Author Contributions
All authors contributed substantially to this body of work, in the form of; Conceptualization, A.U., G.K.K., G.T.H. and B.J.R.; Data Curation, A.U.; Writing-Original Draft Preparation, A.U., G.K.K., G.T.H., J.M.F. and B.J.R.; Writing-Review & Editing, A.U., G.K.K., G.T.H., J.M.F. and B.J.R.; Supervision, G.K.K., G.T.H. and B.J.R.; Project Administration, B.J.R.; Funding Acquisition, B.J.R., G.T.H. and A.U.
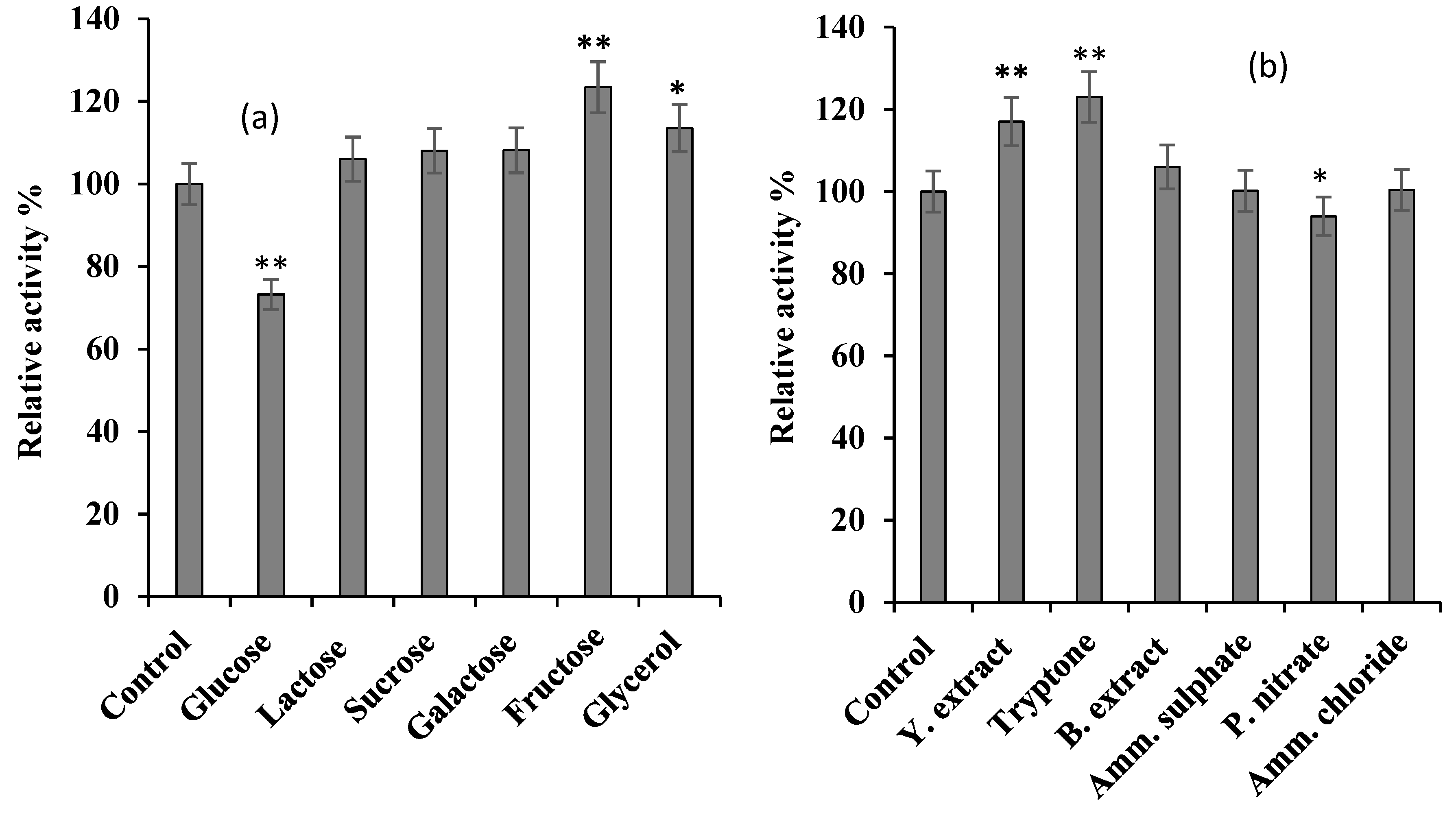
Figure 1.
The effect of various carbon (
a) and nitrogen (
b) sources on
S. griseus recombinant β-glucosidase expression. Recombinant β-glucosidase was expressed by supplementing basal medium with 0.5% (w/v) and 1% (w/v) of different carbon and nitrogen sources, respectively. The control was performed using only basal medium. In each case, cells were grown until OD
600 nm reached 0.5, and then induced with 1 mM IPTG for 6 h at 37 °C, 220 rpm. Enzyme activity of crude lysate was performed using 7 mM
pNPG as the substrate in 50 mM potassium phosphate buffer, pH 7 (see
Section 2.2.2). The data plotted represent the mean of three independent experiments, with standard deviation shown as error bars: *
p-value ≤ 0.05, **
p-value ≤ 0.01 represent a significant and a very significant difference, respectively, based on two-tailed
t-test and in comparison to the control.
Figure 1.
The effect of various carbon (
a) and nitrogen (
b) sources on
S. griseus recombinant β-glucosidase expression. Recombinant β-glucosidase was expressed by supplementing basal medium with 0.5% (w/v) and 1% (w/v) of different carbon and nitrogen sources, respectively. The control was performed using only basal medium. In each case, cells were grown until OD
600 nm reached 0.5, and then induced with 1 mM IPTG for 6 h at 37 °C, 220 rpm. Enzyme activity of crude lysate was performed using 7 mM
pNPG as the substrate in 50 mM potassium phosphate buffer, pH 7 (see
Section 2.2.2). The data plotted represent the mean of three independent experiments, with standard deviation shown as error bars: *
p-value ≤ 0.05, **
p-value ≤ 0.01 represent a significant and a very significant difference, respectively, based on two-tailed
t-test and in comparison to the control.
