.; Garris, R.; Kwari, M.; et al. Results of inotuzumab ozogamicin, a CD22 monoclonal antibody, in refractory and relapsed acute lymphocytic leukemia. Cancer 2013, 119, 2728–2736. [Google Scholar] [CrossRef]
- Thota, S.; Advani, A. Inotuzumab ozogamicin in relapsed B-cell acute lymphoblastic leukemia. Eur. J. Haematol. 2017, 98, 425–434. [Google Scholar] [CrossRef]
- Fry, T.J.; Shah, N.N.; Orentas, R.J.; Stetler-Stevenson, M.; Yuan, C.M.; Ramakrishna, S.; Wolters, P.; Martin, S.; Delbrook, C.; Yates, B.; et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 2018, 24, 20–28. [Google Scholar] [CrossRef]
- Zhu, H.; Deng, H.; Mu, J.; Lyu, C.; Jiang, Y.; Deng, Q. Anti-CD22 CAR-T Cell Therapy as a Salvage Treatment in B Cell Malignancies Refractory or Relapsed After Anti-CD19 CAR-T therapy. Onco Targets Ther. 2021, 14, 4023–4037. [Google Scholar] [CrossRef]
- Yu, X.; Munoz-Sagredo, L.; Streule, K.; Muschong, P.; Bayer, E.; Walter, R.J.; Gutjahr, J.C.; Greil, R.; Concha, M.L.; Müller-Tidow, C.; et al. CD44 loss of function sensitizes AML cells to the BCL-2 inhibitor venetoclax by decreasing CXCL12-driven survival cues. Blood 2021, 138, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, J.; Jiang, B.; Jiang, B.; Liu, H.; Cao, X.; Zhang, M.; Meng, Y.; MA, X.; Jia, Y. A Phase I/IIa Study of Lemzoparlimab, a Monoclonal Antibody Targeting CD47, in Patients with Relapsed and/or Refractory Acute Myeloid Leukemia (AML) and Myelodysplastic Syndrome (MDS): Initial Phase I Results. Blood 2020, 136 (Suppl. 1), 30–31. [Google Scholar] [CrossRef]
- Rodriguez, C.; Jin, T.; Jawde, R.A.; Saber, W.; Baz, R.; His, E.; Kalaycio, M.; Sobecks, R.; Sekeres, M.; Advani, A. c-Kit (CD117) Expression Is a Poor Prognostic Factor for Relapse and Overall Survival in Patients with Newly Diagnosed AML. Blood 2006, 108, 4510. [Google Scholar] [CrossRef]
- Smith, B.D.; Roboz, G.J.; Walter, R.B.; Altman, J.K.; Ferguson, A.; Curcio, T.J.; Orlowski, K.F.; Garrett, L.; Busfield, S.J.; Barnden, M.; et al. First-in Man, Phase 1 Study of CSL362 (Anti-IL3Rα / Anti-CD123 Monoclonal Antibody) in Patients with CD123+ Acute Myeloid Leukemia (AML) in CR at High Risk for Early Relapse. Blood 2014, 124, 120. [Google Scholar] [CrossRef]
- Schumacher, C.E.; Nuebling, T.; Hofmann, M.; Schmiedel, B.J.; Kanz, L.; Jung, G.; Salih, H.R. The Role of OX40 and Its Ligand in Acute Myeloid Leukemia: Expression, Function and Modulation of NK Cell Anti-Leukemia Reactivity. Blood 2012, 120, 3548. [Google Scholar] [CrossRef]
- Byrd, J.C. Targeting CD20 takes the backseat in CLL. Blood 2019, 133, 1003–1004. [Google Scholar] [CrossRef]
- Al-Madhoun, N.Y.; Gadhoum, S.Z.; Merzaban, J.S. ERK1/2 Pathway Is Required for Differentiation of AML Triggered by Anti-CD44 Monoclonal Antibodies. Blood 2012, 120, 4334. [Google Scholar] [CrossRef]
- Sallman, D.A.; Asch, A.S.; Malki, M.M.A.; Lee, D.J.; Donnellan, W.B.; Marcucci, G.; Kambhampati, S.; Daver, N.G.; Garcia-Manero, G.; Komrokji, R.S.; et al. The First-in-Class Anti-CD47 Antibody Magrolimab (5F9) in Combination with Azacitidine Is Effective in MDS and AML Patients: Ongoing Phase 1b Results. Blood 2019, 134 (Suppl. 1), 569. [Google Scholar] [CrossRef]
