Inference of Regulatory System for TAG Biosynthesis in Lipomyces starkeyi
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Data Processing
2.2. Gene Selection
2.3. Factor Analysis
2.4. Stepwise Network Modeling
- STEP 1:
- Initial model assumption of oil productivity group;
- STEP 2:
- Model optimization of oil productivity group;
- STEP 3:
- Definition of pseudo variables from subgroups;
- STEP 4:
- Initial model assumption among pseudo variables;
- STEP 5:
- Model optimization of pseudo variables.
2.4.1. Initial Model Assumption
2.4.2. Network Modeling
3. Results
3.1. Gene Classification by Factor Analysis
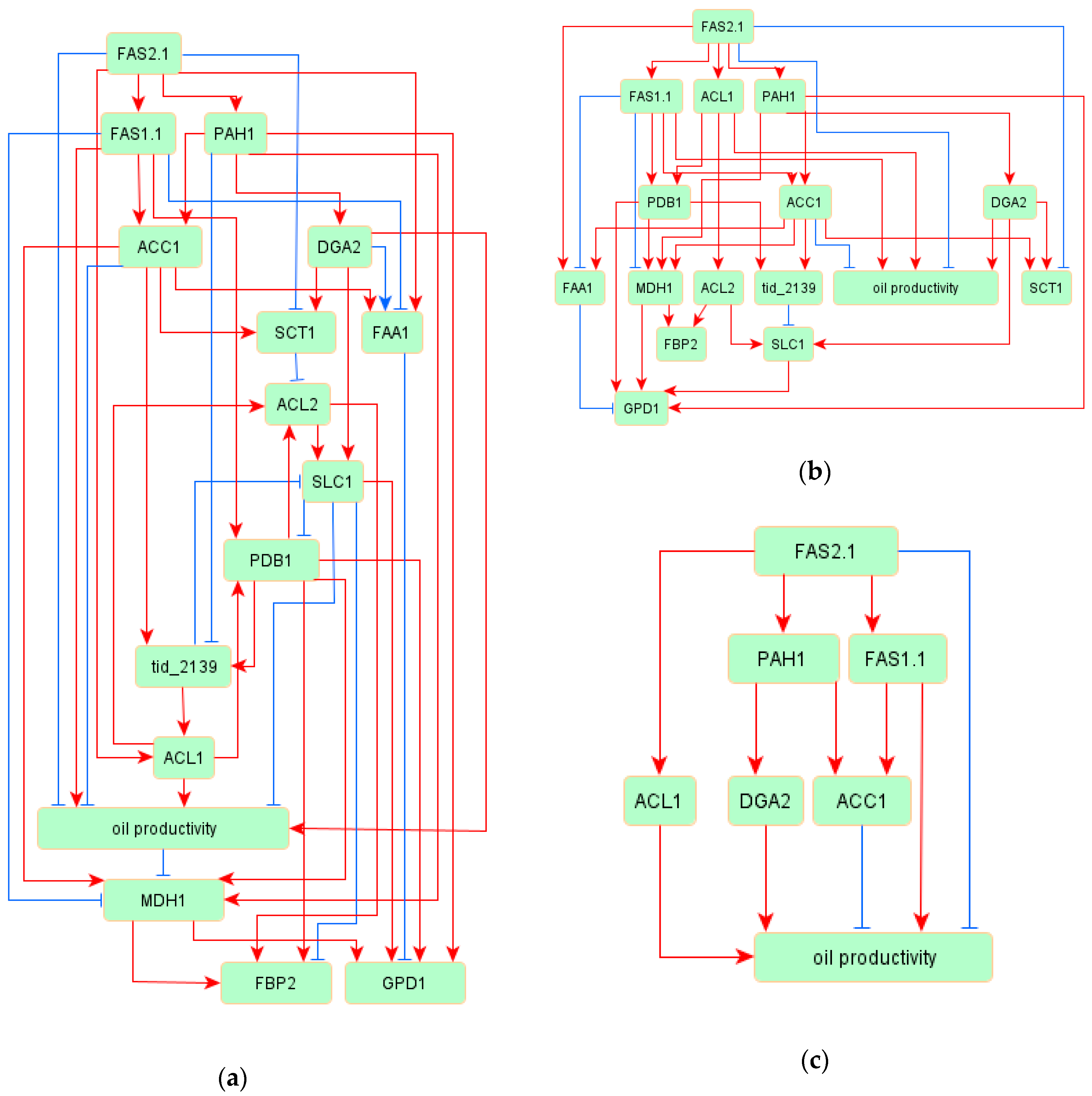
3.2. Oil Productivy Network: Figures, Tables and Schemes
3.3. Regulatory Network of TAG Biosynthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sakaguchi, K.; Okanishi, M. Molecular Breeding and Genetics of Applied Microorganisms; Academic Press: Cambridge, MA, USA, 1980; pp. 139–153. [Google Scholar]
- Zhang, G.Q.; Lin, Y.P.; Qi, X.N.; Wang, L.X.; He, P.; Wang, Q.H.; Ma, Y.H. Genome shuffling of the nonconventional yeast Pichia anomala for improved sugar alcohol production. Microb. Cell Fact. 2015, 14, 112. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.F.; Li, H.P.; Wang, L.Y.; Zhang, C.; Xing, X.H.; Bao, C.Y. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl. Microbiol. Biotechnol. 2014, 98, 5387–5396. [Google Scholar] [CrossRef]
- Liu, Y.N.; Li, Q.G.; Zheng, P.; Zhang, Z.D.; Liu, Y.F.; Sun, C.M.; Cao, G.Q.; Zhou, W.J.; Wang, X.W.; Zhang, D.W.; et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb. Cell Fact. 2015, 14, 121. [Google Scholar] [CrossRef]
- Zhong, W.; Jousset, A. Plant Breeding Goes Microbial. Trends Plant Sci. 2017, 22, 555–558. [Google Scholar]
- Angerbauer, C.; Siebenhofer, M.; Mittelbach, M.; Guebitz, G.M. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour. Technol. 2008, 99, 3051–3056. [Google Scholar] [CrossRef]
- Beligon, V.; Christophe, G.; Fontanille, P.; Larroche, C. Microbial lipids as potential source to food supplements. Curr. Opin. Food. Sci. 2016, 7, 35–42. [Google Scholar] [CrossRef]
- Kosa, M.; Ragauskas, A.J. Lipids from heterotrophic microbes: Advances in metabolism research. Trends Biotechnol. 2011, 29, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Juanssilfero, A.B.; Kahar, P.; Amza, R.L.; Miyamoto, N.; Otsuka, H.; Matsumoto, H.; Kihira, C.; Thontowi, A.; Ogino, C.; Prasetya, B. Effect of inoculum size on single-cell oil production from glucose and xylose using oleaginous yeast Lipomyces starkeyi. J. Biosci. Bioeng. 2018, 125, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Göker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Toya, Y.; Kurokawa, H.; Fukano, Y.; Sato, A.; Umemura, H.; Yamada, K.; Iwasaki, H.; Tobori, N.; Shimizu, H. Characterization of oil-producing yeast Lipomyces starkeyi on glycerol carbon source based on metabolomics and 13C-labeling. Appl. Microbiol. Biotechnol. 2018, 102, 8909–8920. [Google Scholar] [CrossRef] [PubMed]