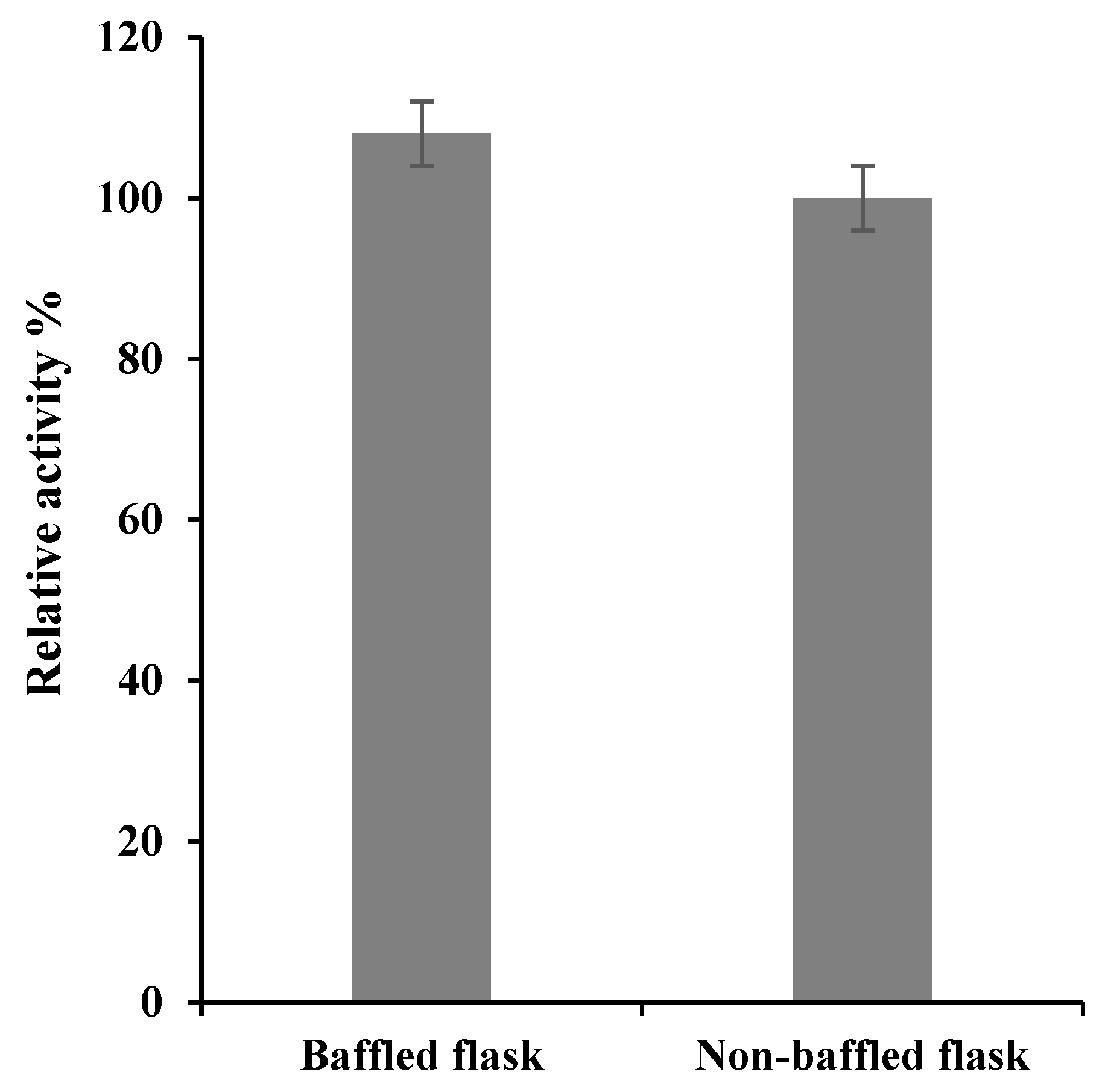
Figure 2.
The effect of baffled versus non-baffled flask on the production of S. griseus recombinant β-glucosidase. Recombinant β-glucosidase was expressed in basal medium supplemented with 0.5% (w/v) fructose, 1% (w/v) yeast extract and tryptone at 37 °C for 6 h, following 1 mM IPTG induction. Enzyme activity was used as a proxy to enzyme production. The data represented are the mean of three independent experiments, with the standard deviations noted as error bars. Both baffled and non-baffled flasks show no significant difference between them (p-value ≥ 0.235) based on two-tailed t-test.
Figure 2.
The effect of baffled versus non-baffled flask on the production of S. griseus recombinant β-glucosidase. Recombinant β-glucosidase was expressed in basal medium supplemented with 0.5% (w/v) fructose, 1% (w/v) yeast extract and tryptone at 37 °C for 6 h, following 1 mM IPTG induction. Enzyme activity was used as a proxy to enzyme production. The data represented are the mean of three independent experiments, with the standard deviations noted as error bars. Both baffled and non-baffled flasks show no significant difference between them (p-value ≥ 0.235) based on two-tailed t-test.