- Melo Garcia, L.; Barabé, F. Harnessing Macrophages through the Blockage of CD47, Implications for Acute Myeloid Leukemia. Cancers 2021, 13, 6258. [Google Scholar] [CrossRef]
- Myburgh, R.; Kiefer, J.D.; Russkamp, N.F.; Magnani, C.F.; Nuñez, N.; Simonis, A.; Pfister, S.; Wilk, C.M.; McHugh, D.; Friemel, J.; et al. Anti-human CD117 CAR T-cells efficiently eliminate healthy and malignant CD117-expressing hematopoietic cells. Leukemia 2020, 34, 2688–2703. [Google Scholar] [CrossRef]
- Yang, M.; Jiang, G.; Li, W.; Qiu, K.; Zhang, M.; Carter, C.M.; Al-Quran, S.Z.; Li, Y. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J. Hematol. Oncol. 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Papayannidis, C.; Paolini, S.; Santoro, A.; Robustelli, V.; Soverini, S.; Benedittis, C.D.; Imbrogno, E.; Terragna, C.; Rorà, A.G.; Parisi, S.; et al. Blinatumomab is safe and effective in relapsed and MRD-positive B-ALL CD19+ patients: The Bologna Compassionate Program Experience. Blood 2016, 128, 5191. [Google Scholar] [CrossRef]
- Lee, D.W., III; Stetler-Stevenson, M.; Yuan, C.M.; Shah, N.N.; Delbrook, C.P.; Yates, B.; Zhang, H.; Zhang, L.; Kochenderfer, J.N.; Rosenberg, S.A.; et al. Long-Term Outcomes Following CD19 CAR T Cell Therapy for B-ALL Are Superior in Patients Receiving a Fludarabine/Cyclophosphamide Preparative Regimen and Post-CAR Hematopoietic Stem Cell Transplantation. Blood 2016, 128, 218. [Google Scholar] [CrossRef]
- Chevallier, P.; Leguay, T.; Doubek, M.; Huguet, F.; Šálek, C.; Cabannes, A.; Wartiovaara-Kautto, U.; Saillard, C.; Raffoux, E.; Cluzeau, T.; et al. Fractionated Inotuzumab Ozogamicin Combined with Low-Intensity Chemotherapy Provides Very Good Outcome in Older Patients with Newly Diagnosed CD22+ Philadelphia Chromosome-Negative B-Cell Precursor Acute Lymphoblastic Leukemia: First Results from the EWALL-INO Study. Blood 2021, 138 (Suppl. 1), 511. [Google Scholar]
- Kreitman, R.J.; Dearden, C.E.; Zinzani, P.L.L.; Delgado, J.; Robak, T.; le Coutre, P.; Gjertsen, B.T.; Troussard, X.; Roboz, G.J.; Karlin, L.; et al. Moxetumomab Pasudotox-Tdfk in Heavily Pretreated Patients with Relapsed/Refractory Hairy Cell Leukemia (HCL): Long-Term Follow-up from the Pivotal Phase 3 Trial. Blood 2019, 134 (Suppl. 1), 2808. [Google Scholar] [CrossRef]
- Wierda, W.G.; Jewell, R.C.; Kipps, T.J.; Dürig, J.; Griškevičius, L.; Stilgenbauer, S.; Smolej, L.; Hess, G.; Hernandez-Ilizaliturri, F.J.; Padmanabhan, S.; et al. Correlations between Ofatumumab Exposure and Treatment Outcomes for Patients with Chronic Lymphocytic Leukemia (CLL) Treated with Frontline Ofatumumab, Fludarabine, and Cyclophosphamide Chemoimmunotherapy. Blood 2011, 118, 1793. [Google Scholar] [CrossRef]