- Pomraning, K.R.; Collett, J.R.; Kim, J.; Panisko, E.A.; Culley, D.E.; Dai, Z.; Deng, S.; Hofstad, B.A.; Butcher, M.G.; Magnuson, J.K. Transcriptomic analysis of the oleaginous yeast Lipomyces starkeyi during lipid accumulation on enzymatically treated corn stover hydrolysate. Biotechnol. Biofuels 2019, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Sutanto, S.; Zullaikah, S.; Tran-Nguyen, P.L.; Ismadji, S.; Ju, Y.-H. Lipomyces starkeyi: Its current status as a potential oil producer. Fuel Process. Technol. 2018, 177, 39–55. [Google Scholar] [CrossRef]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Takaku, H.; Matsuzawa, T.; Yaoi, K.; Yamazaki, H. Lipid metabolism of the oleaginous yeast Lipomyces starkeyi. Appl. Microbiol. Biotechnol. 2020, 104, 6141–6148. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zhang, S.; Wang, Q.; Tan, H.; Zhao, Z.K. The isocitrate dehydrogenase gene of oleaginous yeast Lipomyces starkeyi is linked to lipid accumulation. Can. J. Microbiol. 2009, 55, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Muniraj, I.K.; Uthandi, S.K.; Hu, Z.; Xiao, L.; Zhan, X. Microbial lipid production from renewable and waste materials for second-generation biodiesel feedstock. Environ. Technol. Rev. 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Akutsu, T.; Miyano, S.; Kuhara, S. Algorithms for identifying Boolean networks and related biological networks based on matrix multiplication and fingerprint function. J. Comput. Biol. 2000, 7, 331–343. [Google Scholar] [CrossRef]
- Friedman, N.; Linial, M.; Nachman, I.; Pe’er, D. Using Bayesian networks to analyze expression data. J. Comput. Biol. 2000, 7, 601–620. [Google Scholar] [CrossRef]
- Aburatani, S.; Kuhara, S.; Toh, H.; Horimoto, K. Deduction of a gene regulatory relationship framework from gene expression data by the application of graphical Gaussian modeling. Signal. Process. 2003, 83, 777–788. [Google Scholar] [CrossRef]
- Bollen, K.A. Structural Equations with Latent Variables; Wiley-Interscience: New York, NY, USA, 1989. [Google Scholar]
- Aburatani, S. Application of structure equation modeling for inferring a serial transcriptional regulation in yeast. Gene Regul. Syst. Biol. 2011, 5, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Haavelmo, T. The statistical implications of a system of simultaneous equations. Econometrica 1943, 11, 1–12. [Google Scholar] [CrossRef]
