3.1. Validation of the GRC and GOC Simulations
The LIF models described in the previous section can reproduce the behavior of the GRC and GOC cells, which are the neurons included in the granular layer. It is worth noticing that these neurons can be described by Equations (
3) and (
6), changing the parameters
,
,
,
,
,
and
. The parameters used to describe the GRC and GOC cells have been taken from the NeuroElectro Database [
17]. The parameters values used for each cell type are reported in
Table 1.
Considering the synapses, the excitatory and inhibitory ones are characterized by the parameters
,
,
w and the initial value of
. Again, the parameters values have been taken from the NeuroElectro Database [
17] and reported in
Table 2.
The analysis of the values reported in
Table 1 and
Table 2 allows to compare the behaviors of the GRC and GOC. It is important to notice that the parameter
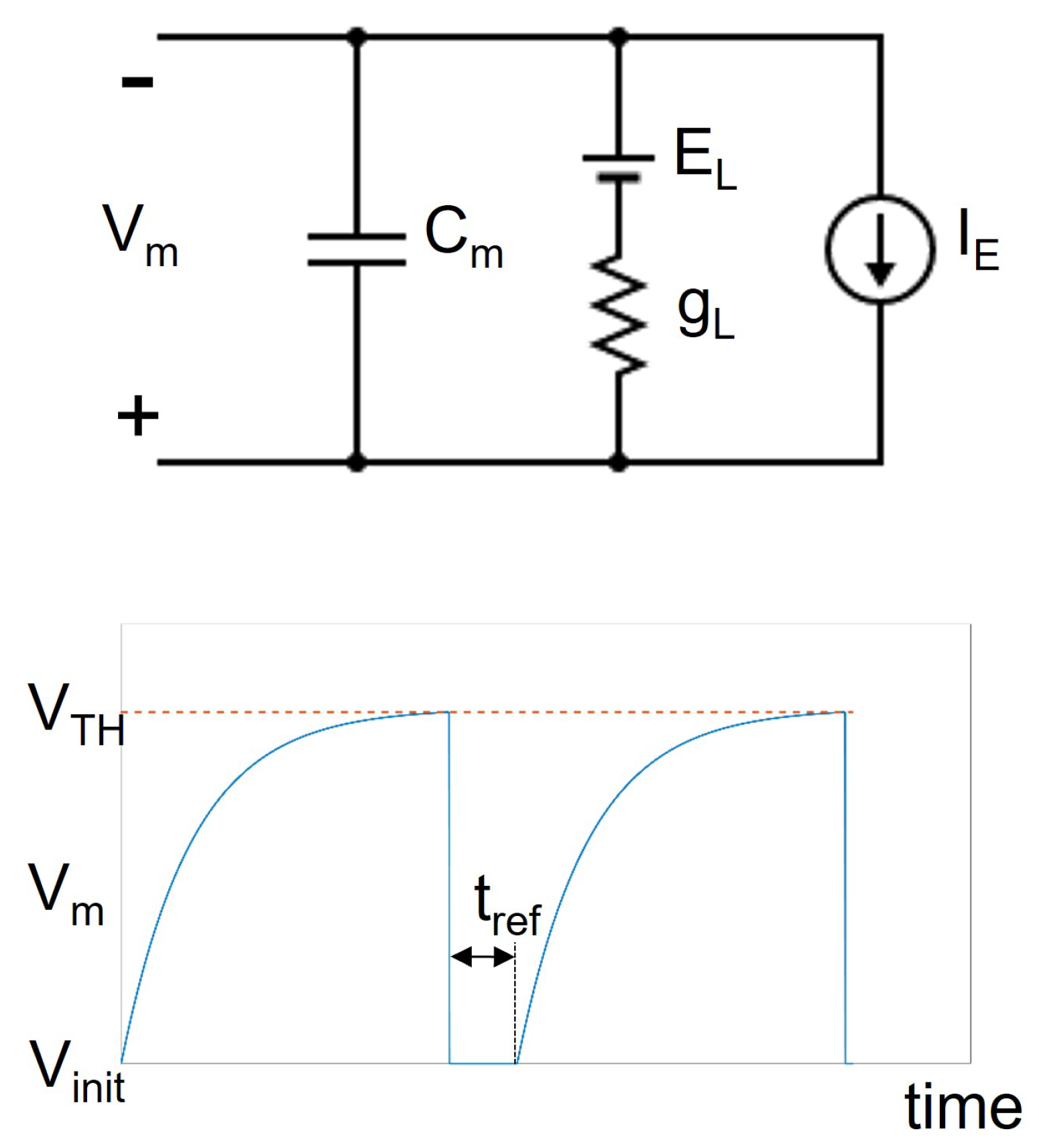
is equal to zero only for the GRC. This means that, in normal conditions, the GRC maintains its resting potential
. On the other hand, the GOC is characterized by a positive value of
. This means that this cell is characterized by a natural peacemaking, i.e., the potential varies as a periodical signal which grows from
to
and then is reset to
. Moreover, the role of the synapses is to instantly change the value of the potential when they receive a spike. In particular, the excitatory synapse increases the potential, while the inhibitory one decreases the value.