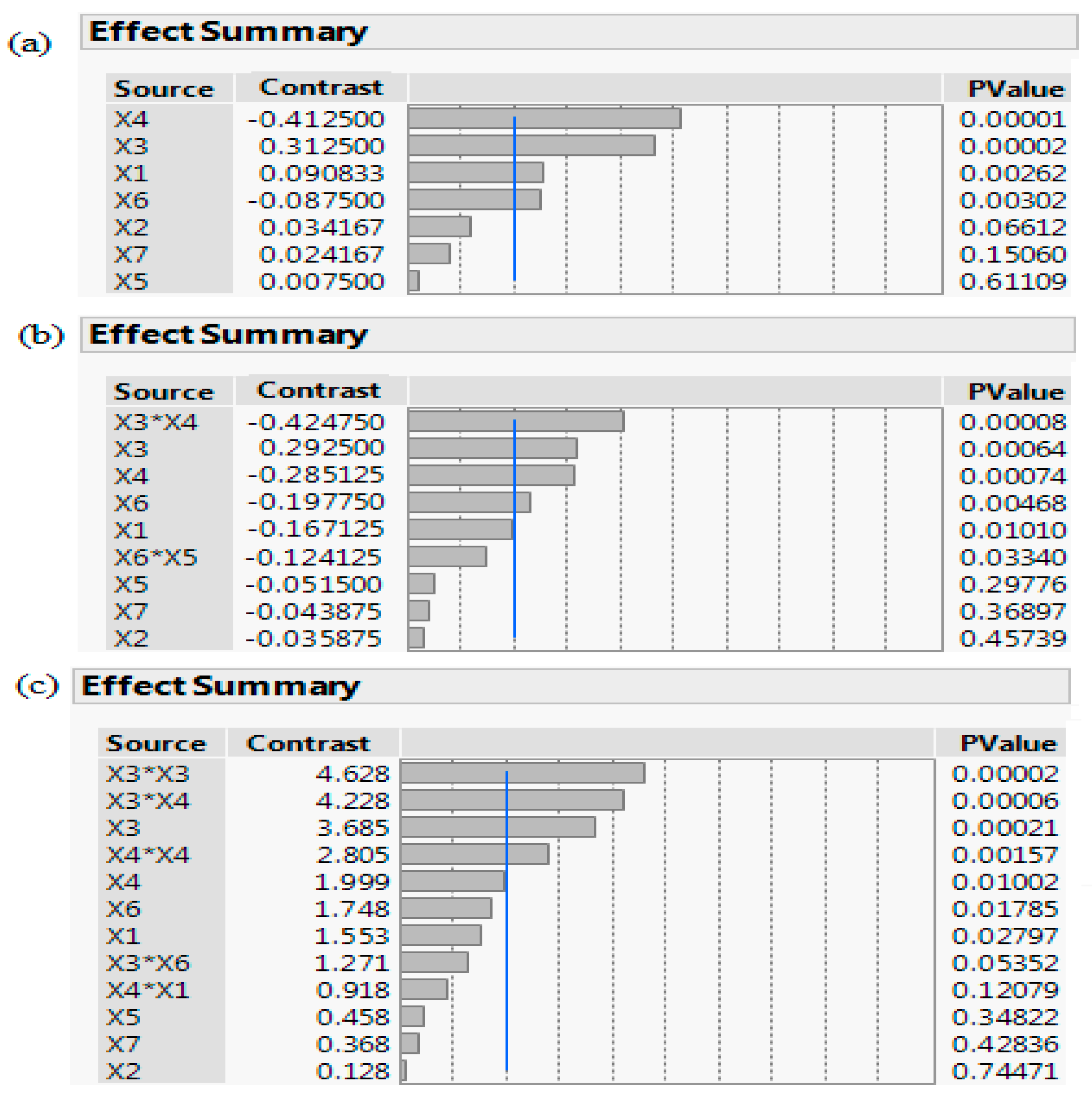
Figure 3.
A pareto chart of the screening processes: (a) PBD, (b) FFD, and (c) DSD. The schematic depicts a scaled summary and corresponding p-value of the seven factors: OD (Abs600 nm) at induction time (X1), IPTG (X2), temperature (X3), incubation time (X4), yeast extract (X5), tryptone (X6), fructose (X7), along with their interactions. Factors with p-value < 0.05 are statistically significant and variables crossing the reference blue line with p-value < 0.001 are considered as highly statistically significant.
Figure 3.
A pareto chart of the screening processes: (a) PBD, (b) FFD, and (c) DSD. The schematic depicts a scaled summary and corresponding p-value of the seven factors: OD (Abs600 nm) at induction time (X1), IPTG (X2), temperature (X3), incubation time (X4), yeast extract (X5), tryptone (X6), fructose (X7), along with their interactions. Factors with p-value < 0.05 are statistically significant and variables crossing the reference blue line with p-value < 0.001 are considered as highly statistically significant.
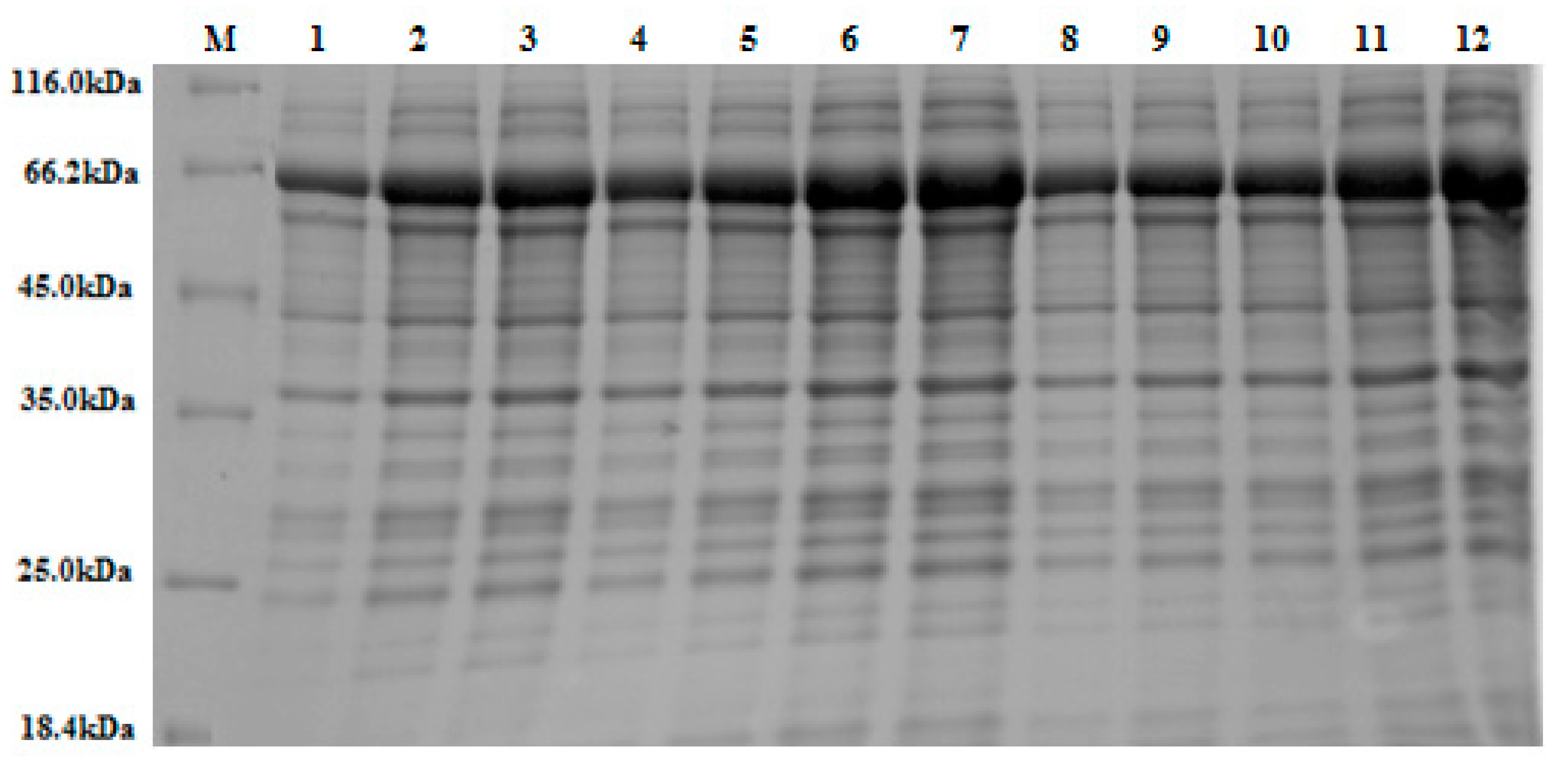
Figure 4.