- Duncan, O.D. Introduction to Structural Equation Models, 3rd ed.; Academic Press: New York, NY, USA, 1975. [Google Scholar]
- Pearl, J. Causality: Models, Reasoning, and Inference, 2nd ed.; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Aburatani, S.; Toh, H. Estimation of Physical Transcriptional Control in Yeast Cell Cycle by Structure Equation Modeling. In Basic Methods in Protein Purification and Analysis; iConcept Press Ltd.: Hong Kong, China, 2012; Volume 14, Chapter 5. [Google Scholar]
- Aburatani, S.; Toh, H. Network inference of AP pattern formation system in D. melanogaster by structural equation modeling. J. Phys. Conf. Ser. 2014, 490, 012145. [Google Scholar] [CrossRef]
- Aburatani, S. Network inference of pal-1 lineage-specific regulation in the C. elegans embryo by structural equation modeling. Bioinformation 2012, 8, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Aburatani, S.; Nagano, R.; Sone, H.; Fujibuchi, W.; Yamane, J.; Imanishi, S.; Ohsako, S. Inference of Gene Regulatory Networks to Detect Toxicity-Specific Effects in Human Embryonic Stem Cells. Int. J. Adv. Life Sci. 2013, 5, 103–114. [Google Scholar]
- Yamazaki, H.; Kobayashi, S.; Ebina, S.; Abe, S.; Ara, S.; Shida, Y.; Ogasawara, W.; Yaoi, K.; Araki, H.; Takaku, H. Highly selective isolation and characterization of Lipomyces starkeyi mutants with increased production of triacylglycerol. Appl. Microbiol. Biotechnol. 2019, 103, 6297–6308. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Nicaud, J.-M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef]
- Spirtes, P.; Glymour, C.; Scheines, R. Causation, Prediction, and Search, 2nd ed.; The MIT Press: Cambridge, UK, 2001. [Google Scholar]
- Aburatani, S.; Shida, Y.; Ogasawara, W.; Yamazaki, H.; Takaku, H. Application of Structural Equation Modelling for Oil Accumulation System Control in Oleaginous yeast. J. Phys. Conf. Ser. 2019, 1391, 012043. [Google Scholar] [CrossRef]
- Wheaton, B.; Muthen, B.; Alwin, D.F.; Summers, G.F. Assessing Reliability and Stability in Panel Models. Sociol. Methodol. 1977, 8, 84–136. [Google Scholar] [CrossRef]
- Joreskog, K.G.; Sorbom, D. LISREL-V1 User’s Guide, 3rd ed.; Scientific Software: Mooresville, NC, USA, 1984. [Google Scholar]
- Browne, M.W.; Cudeck, R. Alternative ways of assessing model fit. In Testing Structural Equation Models; Boolen, K.A., Long, J.S., Eds.; Sage: Beverly Hills, CA, USA, 1993; pp. 136–162. [Google Scholar]