In order to integrate the LIF models of the GRC and GOC into the GES simulator, authors developed a serial C code to solve Equation (
6). The results obtained by these single cells simulators have been compared with the one reported in the NeuroElectro Database, in order to validate the developed serial C codes. Examples of the output produced by these simulators are reported in
Figure 3.
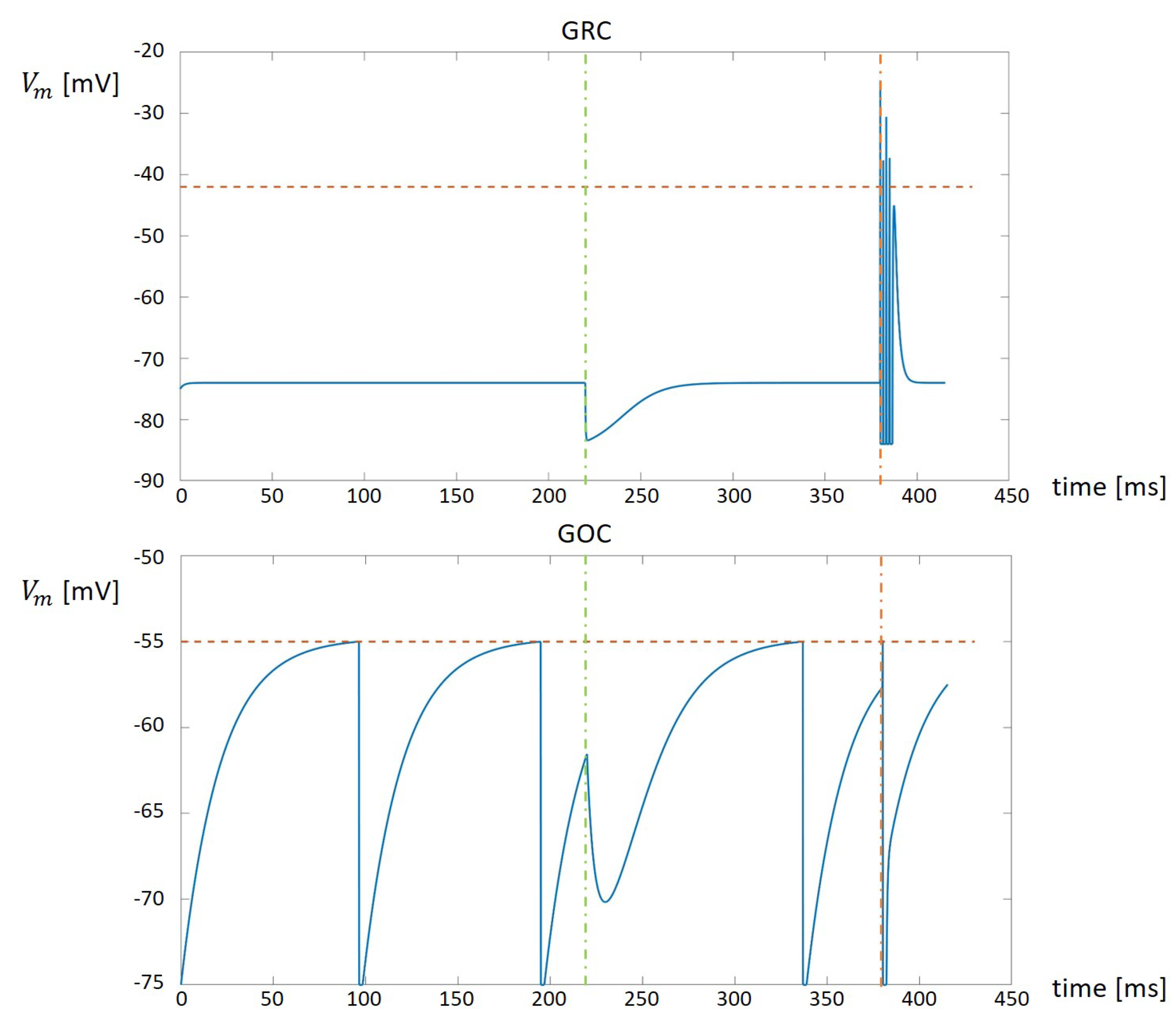
The top part of
Figure 3 shows the result produced by the GRC serial simulator. It is worth noticing that, at the beginning, the potential is fixed to the
value. When an inhibitory synapse is activated by a spike (vertical green dashed line) the potential instantly diminishes and then grows back to the
value. On the other hand, when an excitatory synapse is activated by a spike (vertical red dashed line) the potential is characterized by the presence of spikes. When the excitatory effect ends, the potential comes back again to the
value. The bottom part of
Figure 3 shows the GOC activity. At the beginning, the GOC shows its natural peacemaking. When the inhibitory synapse is activated (vertical green dashed line), the potential is instantly decreased and, then, the cells continues with its natural peacemaking. Considering the excitatory input (vertical red dashed line), it instantly increments the potential. In the case shown in
Figure 3, the excitatory input increases the potential to a value greater then the threshold. The value is then reset to
and, after the refractory time, the cells restart with the natural peacemaking.