A 10% (w/v) SDS-PAGE stained with Coomassie Brilliant Blue. The figure depicts various band sizes of crude β-glucosidase expressed in accordance with the 12 experimental runs, according to Plackett-Burman Design (see
Section 2.2.4). An equal volume of crude extract (15 μL) was loaded into each lane. Lanes 2, 3, 6, 10, 11, and 12 display larger over-expressed protein bands at the expected molecular weight for the recombinant β-glucosidase/GST fusion protein (~65 kDa), as based on ImageJ densitometric analysis. This result mirrors the increased enzyme activity (U/mL) observed in these experimental runs (see
Table 2). Note: M = protein marker (14.4–116 kDa).
Figure 4.
A 10% (w/v) SDS-PAGE stained with Coomassie Brilliant Blue. The figure depicts various band sizes of crude β-glucosidase expressed in accordance with the 12 experimental runs, according to Plackett-Burman Design (see
Section 2.2.4). An equal volume of crude extract (15 μL) was loaded into each lane. Lanes 2, 3, 6, 10, 11, and 12 display larger over-expressed protein bands at the expected molecular weight for the recombinant β-glucosidase/GST fusion protein (~65 kDa), as based on ImageJ densitometric analysis. This result mirrors the increased enzyme activity (U/mL) observed in these experimental runs (see
Table 2). Note: M = protein marker (14.4–116 kDa).
Figure 5.
A plot of (a) actual versus predicted, and (b) predicted versus residual values to estimate the accuracy of the regression model.
Figure 5.
A plot of (a) actual versus predicted, and (b) predicted versus residual values to estimate the accuracy of the regression model.
Figure 6.
Three-dimensional response surface and contour plots for S.griseus recombinant β-glucosidase production using a three level Central Composite Design. The plots depict the interactive effects of: OD600 nm and temperature (a,a’); OD600 nm and incubation time (b,b’); temperature and incubation time (c,c’); and temperature and tryptone (d,d’). The remaining variables remained at constant zero levels.
Figure 6.
Three-dimensional response surface and contour plots for S.griseus recombinant β-glucosidase production using a three level Central Composite Design. The plots depict the interactive effects of: OD600 nm and temperature (a,a’); OD600 nm and incubation time (b,b’); temperature and incubation time (c,c’); and temperature and tryptone (d,d’). The remaining variables remained at constant zero levels.
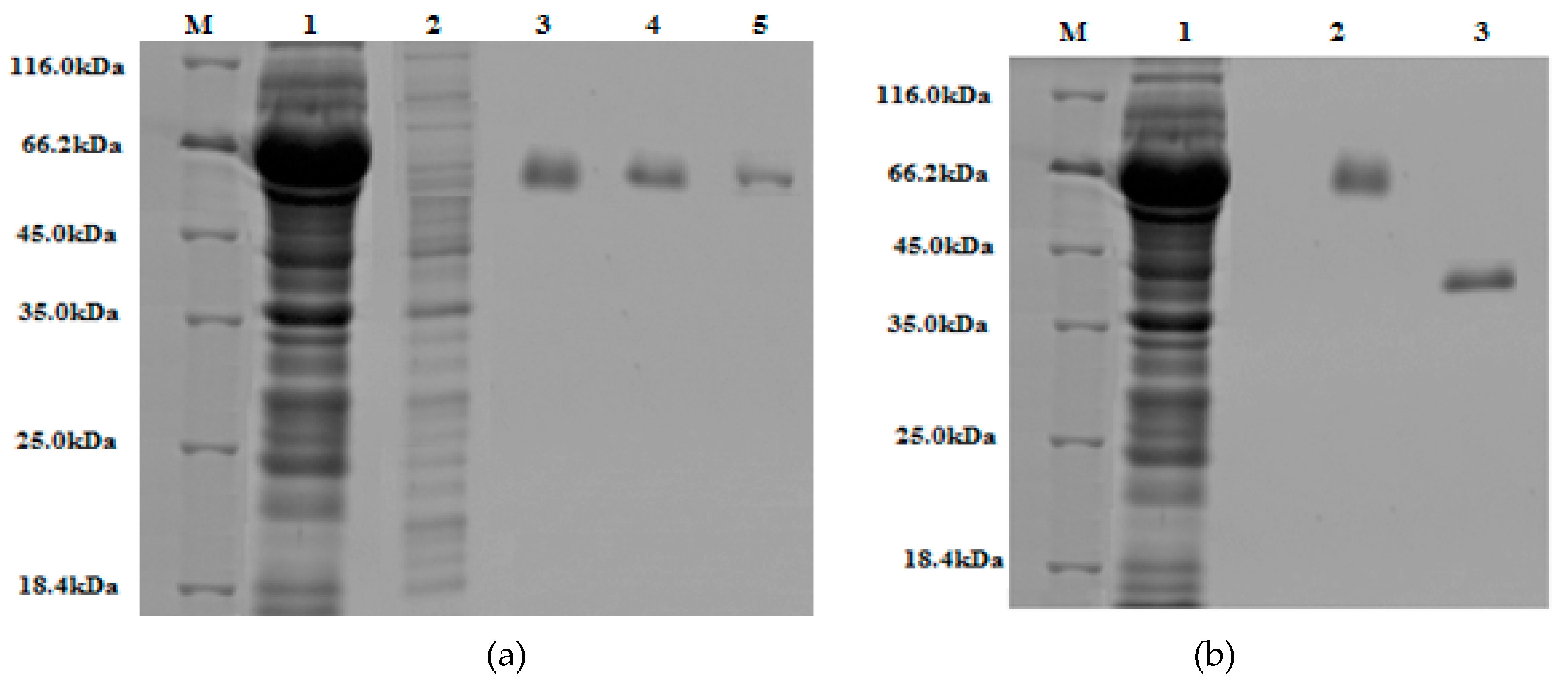
Figure 7.