- Bentler, P.M. Comparative fit indexes in structural models. Psychol. Bull. 1990, 107, 238–246. [Google Scholar] [CrossRef]
- Akaike, H. Factor analysis and AIC. Psychometrika 1987, 52, 317–332. [Google Scholar] [CrossRef]
- Silverman, A.M.; Qiao, K.; Xu, P.; Stephanopoulos, G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2016, 100, 3781–3798. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Marsafari, M.; Deng, L.; Xu, P. Understanding lipogenesis by dynamically profiling transcriptional activity of lipogenic promoters in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2019, 103, 3167–3179. [Google Scholar] [CrossRef] [PubMed]

| Estimated Factor Loadings | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group # | Gene | Communality | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| 1 | LACS1.1 | 0.951 | 1.028 | 0.174 | −0.081 | 0.062 | 0.085 | −0.034 | 0.001 | −0.056 | 0.003 |
| POT2 | 0.953 | 0.984 | 0.125 | −0.060 | 0.333 | 0.012 | −0.129 | 0.010 | 0.102 | 0.042 | |
| POX1 | 0.900 | 0.966 | 0.243 | 0.003 | 0.135 | 0.075 | −0.266 | 0.171 | 0.109 | 0.028 | |
| FOX2 | 0.960 | 0.965 | −0.021 | −0.061 | 0.097 | 0.037 | 0.044 | 0.119 | −0.074 | −0.036 | |
| FBP1 | 0.907 | 0.957 | 0.261 | −0.057 | −0.067 | 0.354 | −0.055 | −0.084 | −0.079 | −0.249 | |
| CIT3 | 0.899 | 0.942 | 0.107 | −0.088 | 0.015 | 0.180 | 0.044 | −0.054 | 0.181 | 0.082 | |
| CIT2 | 0.913 | 0.912 | 0.042 | 0.083 | −0.043 | −0.086 | 0.065 | 0.044 | 0.131 | 0.016 | |
| ALDH | 0.910 | 0.883 | −0.097 | 0.011 | −0.143 | 0.108 | 0.153 | −0.089 | −0.080 | 0.074 | |
| TGL4 | 0.914 | 0.871 | 0.082 | 0.046 | 0.109 | −0.096 | 0.282 | −0.131 | −0.006 | 0.002 | |
| TGL3 | 0.825 | 0.851 | 0.035 | 0.120 | 0.118 | 0.052 | −0.192 | −0.236 | −0.204 | 0.335 | |
| ARE2 | 0.932 | 0.793 | −0.395 | 0.099 | −0.072 | 0.027 | 0.008 | −0.005 | 0.016 | −0.135 | |
| ARE1 | 0.822 | 0.781 | −0.223 | 0.375 | −0.045 | 0.066 | −0.233 | 0.193 | 0.312 | −0.002 | |
| POT1 | 0.899 | 0.720 | −0.038 | −0.021 | −0.085 | −0.050 | 0.174 | −0.169 | 0.037 | 0.397 | |
| ERG13 | 0.905 | −0.688 | 0.129 | 0.441 | 0.029 | 0.002 | −0.048 | −0.063 | −0.030 | 0.060 | |
| PCK1 | 0.782 | 0.686 | −0.282 | −0.082 | −0.130 | 0.510 | 0.037 | −0.043 | 0.182 | 0.074 | |
| ERG10 | 0.915 | −0.649 | 0.127 | 0.372 | 0.132 | 0.042 | 0.044 | 0.150 | −0.012 | −0.013 | |
| ACO2 | 0.847 | −0.641 | 0.024 | −0.110 | 0.030 | 0.109 | 0.046 | 0.565 | 0.133 | −0.077 | |
| LAT1 | 0.929 | −0.605 | −0.001 | 0.342 | −0.043 | 0.324 | 0.089 | 0.252 | −0.102 | 0.135 | |
| PDX1 | 0.962 | −0.595 | 0.283 | 0.461 | −0.181 | 0.067 | −0.013 | 0.161 | −0.015 | 0.048 | |
| PYC2 | 0.787 | 0.584 | −0.047 | 0.255 | −0.199 | 0.033 | 0.279 | −0.306 | 0.323 | 0.094 | |