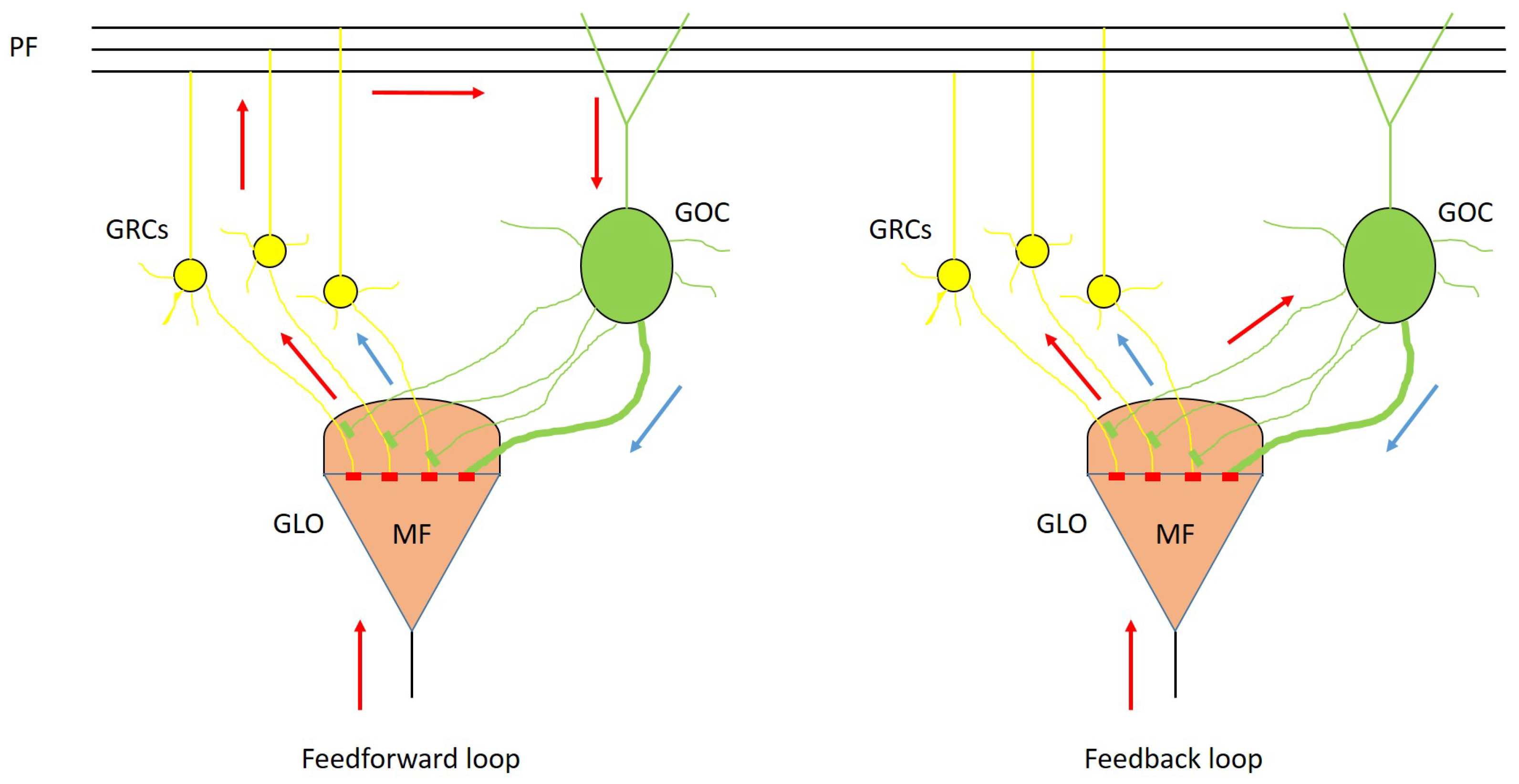
In order to integrate these two serial simulators into the GES software, authors developed a serial simulator of the cerebellar granular layer, exploiting the cells placing and connection module of the GES simulator. This module produces as output a set of connection matrices that are used to route the signals to the cells. For each GRC and GOC, in each synapse, a suitable array is allocated. Thus, these arrays are used to store the spike times related to a synapse and it is removed from the array when a spike is processed by the synapse. Concerning the GRCs, the excitatory inputs are generated by the MF, while for the GOCs they are generated from the MFs or from the GRCs. On the other hand, the inhibition signals are generated by the GOCs for the GRCs. The MFs are initialized using the spike train generator already described in [
1]. The arrays storing the spikes queues not generated by the MFs change their content during the simulation depending on which neurons generate a spike.
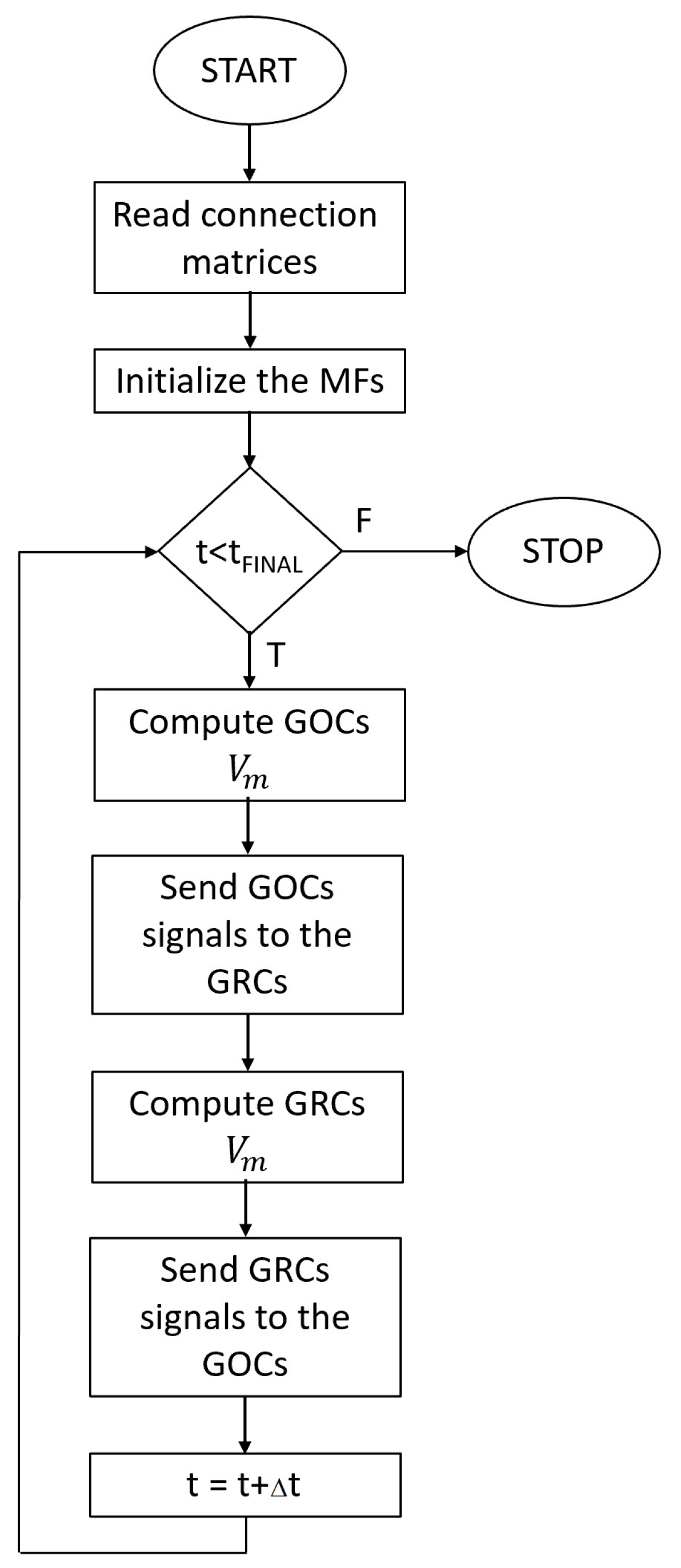
The flowchart of the serial simulator is shown in
Figure 4. After reading the connection matrices and initializing the MFs, the simulator starts to evaluate the main
for loop related to the time. For each time step, the cellular activity of the GOCs is computed (i.e.,
evaluated). If the potential of each GOC reaches the value
, this means that this cell has generated a spike. Therefore, the time stamp of the spike is computed by summing to actual time