The 10% (w/v) SDS-PAGE gels stained with Coomassie Brilliant Blue: (a) Lane M, protein marker (18.4–116 kDa); lane 1, crude extract; lane 2, run-through; lane 3, 4, 5, are elute one, two, and three of purified GST-tagged β-glucosidase, respectively. (b) Lane M, protein marker (18.4–116 kDa); lane 1, crude extract; lane 2, purified GST-tagged β-glucosidase; lane 3, purified β-glucosidase (post GST-tag cleavage).
Figure 7.
The 10% (w/v) SDS-PAGE gels stained with Coomassie Brilliant Blue: (a) Lane M, protein marker (18.4–116 kDa); lane 1, crude extract; lane 2, run-through; lane 3, 4, 5, are elute one, two, and three of purified GST-tagged β-glucosidase, respectively. (b) Lane M, protein marker (18.4–116 kDa); lane 1, crude extract; lane 2, purified GST-tagged β-glucosidase; lane 3, purified β-glucosidase (post GST-tag cleavage).
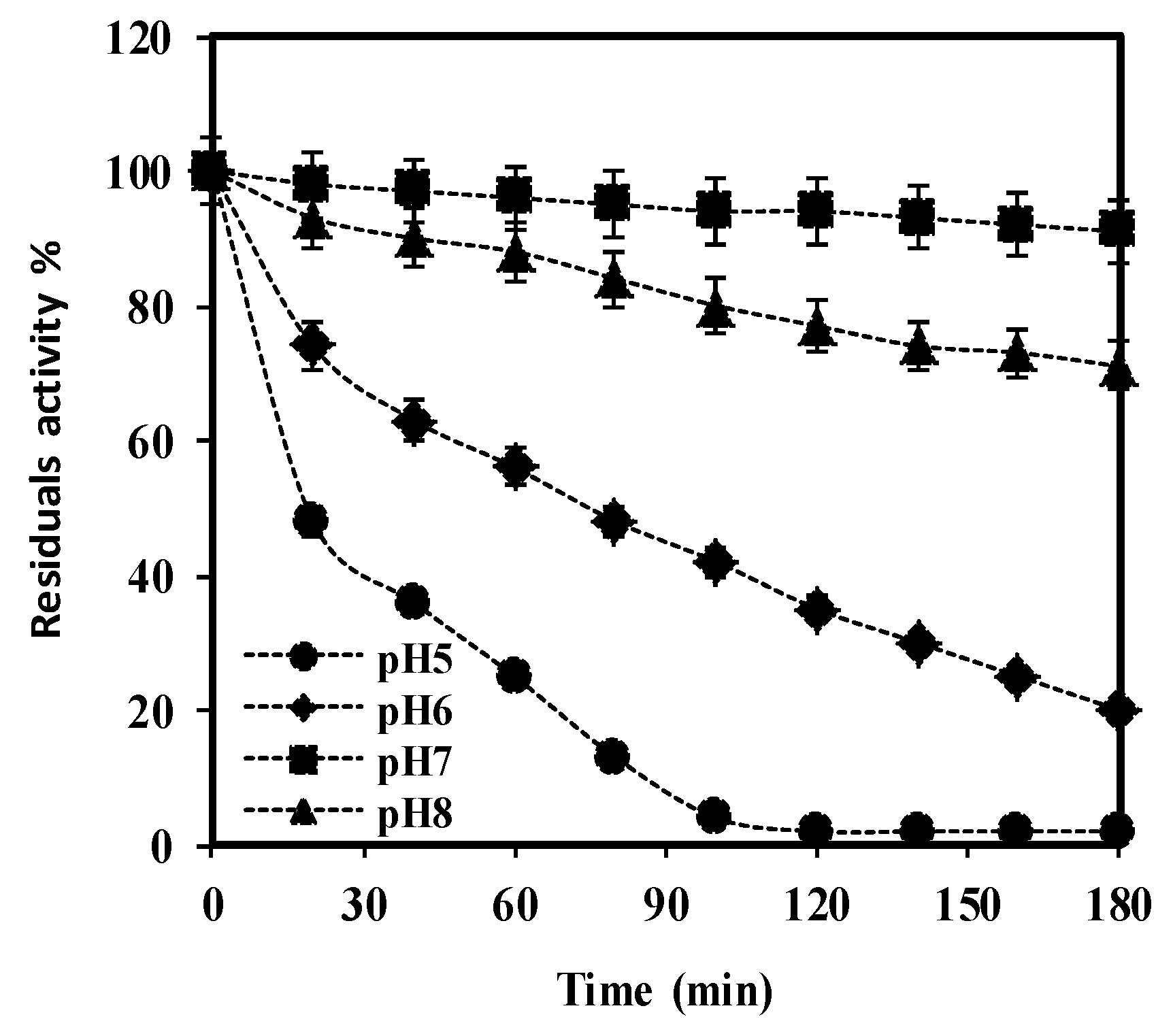
Figure 8.
The pH stability of purified
S. griseus recombinant β-glucosidase was determined by incubating enzyme solutions containing 7 mM pNPG in buffers of various pH, ranging between 5 (
![Bioengineering 06 00061 i001 Bioengineering 06 00061 i001]()
), 6 (
![Bioengineering 06 00061 i002 Bioengineering 06 00061 i002]()
), 7 (
![Bioengineering 06 00061 i003 Bioengineering 06 00061 i003]()
), and 8 (
![Bioengineering 06 00061 i004 Bioengineering 06 00061 i004]()
), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.
Figure 8.