| DGK1 | 0.846 | −0.555 | 0.299 | −0.035 | 0.257 | −0.002 | −0.372 | −0.115 | 0.100 | −0.281 | |
| AYR1 | 0.843 | 0.551 | −0.313 | 0.175 | 0.261 | −0.301 | 0.207 | 0.200 | −0.330 | −0.044 | |
| PDA1 | 0.897 | −0.422 | 0.264 | 0.378 | −0.111 | 0.276 | −0.016 | 0.210 | −0.096 | −0.020 | |
| SDH2 | 0.874 | 0.340 | 0.303 | −0.082 | 0.335 | 0.310 | 0.119 | 0.315 | −0.196 | 0.153 | |
| 2 | PFK2 | 0.837 | −0.118 | 0.949 | 0.001 | 0.028 | −0.140 | 0.017 | −0.040 | 0.171 | 0.027 |
| HXK2 | 0.882 | 0.007 | 0.919 | 0.074 | 0.118 | −0.196 | −0.311 | −0.143 | −0.098 | 0.091 | |
| MDH2 | 0.908 | −0.129 | 0.903 | −0.045 | −0.060 | −0.037 | 0.078 | 0.141 | 0.028 | −0.001 | |
| ACS1 | 0.841 | 0.396 | 0.894 | 0.259 | −0.063 | −0.109 | 0.104 | −0.163 | 0.364 | 0.023 | |
| ZWF1 | 0.907 | 0.050 | 0.868 | 0.044 | 0.104 | −0.197 | 0.097 | −0.062 | −0.270 | −0.105 | |
| PGM1 | 0.891 | 0.474 | 0.854 | 0.075 | 0.230 | −0.330 | −0.092 | −0.031 | −0.212 | 0.218 | |
| LRO1 | 0.600 | −0.235 | 0.806 | −0.186 | −0.324 | −0.195 | 0.237 | −0.251 | 0.135 | 0.112 | |
| FUM1 | 0.882 | −0.109 | 0.794 | −0.244 | −0.104 | 0.071 | −0.196 | 0.362 | 0.044 | 0.135 | |
| PGI1 | 0.891 | 0.327 | 0.792 | 0.092 | 0.195 | 0.217 | 0.025 | −0.276 | −0.109 | −0.132 | |
| KGD2 | 0.947 | −0.299 | 0.779 | −0.199 | −0.203 | 0.321 | 0.025 | −0.207 | 0.018 | −0.031 | |
| LSC2 | 0.976 | −0.386 | 0.761 | −0.091 | 0.048 | 0.126 | −0.077 | −0.087 | −0.052 | 0.009 | |
| CIT1 | 0.885 | 0.326 | 0.743 | 0.292 | −0.178 | 0.277 | 0.003 | 0.041 | −0.022 | 0.083 | |
| IDH2 | 0.900 | −0.186 | 0.740 | −0.285 | −0.071 | 0.355 | −0.066 | 0.042 | 0.052 | −0.011 | |
| SDH1 | 0.957 | 0.415 | 0.716 | −0.383 | 0.156 | 0.041 | 0.003 | 0.334 | 0.060 | −0.225 | |
| GND1 | 0.911 | −0.024 | 0.714 | 0.256 | −0.047 | 0.062 | 0.042 | −0.073 | −0.258 | −0.073 | |
| ENO1 | 0.919 | −0.102 | 0.683 | 0.205 | 0.076 | 0.107 | −0.058 | 0.094 | −0.066 | 0.242 | |
| PGK1 | 0.922 | −0.293 | 0.620 | 0.264 | −0.063 | 0.050 | 0.147 | 0.065 | −0.151 | 0.016 | |
| GUT2 | 0.836 | 0.479 | 0.597 | −0.040 | −0.401 | −0.145 | −0.233 | 0.582 | −0.086 | 0.004 | |
| CDC19 | 0.932 | −0.147 | 0.585 | 0.393 | 0.163 | −0.020 | −0.020 | 0.055 | 0.056 | −0.171 | |
| HMG1 | 0.644 | −0.211 | 0.464 | 0.081 | 0.365 | −0.088 | 0.259 | −0.123 | 0.323 | −0.074 | |
| LSC1 | 0.927 | −0.415 | 0.417 | 0.008 | 0.199 | 0.298 | −0.280 | −0.124 | −0.102 | −0.010 | |
| 3 | PAH1 | 0.840 | 0.234 | 0.039 | 0.931 | −0.245 | −0.069 | −0.010 | 0.006 | 0.034 | 0.084 |
| SCT1 | 0.925 | 0.090 | −0.400 | 0.924 | 0.031 | 0.086 | 0.077 | −0.004 | −0.045 | 0.243 | |
| ACC1 | 0.861 | −0.049 | −0.105 | 0.923 | 0.130 | −0.125 | 0.060 | 0.006 | 0.067 | 0.045 | |