t a pseudo-random value related to the propagation time from the
i-th GOC to the connected cells. This time stamp is then stored in the inhibitory spike queues of all the GRCs connected to the
i-th GOC.
At this point, the GRCs potentials have been updated; a spike evaluation mechanism similar to the one previously described is implemented in order to store the time stamps into the GOCs excitatory queues. The main loop is repeated until the actual simulated time t is lower than the final time .
3.2. Parallel Implementation of the Granular Layer Network
By analyzing the flowchart reported in
Figure 4, the computational complexity of the simulation is related to the main
for loop iterating over the time steps. Moreover, inside this loop, the evaluation of the GOCs and GRCs activities is the most time consuming operations because the simulator should solve Equation (
6) for each cell. Finally, the signal routing to the suitable queues should be performed serially, since more than one cell can write to the same queue at the same iteration
t, but the stored time stamp must be in ascending order to correctly evaluate the spikes. Thus, we developed a CUDA-based algorithm to reproduce in parallel the activity of the GOCs and GRCs. The flowchart reported in
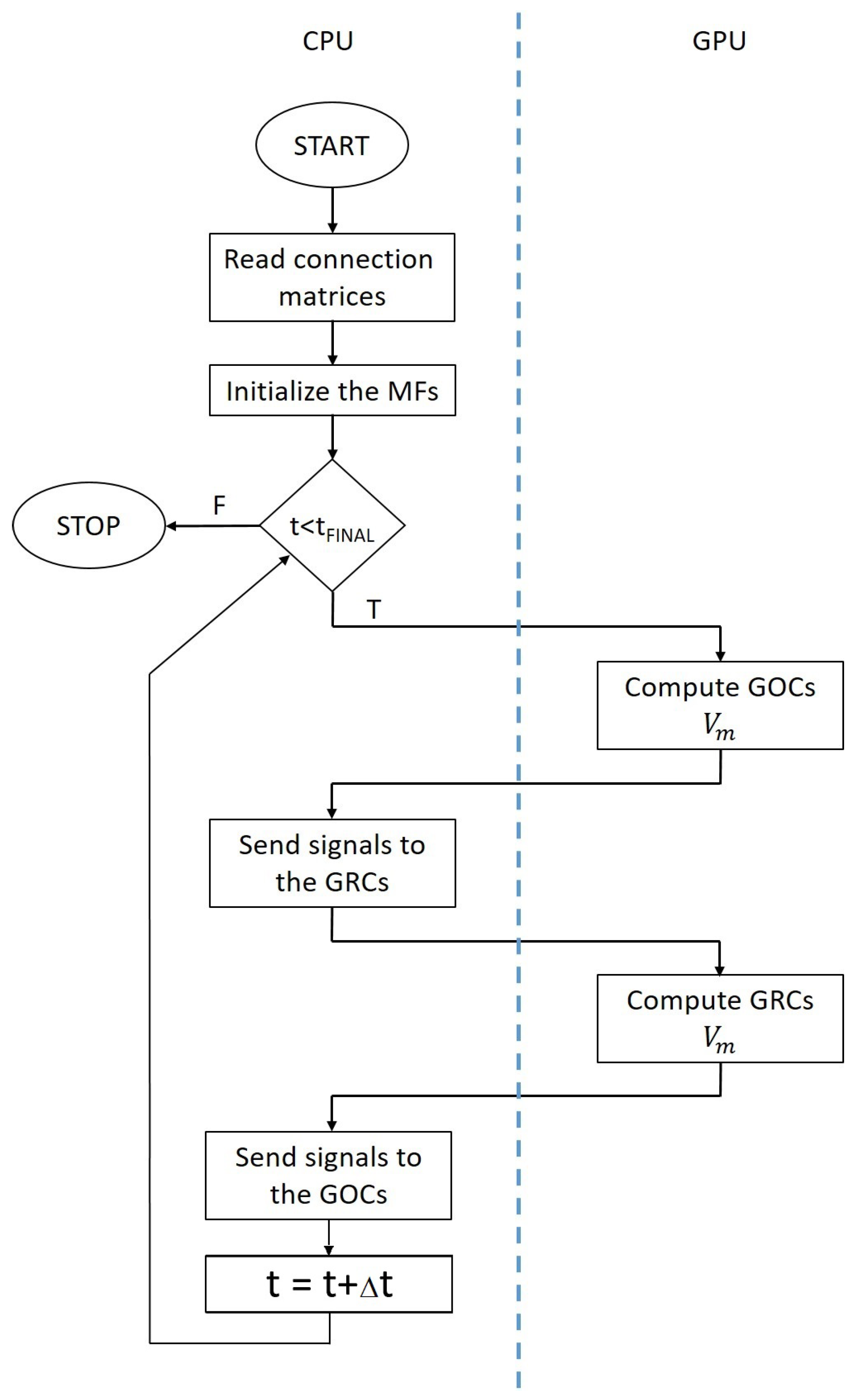
Figure 5 highlights the tasks performed by the CPU in serial and the parallel elaboration carried out by the GPU. Moreover, data transfers are represented by arrows passing through the blue dashed line.