- Brown, J.R.; O′Brien, S.; Kingsley, C.D.; Eradat, H.A.; Pagel, J.M.; Hirata, J.; McIver, T.; Morariu-Zamfir, R.; Kipps, T.J. Durable remissions with obinutuzumab-based chemoimmunotherapy: Long-term follow-up of the phase 1b GALTON trial in CLL. Blood 2019, 133, 990–992. [Google Scholar] [CrossRef]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y.; et al. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Xuan, W.; Peng, Y.; Deng, Z.; Peng, T.; Kuai, H.; Li, Y.; He, J.; Jin, C.; Liu, Y.; Wang, R.; et al. A basic insight into aptamer-drug conjugates (ApDCs). Biomaterials 2018, 182, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, J.; Wu, M.; Zhao, J.X. Aptamers: Active targeting ligands for cancer diagnosis and therapy. Theranostics 2015, 5, 322–344. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.L.; Zhu, G.; Xiao, X.; Puszyk, W.M.; Sefah, K.; Wu, Q.; Tan, W.; Liu, C. A Synthetic Aptamer-Drug Adduct for Targeted Liver Cancer Therapy. PLoS ONE 2015, 10, e0136673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, R.; Chen, F.; Chen, M.; Wang, Y. Nucleolin targeting AS1411 aptamer modified pH-sensitive micelles: A dual-functional strategy for paclitaxel delivery. J. Control. Release 2015, 213, e137–e138. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Cho, Y.L.; Chae, J.R.; Moon, S.H.; Cho, W.G.; Choi, Y.; Lee, S.J.; Kang, W.J. Gemcitabine-Incorporated G-Quadruplex Aptamer for Targeted Drug Delivery into Pancreas Cancer. Mol. Ther. Nucleic Acids 2018, 12, 543–553. [Google Scholar] [CrossRef]
- Yoon, S.; Huang, K.W.; Reebye, V.; Spalding, D.; Przytycka, T.M.; Wang, Y.; Swiderski, P.M.; Li, L.; Armstrong, B.; Reccia, I.; et al. Aptamer-Drug Conjugates of Active Metabolites of Nucleoside Analogs and Cytotoxic Agents Inhibit Pancreatic Tumor Cell Growth. Mol. Ther. Nucleic Acids 2017, 6, 80–88. [Google Scholar] [CrossRef]
- Kratschmer, C.; Levy, M. Targeted Delivery of Auristatin-Modified Toxins to Pancreatic Cancer Using Aptamers. Mol. Ther. Nucleic Acids 2018, 10, 227–236. [Google Scholar] [CrossRef]
- Ray, P.; Cheek, M.A.; Sharaf, M.L.; Li, N.; Ellington, A.D.; Sullenger, B.A.; Shaw, B.R.; White, R.R. Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid Ther. 2012, 22, 295–305. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y.; Ge, M.H.; Fu, Y.; Hao, R.; Islam, K.; Huang, P.; Chen, F.; Sun, J.; Hong, D.; et al. Rapid identification of specific DNA aptamers precisely targeting CD33 positive leukemia cells through a paired cell-based approach. Biomater. Sci. 2019, 7, 938–950. [Google Scholar] [CrossRef]
- Huang, Y.F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem 2009, 10, 862–868. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, G.; Mei, L.; Xie, Y.; Ma, H.; Ye, M.; Qing, F.; Tan, W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J. Am. Chem. Soc. 2014, 136, 2731–2734. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Raghunathan, V.; Kanwar, J.R.; Kanwar, R.K.; Elchuri, S.V.; Khetan, V.; Krishnakumar, S. Target-specific delivery of doxorubicin to retinoblastoma using epithelial cell adhesion molecule aptamer. Mol. Vis. 2012, 18, 2783–2795. [Google Scholar]