| SLC1 | 0.921 | 0.121 | −0.092 | 0.899 | −0.238 | −0.127 | −0.361 | 0.007 | −0.103 | −0.139 | |
| DGA2 | 0.851 | 0.104 | −0.113 | 0.882 | −0.158 | −0.085 | −0.209 | −0.023 | −0.077 | 0.239 | |
| FAS1.1 | 0.865 | −0.328 | 0.016 | 0.833 | 0.017 | −0.116 | −0.013 | −0.081 | 0.012 | 0.038 | |
| ACL1 | 0.974 | −0.182 | 0.313 | 0.793 | 0.089 | −0.228 | −0.013 | −0.014 | 0.042 | −0.021 | |
| GPD1 | 0.878 | 0.240 | 0.287 | 0.728 | −0.300 | 0.316 | −0.328 | 0.087 | 0.164 | −0.037 | |
| FAS2.1 | 0.865 | −0.165 | 0.409 | 0.721 | −0.073 | −0.298 | 0.017 | −0.024 | 0.050 | 0.047 | |
| ACL2 | 0.971 | −0.254 | 0.421 | 0.692 | 0.065 | −0.222 | 0.028 | −0.027 | 0.045 | −0.005 | |
| MDH1 | 0.882 | 0.458 | 0.270 | 0.673 | −0.090 | 0.309 | 0.123 | 0.042 | 0.017 | 0.065 | |
| FAA1 | 0.728 | 0.130 | 0.363 | 0.578 | 0.031 | −0.349 | 0.149 | 0.226 | 0.050 | −0.064 | |
| FBP2 | 0.913 | 0.008 | 0.204 | 0.568 | 0.196 | 0.325 | 0.321 | −0.134 | 0.055 | −0.009 | |
| oil productivity | 0.382 | −0.252 | −0.105 | 0.556 | −0.059 | 0.032 | −0.062 | −0.008 | 0.053 | 0.111 | |
| tid_2139 | 0.645 | −0.433 | −0.173 | 0.532 | 0.430 | 0.167 | −0.055 | 0.181 | 0.097 | −0.078 | |
| PDB1 | 0.891 | −0.300 | 0.296 | 0.421 | −0.013 | 0.324 | 0.086 | 0.103 | −0.038 | −0.039 | |
| 4 | YEH2 | 0.729 | −0.095 | 0.117 | 0.153 | −0.962 | 0.060 | −0.147 | 0.223 | 0.054 | 0.117 |
| K_6707 | 0.956 | 0.200 | 0.023 | 0.243 | −0.931 | −0.083 | 0.059 | 0.329 | 0.001 | −0.025 | |
| PYC1 | 0.786 | 0.040 | 0.124 | −0.071 | 0.820 | 0.004 | 0.169 | −0.023 | −0.014 | −0.057 | |
| LACS1.2 | 0.734 | −0.308 | −0.087 | −0.193 | −0.799 | 0.192 | 0.459 | 0.157 | −0.474 | −0.016 | |
| FAS2.2 | 0.821 | 0.138 | 0.115 | −0.186 | −0.722 | −0.304 | 0.151 | 0.100 | 0.188 | 0.025 | |
| ACO1 | 0.889 | 0.317 | 0.219 | −0.255 | 0.624 | 0.165 | −0.059 | 0.308 | −0.030 | −0.029 | |
| MAE1 | 0.702 | 0.155 | −0.558 | 0.098 | 0.564 | 0.115 | 0.072 | 0.006 | 0.057 | 0.262 | |
| SDH3 | 0.746 | 0.000 | 0.370 | −0.109 | 0.538 | −0.044 | 0.202 | 0.144 | −0.129 | −0.105 | |
| tid_69043 | 0.708 | −0.059 | 0.194 | 0.255 | 0.382 | 0.169 | 0.313 | −0.046 | −0.264 | 0.179 | |
| 5 | KGD1 | 0.656 | 0.206 | −0.199 | −0.016 | 0.070 | 0.824 | 0.170 | −0.040 | 0.085 | 0.025 |
| IDH1 | 0.900 | −0.140 | 0.277 | −0.367 | 0.129 | 0.664 | 0.179 | 0.078 | 0.053 | 0.099 | |
| TPI1 | 0.775 | −0.052 | 0.061 | 0.201 | 0.214 | 0.619 | 0.338 | −0.213 | 0.078 | −0.022 | |
| IDP1 | 0.693 | 0.199 | 0.403 | −0.346 | 0.184 | 0.605 | 0.092 | −0.106 | 0.045 | 0.054 | |
| LPD1 | 0.819 | −0.306 | 0.482 | −0.096 | −0.249 | 0.515 | 0.219 | −0.248 | 0.115 | −0.013 | |