As for the serial network simulator, the first tasks are related to the acquisition of the connection matrices and the generation of the spike queues related to the MFs. Then, the main
for loop can begin. The first task checks if each queue related to each GOC contains a spike at the actual simulation time. If this condition is true, a flag is set into a suitable array in the
i-th position to indicate that the GOC connected to the
i-th synapse is receiving a stimulus. Thus, after evaluating each queue, an array of flags is transferred to the GPU memory. Then, a number of parallel threads equal to the number of GOC is used by the GPU to carry out the cellular activity simulation. Thus, each GPU thread solves Equation (
6), considering the presence of a spike in each synapse by checking the flags array portion related to the considered neuron. It is worth noticing that the parameters and variables values needed to solve Equation (
3) are stored in suitable arrays allocated on the GPU memory before the main loop. On the other hand, the constant parameters are defined as macro and then substituted with the corresponding value at compilation time. If the
i-th GOC has produced a spike at the current time (i.e., its potential is equal or grater than
), a flag is set in the
i-th position of an array, then is transferred back to the CPU memory. The information stored in this array are used by the CPU to set flags into the GRCs inhibitory queues. In particular, if the
i-th GOC is connected to the
j-th GRC and this GOC produces a spike, the flag array related to the inhibitory queue of the
j-th GRC is set true. Moreover, the spike queues related to the MFs are checked in order to correctly set the flag array related to the excitatory synapses. These flag arrays are then transferred to the GPU memory, as for the flag array related to the GOC. On the GPU, a set of threads equal to the number of GOCs is created to evaluate in parallel the activity of these neurons. After updating the GRC potential value, each thread checks if the assigned cell produced a spike at the current time. Again, the memory addresses of the flag array related to the GRC that generated a spike are set to the true value. This array is transferred back to the CPU memory, where it is used, together with the MFs queues to generate the signals for the GOC of the next iteration of the main loop.
This simulation framework can be generalized to exploit multi-GPU systems by using CUDA streams, which allow asynchronous data transfers and GPU kernel execution. In particular, a stream is associated to each GPU of the system. Thus, each GPU elaborates data independently from the other devices. This strategy can be applied only to the GOCs and GRCs activity evaluation, since, as explained before, the queues should be serially managed. Therefore, a synchronization barrier is placed before each serial code block. Notice that all the other GPU related operations (memory transfers and kernels executions) are managed through CUDA streams; therefore, the data transfers and kernel executions of different devices are overlapped.