- Ge, M.H.; Zhu, X.H.; Shao, Y.M.; Wang, C.; Huang, P.; Wang, Y.; Jiang, Y.; Maimaitiyiming, Y.; Chen, E.; Yang, C.; et al. Synthesis and characterization of CD133 targeted aptamer-drug conjugates for precision therapy of anaplastic thyroid cancer. Biomater. Sci. 2021, 9, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Duan, J.; Song, Y.; Ma, J.; Wang, F.; Lu, X.; Yang, X. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells In Vitro. J. Transl. Med. 2012, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Duan, J.; Zhan, Q.; Wang, F.; Lu, X.; Yang, X. Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells In Vitro. PLoS ONE 2012, 7, e31970. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Niu, G.; Chen, X. Aptamer-Drug Conjugates. Bioconjugate Chem. 2015, 26, 2186–2197. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. A review of therapeutic aptamer conjugates with emphasis on new approaches. Pharmaceuticals 2013, 6, 340–357. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zeng, Z.; Chen, Z.; Nipper, C.; Liu, X.; Wan, Q.; Chen, J.; Tung, C.; Zu, Y. Aptamer-Gemcitabine Conjugates with Enzymatically Cleavable Linker for Targeted Delivery and Intracellular Drug Release in Cancer Cells. Pharmaceuticals 2022, 15, 558. [Google Scholar] [CrossRef]
- Bargh, J.D.; Isidro-Llobet, A.; Parker, J.S.; Spring, D.R. Cleavable linkers in antibody–drug conjugates. Chem. Soc. Rev. 2019, 48, 4361–4374. [Google Scholar] [CrossRef]
- Sheyi, R.; de la Torre, B.G.; Albericio, F. Linkers: An Assurance for Controlled Delivery of Antibody-Drug Conjugate. Pharmaceutics 2022, 14, 396. [Google Scholar] [CrossRef]
- McCombs, J.R.; Owen, S.C. Antibody drug conjugates: Design and selection of linker, payload and conjugation chemistry. AAPS J. 2015, 17, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Kostova, V.; Désos, P.; Starck, J.B.; Kotschy, A. The Chemistry Behind ADCs. Pharmaceuticals 2021, 14, 442. [Google Scholar] [CrossRef] [PubMed]
- van de Donk, N.W.; Dhimolea, E. Brentuximab vedotin. Mabs-Austin 2012, 4, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Poreba, M. Protease-activated prodrugs: Strategies, challenges, and future directions. FEBS J. 2020, 287, 1936–1969. [Google Scholar] [CrossRef] [PubMed]
- Hamadani, M.; Radford, J.; Carlo-Stella, C.; Caimi, P.F.; Reid, E.G.; O’Connor, O.A.; Feingold, J.M.; Ardeshna, K.; Townsend, W.M.; Solh, M.M.; et al. Final results of a phase 1 study of loncastuximab tesirine in relapsed/refractory B-cell non-Hodgkin lymphoma. Blood 2021, 137, 2634–2645. [Google Scholar] [CrossRef]
- Su, Z.; Xiao, D.; Xie, F.; Liu, L.; Wang, Y.; Fan, S.; Zhou, X.; Li, S. Antibody–drug conjugates: Recent advances in linker chemistry. Acta Pharm. Sin. B 2021, 11, 3889–3907. [Google Scholar] [CrossRef]
- Vollmar, B.S.; Frantz, C.; Schutten, M.M.; Zhong, F.; del Rosario, G.; Go, M.; Yu, S.; Leipold, D.D.; Kamath, A.V.; Ng, C.; et al. Calicheamicin Antibody-Drug Conjugates with Improved Properties. Mol. Cancer Ther. 2021, 20, 1112–1120. [Google Scholar] [CrossRef]