| 6 | CDS1 | 0.760 | −0.034 | −0.049 | 0.125 | −0.034 | −0.243 | −0.815 | 0.256 | −0.028 | −0.013 |
| ALE1 | 0.796 | −0.056 | 0.244 | 0.219 | −0.196 | −0.384 | −0.648 | 0.105 | −0.132 | −0.072 | |
| K_291711 | 0.815 | 0.425 | 0.288 | −0.016 | −0.120 | −0.036 | 0.574 | 0.003 | −0.283 | −0.160 | |
| GAP1 | 0.712 | −0.184 | −0.212 | 0.297 | 0.243 | 0.133 | 0.544 | 0.067 | −0.041 | 0.197 | |
| EMI2 | 0.766 | 0.223 | 0.272 | 0.293 | 0.283 | 0.066 | 0.478 | −0.089 | −0.084 | 0.143 | |
| SHH4 | 0.882 | 0.431 | −0.099 | −0.111 | 0.417 | 0.347 | 0.439 | 0.071 | 0.147 | −0.339 | |
| 7 | TGL1 | 0.611 | 0.157 | 0.135 | −0.052 | 0.209 | 0.151 | 0.201 | −0.828 | 0.219 | 0.192 |
| 8 | TPI2.2 | 0.730 | 0.377 | −0.038 | 0.033 | −0.237 | 0.273 | 0.093 | −0.269 | 0.616 | −0.082 |
| TPI2.1 | 0.792 | 0.414 | −0.428 | 0.231 | −0.008 | 0.122 | −0.064 | −0.079 | 0.603 | 0.004 | |
| FAS1.2 | 0.560 | −0.460 | 0.009 | −0.305 | −0.059 | 0.066 | 0.059 | −0.114 | 0.478 | −0.008 | |
| 9 | PDC1 | 0.851 | 0.167 | 0.030 | 0.426 | −0.068 | 0.179 | 0.246 | −0.272 | 0.022 | 0.665 |
| DGA1 | 0.684 | 0.357 | 0.044 | 0.427 | −0.183 | −0.063 | −0.103 | 0.099 | −0.235 | 0.554 | |
| PDC2 | 0.846 | 0.043 | −0.404 | 0.160 | −0.179 | 0.022 | 0.021 | −0.318 | 0.209 | 0.518 | |
| SOL3 | 0.768 | −0.153 | 0.166 | 0.298 | 0.461 | 0.089 | −0.134 | −0.300 | 0.028 | 0.497 | |
| CMIN (P) | GFI | AGFI | CFI | RMSEA | AIC | |
|---|---|---|---|---|---|---|
| Estimated model | 0.032 | 0.958 | 0.892 | 0.996 | 0.043 | 239.68 |
| Saturated model | 1 | 1 | 272 | |||
| Independent model | 0 | 0.151 | 0.038 | 0 | 0.447 | 5166.72 |
| Source | Target | Standardized Regression Weight | p Values |
|---|---|---|---|
| FAS2.1 | FAS1.1 | 0.894 | *** |
| FAS2.1 | PAH1 | 0.695 | *** |
| FAS1.1 | ACC1 | 0.668 | *** |
| PAH1 | DGA2 | 0.827 | *** |
| PAH1 | ACC1 | 0.339 | *** |
| ACC1 | SCT1 | 1.148 | *** |
| DGA2 | SCT1 | 0.306 | *** |
| FAS2.1 | SCT1 | −0.555 | *** |
| FAS2.1 | ACL1 | 0.765 | *** |
| DGA2 | SLC1 | 0.572 | *** |
| ACC1 | tid_2139 | 0.443 | *** |
| SCT1 | ACL2 | −0.112 | *** |
| PAH1 | tid_2139 | −0.193 | 0.002 |
| FAS1.1 | PDB1 | 0.436 | *** |
| ACL1 | oil productivity | 0.487 | *** |
| ACC1 | oil productivity | −0.36 | 0.007 |
| FAS1.1 | oil productivity | 0.687 | *** |
| FAS2.1 | oil productivity | −0.479 | 0.002 |
| SLC1 | oil productivity | −0.254 | 0.007 |
| DGA2 | oil productivity | 0.488 | *** |
| FAS2.1 | FAA1 | 1.117 | *** |
| PAH1 | MDH1 | 0.619 | *** |
| PDB1 | MDH1 | 0.615 | *** |
| ACC1 | FAA1 | 0.786 | *** |
| oil productivity | MDH1 | −0.107 | 0.005 |
| FAS1.1 | MDH1 | −0.802 | *** |
| FAS1.1 | FAA1 | −0.98 | *** |
| DGA2 | FAA1 | −0.175 | *** |