The pH stability of purified
S. griseus recombinant β-glucosidase was determined by incubating enzyme solutions containing 7 mM pNPG in buffers of various pH, ranging between 5 (
![Bioengineering 06 00061 i001 Bioengineering 06 00061 i001]()
), 6 (
![Bioengineering 06 00061 i002 Bioengineering 06 00061 i002]()
), 7 (
![Bioengineering 06 00061 i003 Bioengineering 06 00061 i003]()
), and 8 (
![Bioengineering 06 00061 i004 Bioengineering 06 00061 i004]()
), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.
Table 1.
The minimum and maximum ranges for different factors studied during screening process.
Table 1.
The minimum and maximum ranges for different factors studied during screening process.
| Factors Types | Factors Codes | Factors | Levels |
|---|
| −1 (Low) | +1 (High) |
|---|
| Induction condition | X1 | OD (Abs600 nm) | 0.3 | 0.9 |
| X2 | IPTG (mM) | 0.1 | 1.0 |
| X3 | Temperature (°C) | 18 | 36 |
| X4 | Incubation time (h) | 4.0 | 20 |
| Media Composition | X5 | Yeast Extract (g/L) | 5.0 | 10 |
| X6 | Tryptone (g/L) | 10 | 20 |
| X7 | Fructose (g/L) | 1.0 | 5.0 |
Table 2.
Plackett-Burman Design. Multiple screening designs for identification of most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
Table 2.
Plackett-Burman Design. Multiple screening designs for identification of most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
| | Coded Values | Response |
|---|
| Runs | X1 | X2 | X3 | X4 | X5 | X6 | X7 | BGL (U/mL) |
|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 39.301 |
| 2 | −1 | −1 | 1 | −1 | −1 | 1 | −1 | 39.781 |
| 3 | −1 | 1 | −1 | −1 | 1 | −1 | 1 | 40.681 |
| 4 | 1 | −1 | −1 | 1 | −1 | 1 | 1 | 39.761 |
| 5 | −1 | −1 | 1 | −1 | 1 | 1 | 1 | 39.801 |
| 6 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 40.631 |
| 7 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | 39.271 |
| 8 | −1 | 1 | −1 | 1 | 1 | 1 | −1 | 39.621 |
| 9 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 39.181 |
| 10 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | 40.211 |
| 11 | −1 | −1 | −1 | 1 | −1 | −1 | 1 | 39.811 |
| 12 | 1 | −1 | −1 | −1 | 1 | −1 | −1 | 40.791 |
Table 3.
Fractional Factorial Design. Multiple screening designs for identification of the most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
Table 3.
Fractional Factorial Design. Multiple screening designs for identification of the most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
| | Coded Values | Response |
|---|
| Runs | X1 | X2 | X3 | X4 | X5 | X6 | X7 | BGL (U/mL) |
|---|
| 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 41.641 |
| 2 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 40.471 |
| 3 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 40.571 |
| 4 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 39.821 |
| 5 | 1 | −1 | 1 | −1 | 1 | −1 | 1 | 41.601 |
| 6 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 40.561 |
| 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 39.411 |
| 8 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 40.388 |
| 9 | 1 | −1 | 1 | 1 | −1 | 1 | −1 | 40.281 |
| 10 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 39.631 |
| 11 | −1 | 1 | −1 | 1 | −1 | 1 | −1 | 40.421 |
| 12 | 1 | 1 | −1 | 1 | −1 | −1 | 1 | 40.311 |
| 13 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 41.379 |
| 14 | −1 | 1 | −1 | −1 | 1 | −1 | 1 | 40.651 |
| 15 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 41.691 |
| 16 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | 40.380 |
Table 4.
Definitive Screening Design. Multiple screening designs for identification of most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
Table 4.
Definitive Screening Design. Multiple screening designs for identification of most influential factors on the production of S.griseus recombinant β-glucosidase. It depicts coded values of the designs, along with the observed experimental response (β-glucosidase activity (BGL); U/mL).
| | Coded Values | Response |
|---|
| Runs | X1 | X2 | X3 | X4 | X5 | X6 | X7 | BGL (U/mL) |
|---|
| 1 | −1 | 1 | 1 | 0 | −1 | −1 | −1 | 40.111 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41.221 |
| 3 | −1 | 0 | 1 | 1 | 1 | 1 | −1 | 39.033 |
| 4 | −1 | 1 | 0 | 1 | −1 | −1 | 1 | 40.621 |
| 5 | 1 | −1 | 1 | 1 | 0 | −1 | −1 | 39.121 |
| 6 | −1 | −1 | 1 | −1 | −1 | 1 | 1 | 39.961 |
| 7 | 1 | −1 | −1 | 0 | 1 | 1 | 1 | 40.511 |
| 8 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 40.611 |
| 9 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 40.261 |
| 10 | 1 | 1 | −1 | 1 | −1 | 1 | 0 | 40.011 |
| 11 | 1 | 0 | −1 | −1 | −1 | −1 | 1 | 40.561 |
| 12 | −1 | 1 | −1 | −1 | 0 | 1 | 1 | 39.451 |
| 13 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 40.651 |
| 14 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 39.106 |
| 15 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 39.981 |
| 16 | −1 | 1 | −1 | −1 | 1 | 0 | −1 | 39.791 |
| 17 | 1 | 1 | 1 | −1 | −1 | 1 | −1 | 40.021 |
| 18 | 1 | −1 | 1 | 1 | −1 | 0 | 1 | 39.190 |
| 19 | −1 | −1 | 1 | −1 | 1 | −1 | 0 | 39.841 |
| 20 | 1 | −1 | 0 | −1 | 1 | 1 | −1 | 41.211 |
| 21 | 0 | −1 | −1 | −1 | −1 | −1 | −1 | 39.781 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41.231 |
Table 5.
The most influential factors (X1, X3, X4, X6), as identified through the screening process, were examined at three levels; low (−1), central (0), and high (+1), for the optimisation of S. griseus recombinant β-glucosidase expression in E. coli BL21 (DE3).
Table 5.
The most influential factors (X1, X3, X4, X6), as identified through the screening process, were examined at three levels; low (−1), central (0), and high (+1), for the optimisation of S. griseus recombinant β-glucosidase expression in E. coli BL21 (DE3).
| Factor code | Factors (unit) | Levels |
|---|
| −1 | 0 | +1 |
|---|
| X1 | OD (Abs600 nm) | 0.3 | 6.0 | 0.9 |
| X3 | Temperature (°C) | 18 | 27 | 36 |
| X4 | Incubation time (h) | 4 | 12 | 20 |
| X6 | Tryptone (g/L) | 10 | 15 | 20 |
Table 6.