- Cordo′, V.; van der Zwet, J.C.G.; Canté-Barrett, K.; Pieters, R.; Meijerink, J. T-cell Acute Lymphoblastic Leukemia: A Roadmap to Targeted Therapies. Blood Cancer Discov. 2020, 2, 19–31. [Google Scholar] [CrossRef]
- Kaushal, A.; Nooka, A.K.; Carr, A.R.; Pendleton, K.E.; Barwick, B.G.; Manalo, J.; McCachren, S.S.; Gupta, V.A.; Joseph, N.S.; Hofmeister, C.C.; et al. Aberrant Extrafollicular B Cells, Immune Dysfunction, Myeloid Inflammation, and MyD88-Mutant Progenitors Precede Waldenstrom Macroglobulinemia. Blood Cancer Discov. 2021, 2, 600–615. [Google Scholar] [CrossRef]
- Lyon, R.P.; Setter, J.R.; Bovee, T.D.; Doronina, S.O.; Hunter, J.H.; Anderson, M.E.; Balasubramanian, C.L.; Duniho, S.M.; Leiske, C.I.; Li, F.; et al. Self-hydrolyzing maleimides improve the stability and pharmacological properties of antibody-drug conjugates. Nat. Biotechnol. 2014, 32, 1059–1062. [Google Scholar] [CrossRef]
- Chen, Q.; Gabathuler, R. Efficient Synthesis of Doxorubicin Melanotransferrin p97 Conjugates Through SMCC Linker. Synth. Commun. 2004, 34, 2407–2414. [Google Scholar] [CrossRef]
- Barok, M.; Joensuu, H.; Isola, J. Trastuzumab emtansine: Mechanisms of action and drug resistance. Breast Cancer Res. 2014, 16, 209. [Google Scholar] [CrossRef]
- Palomba, M.L.; Younes, A. In the spotlight: A novel CD37 antibody-drug conjugate. Blood 2013, 122, 3397–3398. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.; Filip, I.; Gomez, K.; Engelbrecht, D.; Meer, S.; Lalloo, P.N.; Patel, P.; Perner, Y.; Zhao, J.; Wang, J.; et al. Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK-STAT pathway. Blood Cancer Discov. 2020, 1, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Belantamab Mafodotin: First Approval. Drugs 2020, 80, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Q.; Qiu, L. Smart ligand: Aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J. Control. Release 2013, 171, 152–162. [Google Scholar] [CrossRef]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006, 45, 8149–8152. [Google Scholar] [CrossRef]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23–42. [Google Scholar] [CrossRef]
- Macdonald, J.; Denoyer, D.; Henri, J.; Jamieson, A.; Burvenich, I.; Pouliot, N.; Shigdar, S. Bifunctional Aptamer-Doxorubicin Conjugate Crosses the Blood-Brain Barrier and Selectively Delivers Its Payload to EpCAM-Positive Tumor Cells. Nucleic Acid Ther. 2020, 30, 117–128. [Google Scholar] [CrossRef]
- Yang, C.; Jiang, Y.; Hao, S.H.; Yan, X.Y.; Hong, D.F.; Naranmandura, H. Aptamers: An emerging navigation tool of therapeutic agents for targeted cancer therapy. J. Mater. Chem. B 2021, 10, 20–33. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, J.; Ma, Y.; Zhu, X.; Zhang, C. Aptamers Entirely Built from Therapeutic Nucleoside Analogues for Targeted Cancer Therapy. J. Am. Chem. Soc. 2022, 144, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.; Padilla, R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995, 14, 4609–4621. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Larrea, C.; Staehr, M.; Lopez, A.V.; Ng, K.Y.; Chen, Y.; Godfrey, W.D.; Purdon, T.J.; Ponomarev, V.; Wendel, H.G.; Brentjens, R.J.; et al. Defining an Optimal Dual-Targeted CAR T-cell Therapy Approach Simultaneously Targeting BCMA and GPRC5D to Prevent BCMA Escape-Driven Relapse in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 146–154. [Google Scholar] [CrossRef]