| ACC1 | MDH1 | 0.608 | *** |
| MDH1 | FBP2 | 0.486 | *** |
| MDH1 | GPD1 | 0.327 | *** |
| SLC1 | GPD1 | 0.3 | *** |
| PDB1 | GPD1 | 0.422 | *** |
| PDB1 | FBP2 | 0.254 | *** |
| SLC1 | FBP2 | −0.182 | *** |
| ACL2 | FBP2 | 0.3 | *** |
| FAA1 | GPD1 | −0.398 | *** |
| PAH1 | GPD1 | 0.362 | *** |
| ACL1 | ACL2 | 0.991 | *** |
| PDB1 | tid_2139 | 0.504 | *** |
| ACL1 | PDB1 | 0.429 | *** |
| PDB1 | ACL2 | 0.081 | *** |
| tid_2139 | SLC1 | −0.342 | *** |
| tid_2139 | ACL1 | 0.26 | *** |
| SLC1 | PDB1 | −0.225 | *** |
| ACL2 | SLC1 | 0.545 | *** |
| CMIN (P) | GFI | AGFI | CFI | RMSEA | AIC | |
|---|---|---|---|---|---|---|
| Estimated model | 0.027 | 0.973 | 0.913 | 0.981 | 0.064 | 87.91 |
| Saturated model | 1 | 1 | 90 | |||
| Independent model | 0 | 0.592 | 0.49 | 0 | 0.289 | 683.446 |
| Source | Target | Standardized Regression Weight | p Values |
|---|---|---|---|
| Group4 | Group2 | 0.353 | *** |
| Group4 | Group7 | −0.285 | *** |
| Group4 | Group6 | 0.472 | *** |
| Group2 | Group6 | 0.249 | *** |
| Group2 | Group8 | −0.383 | *** |
| Group7 | Group8 | 0.178 | 0.004 |
| Group6 | Group9 | 0.364 | *** |
| Group2 | Group9 | −0.177 | 0.009 |
| Group7 | Group9 | 0.362 | *** |
| Group6 | Group1 | 0.549 | *** |
| Group2 | Group5 | 0.557 | *** |
| Group7 | Group5 | 0.304 | *** |
| Group4 | Group5 | 0.24 | *** |
| Group9 | Group1 | 0.157 | 0.006 |
| Group2 | Group3 | 0.462 | *** |
| Group8 | Group1 | 0.233 | *** |
| Group9 | Group3 | 0.476 | *** |
| Group8 | Group5 | 0.215 | *** |
| Group8 | Group3 | −0.152 | 0.001 |
| Group6 | Group3 | 0.241 | *** |
| Group4 | Group3 | −0.189 | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aburatani, S.; Ishiya, K.; Itoh, T.; Hayashi, T.; Taniguchi, T.; Takaku, H. Inference of Regulatory System for TAG Biosynthesis in Lipomyces starkeyi. Bioengineering 2020, 7, 148. https://doi.org/10.3390/bioengineering7040148
Aburatani S, Ishiya K, Itoh T, Hayashi T, Taniguchi T, Takaku H. Inference of Regulatory System for TAG Biosynthesis in Lipomyces starkeyi. Bioengineering. 2020; 7(4):148. https://doi.org/10.3390/bioengineering7040148
Chicago/Turabian StyleAburatani, Sachiyo, Koji Ishiya, Toshikazu Itoh, Toshihiro Hayashi, Takeaki Taniguchi, and Hiroaki Takaku. 2020. "Inference of Regulatory System for TAG Biosynthesis in Lipomyces starkeyi" Bioengineering 7, no. 4: 148. https://doi.org/10.3390/bioengineering7040148
APA StyleAburatani, S., Ishiya, K., Itoh, T., Hayashi, T., Taniguchi, T., & Takaku, H. (2020). Inference of Regulatory System for TAG Biosynthesis in Lipomyces starkeyi. Bioengineering, 7(4), 148. https://doi.org/10.3390/bioengineering7040148