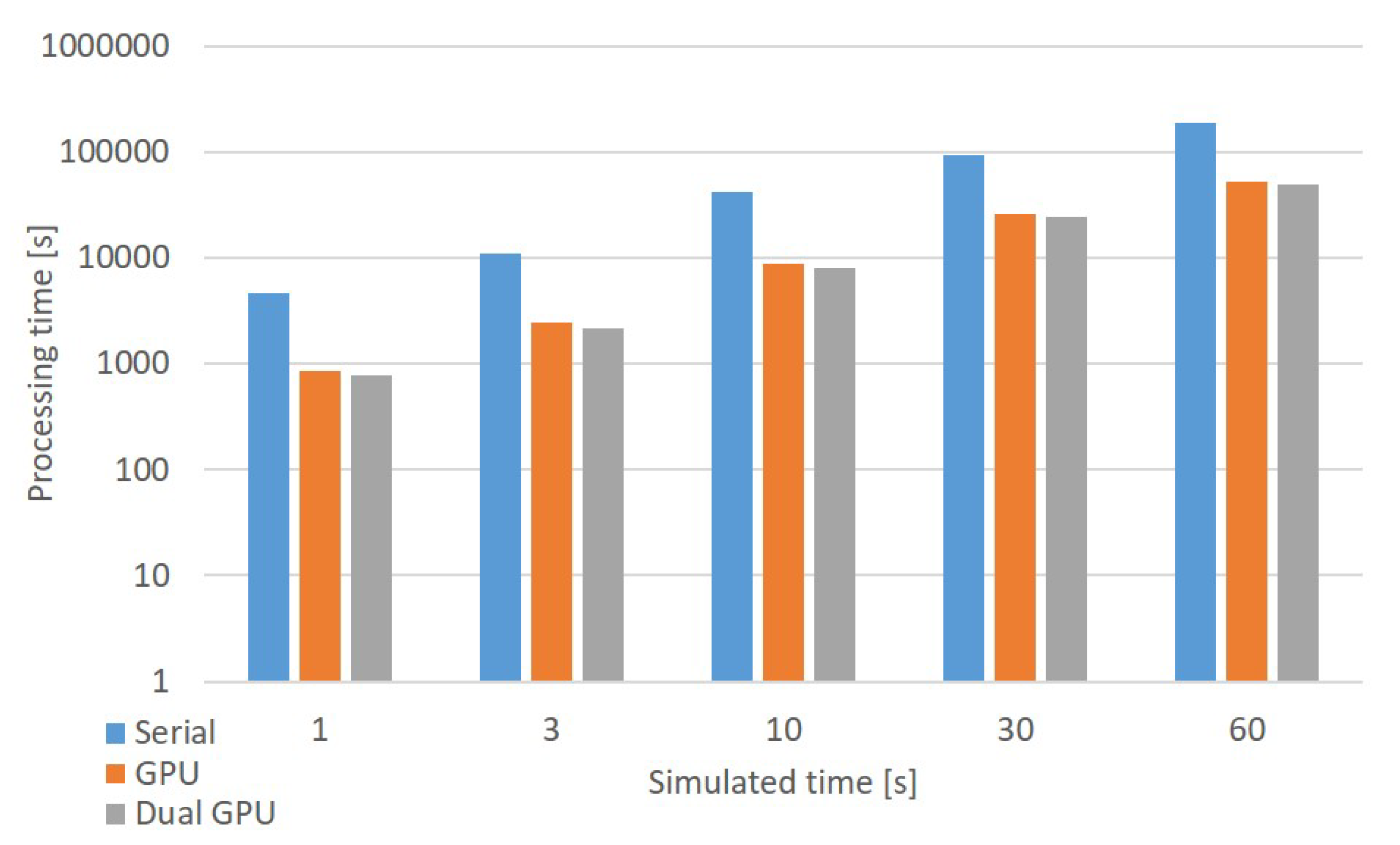
In general, considering a system equipped with N GPUs, each device performs the activity simulation of an equal number of neurons. If denotes the total number of GOCs and is the total number of GRCs, each GPU elaborates and neurons.