- Silvestri, G.; Trotta, R.; Stramucci, L.; Ellis, J.J.; Harb, J.G.; Neviani, P.; Wang, S.; Eisfeld, A.K.; Walker, C.J.; Zhang, B.; et al. Persistence of Drug-Resistant Leukemic Stem Cells and Impaired NK Cell Immunity in CML Patients Depend on MIR300 Antiproliferative and PP2A-Activating Functions. Blood Cancer Discov. 2020, 1, 48–67. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Gu, X.; Tsao, S.T.; Zhang, Q.J.; Liu, Y.; Yaxian, J.; Yan, L.; Chun, J.; Zhang, R.; Du, Y.; et al. Double CD19/CD22 Chimeric Antigen Receptor-Modified T Cells for the Treatment of Stage IV Relapsed and Refractory Follicular Lymphoma. Blood 2017, 130 (Suppl. 1), 5154. [Google Scholar]
- van de Donk, N.W.C.J.; Themeli, M.; Usmani, S.Z. Determinants of Response and Mechanisms of Resistance of CAR T-cell Therapy in Multiple Myeloma. Blood Cancer Discov. 2021, 2, 302–318. [Google Scholar] [CrossRef]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef]
- Kacherovsky, N.; Cardle, I.I.; Cheng, E.L.; Yu, J.L.; Baldwin, M.; Salipante, S.J.; Jensen, M.C.; Pun, S.H. Traceless aptamer-mediated isolation of CD8(+) T cells for chimeric antigen receptor T-cell therapy. Nat. Biomed. Eng. 2019, 3, 783–795. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, Y.; Liu, P.; Yao, Q.; Zhou, Y.; Li, C.; Zhao, Q.; Liu, G.; Zhang, X. Aptamer-T Cell Targeted Therapy for Tumor Treatment Using Sugar Metabolism and Click Chemistry. ACS Chem. Biol. 2020, 15, 1554–1565. [Google Scholar] [CrossRef]
- Wang, H.; Mooney, D.J. Metabolic glycan labelling for cancer-targeted therapy. Nat. Chem. 2020, 12, 1102–1114. [Google Scholar] [CrossRef]
- Liu, X.; Yan, H.; Liu, Y.; Chang, Y. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small (Weinh. Der Bergstr. Ger.) 2011, 7, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, M.M.; Villanueva, H.; Casares, N.; Lasarte, J.J.; Bendandi, M.; Inogés, S.; de Cerio, A.L.; Pastor, F. MRP1-CD28 bi-specific oligonucleotide aptamers: Target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget 2016, 7, 23182–23196. [Google Scholar] [CrossRef] [PubMed]
- Lancman, G.; Sastow, D.L.; Cho, H.J.; Jagannath, S.; Madduri, D.; Parekh, S.; Richard, S.; Richter, J.; Sanchez, L.; Chari, A. Bispecific Antibodies in Multiple Myeloma: Present and Future. Blood Cancer Discov. 2021, 2, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, X.; Xu, J.; Cui, C.; Safari Yazd, H.; Pan, X.; Zhu, Y.; Chen, X.; Li, X.; Li, J.; et al. Circular Bispecific Aptamer-Mediated Artificial Intercellular Recognition for Targeted T Cell Immunotherapy. ACS Nano 2020, 14, 9562–9571. [Google Scholar] [CrossRef]
- Maimaitiyiming, Y.; Wang, Q.Q.; Yang, C.; Ogra, Y.; Lou, Y.; Smith, C.A.; Hussain, L.; Shao, Y.M.; Lin, J.; Liu, J.; et al. Hyperthermia Selectively Destabilizes Oncogenic Fusion Proteins. Blood Cancer Discov. 2021, 2, 388–401. [Google Scholar] [CrossRef]