CCD for production optimisation of S. griseus recombinant β-glucosidase. The table depicts coded values, along with the experimental response: BGL (U/mL), actual, predicted, and residuals (the difference between the actual and predicted values).
Table 6.
CCD for production optimisation of S. griseus recombinant β-glucosidase. The table depicts coded values, along with the experimental response: BGL (U/mL), actual, predicted, and residuals (the difference between the actual and predicted values).
| | Coded values | Response: BGL (U/mL) |
|---|
| Runs | X1 | X3 | X4 | X6 | Actual | Predicted | Residuals |
|---|
| 1 | 0 | 0 | 0 | −1 | 41.633 | 41.583 | 0.050 |
| 2 | −1 | 1 | 1 | −1 | 39.072 | 39.049 | 0.023 |
| 3 | 1 | −1 | −1 | 1 | 39.899 | 39.903 | −0.004 |
| 4 | −1 | 0 | 0 | 0 | 41.263 | 41.313 | −0.050 |
| 5 | 1 | 1 | 1 | 1 | 39.089 | 39.052 | 0.037 |
| 6 | 0 | 0 | 1 | 0 | 41.213 | 41.230 | −0.017 |
| 7 | −1 | −1 | 1 | 1 | 40.433 | 40.354 | 0.079 |
| 8 | −1 | −1 | −1 | 1 | 39.988 | 40.043 | −0.055 |
| 9 | 1 | −1 | 1 | −1 | 39.804 | 39.907 | −0.103 |
| 10 | 1 | −1 | 1 | 1 | 40.232 | 40.218 | 0.014 |
| 11 | 1 | 1 | −1 | −1 | 39.824 | 39.883 | −0.059 |
| 12 | 0 | 1 | 0 | 0 | 40.430 | 40.558 | −0.128 |
| 13 | 0 | 0 | 0 | 0 | 41.804 | 41.789 | 0.015 |
| 14 | 1 | 1 | 1 | −1 | 39.023 | 38.963 | 0.060 |
| 15 | 0 | 0 | 0 | 1 | 41.587 | 41.734 | −0.147 |
| 16 | 0 | 0 | 0 | 0 | 41.900 | 41.789 | 0.111 |
| 17 | −1 | 1 | −1 | −1 | 39.965 | 39.974 | −0.009 |
| 18 | 0 | −1 | 0 | 0 | 41.100 | 41.069 | 0.031 |
| 19 | 1 | 0 | 0 | 0 | 41.153 | 41.200 | −0.047 |
| 20 | −1 | −1 | 1 | −1 | 40.024 | 40.045 | −0.021 |
| 21 | −1 | 1 | −1 | 1 | 40.087 | 39.964 | 0.123 |
| 22 | 0 | 0 | 0 | 0 | 41.890 | 41.789 | 0.101 |
| 23 | −1 | −1 | −1 | −1 | 39.813 | 39.830 | −0.017 |
| 24 | 1 | 1 | −1 | 1 | 39.902 | 39.877 | 0.025 |
| 25 | 1 | −1 | −1 | −1 | 39.763 | 39.686 | 0.077 |
| 26 | 0 | 0 | −1 | 0 | 41.455 | 41.535 | −0.080 |
| 27 | 0 | 0 | 0 | 0 | 41.852 | 41.789 | 0.063 |
| 28 | -1 | 1 | 1 | 1 | 39.062 | 39.134 | -0.072 |
Table 7.
Analysis of Variance was used to confirm the adequacy of the model used in this study.
Table 7.
Analysis of Variance was used to confirm the adequacy of the model used in this study.
| Source | DF | Adj SS | Adj MS | F-Value | p-Value |
|---|
| Model | 14 | 24.5007 | 1.75005 | 167.32 | <0.001 |
| Residuals (error) | 13 | 0.1360 | 0.01046 | | |
| Lack-of-Fit | 10 | 0.1303 | 0.01303 | 6.87 | 0.070 |
| Pure Error | 3 | 0.0057 | 0.00190 | | |
| Total | 27 | 24.6366 | | | |
Table 8.
Regression coefficients significance: Coef, SE Coef, t-value, and p-value of the model terms (X1, X2, X3, X4) and their interactions are noted. The t- and p-values were determined by using JMP 13 (SAS Institute, Wittington House, UK).
Table 8.
Regression coefficients significance: Coef, SE Coef, t-value, and p-value of the model terms (X1, X2, X3, X4) and their interactions are noted. The t- and p-values were determined by using JMP 13 (SAS Institute, Wittington House, UK).
| Model Term | Coef | SE Coef | t-Value | p-Value |
|---|
| Constant | 41.7891 | 0.0354 | 1181.70 | <0.001 |
| X1 | −0.0566 | 0.0241 | −2.35 | 0.035 |
| X3 | −0.2557 | 0.0241 | −10.61 | <0.001 |
| X4 | −0.1524 | 0.0241 | −6.32 | <0.001 |
| X6 | 0.0754 | 0.0241 | 3.13 | 0.008 |
| X1*X1 | −0.5328 | 0.0637 | −8.37 | <0.001 |
| X3*X3 | −0.9758 | 0.0637 | −15.32 | <0.001 |
| X4*X4 | −0.4068 | 0.0637 | −6.39 | <0.001 |
| X6*X6 | −0.1308 | 0.0637 | −2.05 | 0.061 |
| X1*X3 | 0.0133 | 0.0256 | 0.52 | 0.613 |
| X1*X4 | 0.0014 | 0.0256 | 0.05 | 0.958 |
| X1*X6 | 0.0008 | 0.0256 | 0.03 | 0.977 |
| X3*X4 | −0.2851 | 0.0256 | −11.15 | <0.001 |
| X3*X6 | −0.0557 | 0.0256 | −2.18 | 0.048 |
| X4*X6 | 0.0239 | 0.0256 | 0.93 | 0.367 |
Table 9.
Comparison of S. griseus recombinant β-glucosidase production under optimised and pre-optimised conditions. The non-optimised expression was carried out using basal medium (LB broth) overnight at 37 °C, 220 rpm and cells were induced with 1 mM IPTG when OD600 nm reached 0.6.
Table 9.