- Rosselló-Tortella, M.; Ferrer, G.; Esteller, M. Epitranscriptomics in Hematopoiesis and Hematologic Malignancies. Blood Cancer Discov. 2020, 1, 26–31. [Google Scholar] [CrossRef]
- Romine, K.A.; Nechiporuk, T.; Bottomly, D.; Jeng, S.; McWeeney, S.K.; Kaempf, A.J.; Corces, M.R.; Majeti, R.; Tyner, J.W. Monocytic Differentiation and AHR Signaling as Primary Nodes of BET Inhibitor Response in Acute Myeloid Leukemia. Blood Cancer Discov. 2021, 2, 518–531. [Google Scholar] [CrossRef]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Schmidt, C.R.; Achille, N.J.; Kuntimaddi, A.; Boulton, A.M.; Leach, B.I.; Zhang, S.; Zeleznik-Le, N.J.; Bushweller, J.H. BCOR Binding to MLL-AF9 Is Essential for Leukemia via Altered EYA1, SIX, and MYC Activity. Blood Cancer Discov. 2020, 1, 162–177. [Google Scholar] [CrossRef]
- Ogawa, S. Deciphering the Clonal Origin of Relapsed Acute Lymphoblastic Leukemia in Children. Blood Cancer Discov. 2020, 1, 21–22. [Google Scholar] [CrossRef]
- Khan, S.; He, Y.; Zhang, X.; Yuan, Y.; Pu, S.; Kong, Q.; Zheng, G.; Zhou, D. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene 2020, 39, 4909–4924. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, S.D.; Yang, B.; Fallan, C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: Recent progress and future challenges. Bioorganic Med. Chem. Lett. 2019, 29, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Bricelj, A.; Steinebach, C.; Kuchta, R.; Gütschow, M.; Sosič, I. E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points. Front. Chem. 2021, 9, 707317. [Google Scholar] [CrossRef]
- He, Y.; Khan, S.; Huo, Z.; Lv, D.; Zhang, X.; Liu, X.; Yuan, Y.; Hromas, R.; Xu, M.; Zheng, G.; et al. Proteolysis targeting chimeras (PROTACs) are emerging therapeutics for hematologic malignancies. J. Hematol. Oncol. 2020, 13, 103. [Google Scholar] [CrossRef]
- He, S.; Gao, F.; Ma, J.; Ma, H.; Dong, G.; Sheng, C. Aptamer-PROTAC Conjugates (APCs) for Tumor-Specific Targeting in Breast Cancer. Angew. Chem. Int. Ed. Engl. 2021, 60, 23299–23305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, L.; Wang, X.; Liu, H.; Zhang, Y.; Xie, T.; Zhang, H.; Li, X.; Peng, T.; Sun, X.; et al. Development of a Novel PROTAC using the Nucleic Acid Aptamer as a Targeting Ligand for Tumor Selective Degradation of Nucleolin. Mol. Ther.-Nucleic Acids 2022, 30, 66–79. [Google Scholar] [CrossRef] [PubMed]


| Classification | Biomarker | Description | Agent | Ref. | |
|---|---|---|---|---|---|
| CD33 | Belongs to Siglecs family; in approximately 85% to 90% AML patients. | Gemtuzumab ozogamicin CAR-T (phase 1) | [32] | ||
| CD44 | Strongly expressed on all AML cells. | RO5429083 with cytarabine (phase 1), CAR-T (phase 1/2) | [52,58] | ||
| CD47 | Overexpressed in leukemic blasts and progenitors, a macrophage immune checkpoint, protects cells from phagocytosis. | Lemzoparlima (phase 1/2a), magrolimab (5F9) with azacitidine (phase 1b) | [53,59,60] | ||
| Acute Myeloid Leukemia (AML) | CD117 | Also named C-kit, a tyrosine kinase receptor, expressed in more than 90% of AML patients with physiological HSPC and leukemic blasts. | MGTA-117 (phase 1) | [54,61] | |
| Acute Leukemia | CD123 | Mainly expressed on AML leukemic stem cells. | CSL362 (phase 1), flotetuzumab (phase 1) CAR-T (phase 2) | [32,55] | |
| CD134 | Also named OX40, belongs to NGFR/TNFR superfamily, mainly expressed on Teffs and Tregs. OX40–OX40L interaction promotes NK cells in AML. | n.a. | [56] | ||
| CD170 | Also named siglec-5, upregulated during granulocyte maturation, overexpressed on the AML non-M3 phenotypes. | n.a. | [62] | ||
| Acute Lymphocytic Leukemia (ALL) | CD19 | 80% of ALL expressed moderate to high levels of CD19. | Blinatumomab | [63,64] | |
| CD22 | Highly expressed on leukemic cells from most R/R B-ALL patients. | Inotuzumab ozogamicin (phase 2), moxetumomab pasudotox-tdfk | [65,66] | ||
| Chronic Leukemia | Chronic Lymphocytic Leukemia (CLL) | CD20 | Expressed in B-cell-derived tumor cells, such as CLL. | Ofatumumab (phase 2), obinutuzumab (phase 2) | [57,67,68] |
| Aptamer | Target | Drug | Cancer | Reference |
|---|---|---|---|---|
| AS1411 | Nucleolin | Dox | Liver Cancer | [74] |
| Pacitaxel | Ovarian Cancer | [75] | ||
| Gemcitabine | Pancreatic Cancer | [76] | ||
| P19 | PANC-1 cell | MMAE | Pancreatic Cancer | [77] |
| DM1 | Pancreatic Cancer | [77] | ||
| E07 | EGFR | MMAE | Pancreatic Cancer | [78] |
| MMAF | Pancreatic Cancer | [78] | ||
| Gemcitabine | Pancreatic Cancer | [79] | ||
| Waz | Transferrin | MMAE | Pancreatic Cancer | [78] |
| MMAF | Pancreatic Cancer | [78] | ||
| S30-T1 | CD33 | Dox | Acute Myeloid Leukemia | [80] |
| Sgc8 | PTK7 | Dox | Acute Lymphoblastic Leukemia | [81] |
| 5-FU | Colorectal Cancer | [82] | ||
| EpDT3 | EpCAM | Dox | Colorectal Cancer | [83] |
| AP-1 | CD133 | Dox | Anaplastic Thyroid Cancer | [84] |
| HB-5 | HER-2 | Dox | Breast Cancer | [85] |
| MA-3 | MUC-1 | Dox | Lung CancerBreast Cancer | [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.C.; Yan, X.Y.; Yang, C.; Naranmandura, H. The Landscape of Nucleic-Acid-Based Aptamers for Treatment of Hematologic Malignancies: Challenges and Future Directions. Bioengineering 2022, 9, 635. https://doi.org/10.3390/bioengineering9110635
Wang SC, Yan XY, Yang C, Naranmandura H. The Landscape of Nucleic-Acid-Based Aptamers for Treatment of Hematologic Malignancies: Challenges and Future Directions. Bioengineering. 2022; 9(11):635. https://doi.org/10.3390/bioengineering9110635
Chicago/Turabian StyleWang, Si Chun, Xing Yi Yan, Chang Yang, and Hua Naranmandura. 2022. "The Landscape of Nucleic-Acid-Based Aptamers for Treatment of Hematologic Malignancies: Challenges and Future Directions" Bioengineering 9, no. 11: 635. https://doi.org/10.3390/bioengineering9110635
APA StyleWang, S. C., Yan, X. Y., Yang, C., & Naranmandura, H. (2022). The Landscape of Nucleic-Acid-Based Aptamers for Treatment of Hematologic Malignancies: Challenges and Future Directions. Bioengineering, 9(11), 635. https://doi.org/10.3390/bioengineering9110635