Comparison of S. griseus recombinant β-glucosidase production under optimised and pre-optimised conditions. The non-optimised expression was carried out using basal medium (LB broth) overnight at 37 °C, 220 rpm and cells were induced with 1 mM IPTG when OD600 nm reached 0.6.
| Production Method | Fraction | Fraction Volume (mL) | Fraction Volume (mL) | Total Protein (mg) | Enzyme Activity U/mL | Total Activity (IU) |
|---|
| Optimised | Crude extract | 10 | 9.94 | 99.40 | 42.00 | 420 |
| Non-optimised | Crude extract | 10 | 3.80 | 38.00 | 16.06 | 160 |
Table 10.
S. griseus recombinant β-glucosidase purification table.
Table 10.
S. griseus recombinant β-glucosidase purification table.
| Purification Step | Total Protein (mg) | Total Activity (IU) | Specific Activity (IU/mg) | Yield (%) | Purification (Fold) |
|---|
| Crude extract | 99.40 | 420 | 4.23 | 100 | 1.00 |
| Ultrafiltration | 71.82 | 392 | 5.46 | 93 | 1.29 |
| Affinity chromatography | 31.70 | 259 | 8.17 | 62 | 1.93 |
| GST-tag cleavage | 21.47 | 196 | 9.13 | 47 | 2.16 |
Table 11.
The effect of metal ions or additive on purified S.griseus recombinant β-glucosidase was determined spectrometrically after 1-h and 6-h of incubation in the presence of 1 mM of each ion in potassium phosphate buffer, pH 7. The residual activity (%) was calculated in comparison to the activity obtained from enzyme in the same condition, but in the absence of any metal ion or additive. The results are the average of three independent experiments with standard derivation (±SD) noted (* p-value ≤ 0.05 and ** p-value ≤ 0.01 represent significant and very significant difference, respectively, based on two-tailed t-test).
Table 11.
The effect of metal ions or additive on purified S.griseus recombinant β-glucosidase was determined spectrometrically after 1-h and 6-h of incubation in the presence of 1 mM of each ion in potassium phosphate buffer, pH 7. The residual activity (%) was calculated in comparison to the activity obtained from enzyme in the same condition, but in the absence of any metal ion or additive. The results are the average of three independent experiments with standard derivation (±SD) noted (* p-value ≤ 0.05 and ** p-value ≤ 0.01 represent significant and very significant difference, respectively, based on two-tailed t-test).
| Compounds | Residual Activity % ± SD (1 h) | Residual Activity % ± SD (6 h) |
|---|
| Control | 100 ± 0.044 | 100 ± 0.074 |
| Ca2+ | 105.57 ± 2.91 | 116.28 ± 3.90 ** |
| Mg2+ | 106.55 ± 2.57 | 118.45 ± 4.11** |
| N+ | 101.98 ± 2.91 | 116.57 ± 1.54 ** |
| K+ | 101.01 ± 1.65 | 113.53 ± 3.38 * |
| ZnSO4 | 86.99 ± 2.01 * | 11.56 ± 0.58 ** |
| (NH4)2S4 | 86.27 ± 1.36 * | 99.74 ± 2.69 |
Table 12.
Kinetic constants determined for purified β-glucosidase activity towards pNPG and cellobiose, as determined by non-linear regression analysis, using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
Table 12.
Kinetic constants determined for purified β-glucosidase activity towards pNPG and cellobiose, as determined by non-linear regression analysis, using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
| Substrate | Km (mM) | Vmax (μmol·min−1·mg−1) |
|---|
| pNPG | 8.7 ± 0.42 | 243 ± 6.22 |
| Cellobiose | 15.8 ± 0.62 | 275 ± 7.12 |
Table 13.
Kinetic parameters for β-glucosidases from various sources towards pNPG and cellobiose; n/d is not defined.
Table 13.
Kinetic parameters for β-glucosidases from various sources towards pNPG and cellobiose; n/d is not defined.
| Microorganism | Substrate | Vmax | Km | Optimum Temp. and pH | References |
|---|
| Thermoanaerobacterium thermosaccharolyticum | pNPG | 64 U/mg | 0.62 mM | 70 °C, pH 6.4 | [56] |
| cellobiose | 120 U/mg | 7.9 mM |
| Phoma sp. KCTC11825BP | pNPG | n/d | 0.3 mM | 60 °C, pH 4.5 | [61] |
| cellobiose | n/d | 3.2 mM, |
| Pyrococcus furiosus | pNPG | 700 U/mg | 0.15 mM | 105 °C, pH 5 | [64] |
| Celobiose | 470 U/mg | 20 mM |
| Thermoascus aurantiacus | pNPG | n/d | 0.1137 mM | 80 °C, pH 4.5 | [65] |
| cellobiose | n/d | 0.6370 mM |
| Aureobasidium pullulans (NRRL Y-1297) | pNPG | 897 μmol·min−1·mg−1 | 1.17 mM | 75 °C, pH 4.5 | [66] |
| cellobiose | 800 μmol·min−1·mg−1 | 1.00 mM |
| Monascus purpureus NRRL1992 | pNPG | 6.51 U/mg | 0.39 mM | 50 °C, pH 5.5 | [47] |
| cellobiose | 4.71 U/mg | 2.86 mM |
| Aspergillus fumigatus Z5 | pNPG | 141.60 μmol·min−1·mg−1 | 1.73 mM | 60 °C, pH 6 | [67] |
| cellobiose | 52.37 μmol·min−1·mg−1 | 1.75 mM |
| Stachybotrys strain | pNPG | 78 U/mg | 0.27 mM | 50 °C, pH 5 | [68] |
| cellobiose | 59.4 U/mg | 2.22 mM |
| Neosartorya fischeri NRRL181 | pNPG | 886 μmol·min−1·mg−1 | 68 mM | 40 °C, pH 6 | [69] |
| Aspergillus niger | pNPG | 166 μmol·min−1·mg−1 | 8.0 mM | 50 °C, pH 8 | [70] |



 ), 6 (
), 6 ( ), 7 (
), 7 ( ), and 8 (
), and 8 ( ), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.
), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.
 ), 6 (
), 6 ( ), 7 (
), 7 ( ), and 8 (
), and 8 ( ), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.
), over a period of 180 min at 37 °C. Residual activity (%) at each time point was calculated considering the initial activity, at zero time as 100%. Data represent the mean of three independent experiments, with standard deviations noted as error bars.