Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors †
Abstract
1. Introduction
1.1. The Relationship between Oxidative Phosphorylation and Aerobic Glycolysis
1.2. Are Growth Rates Inversely Related to Functionality?
2. Materials and Methods
2.1. Cell Culture
2.2. Determination of Protein Content of Spheroids
2.3. Determination of Glucose and Glycogen Content of Spheroids
2.4. Mass Spectroscopy
3. Results and Discussion
3.1. Adaptation to Growth in 3D Culture
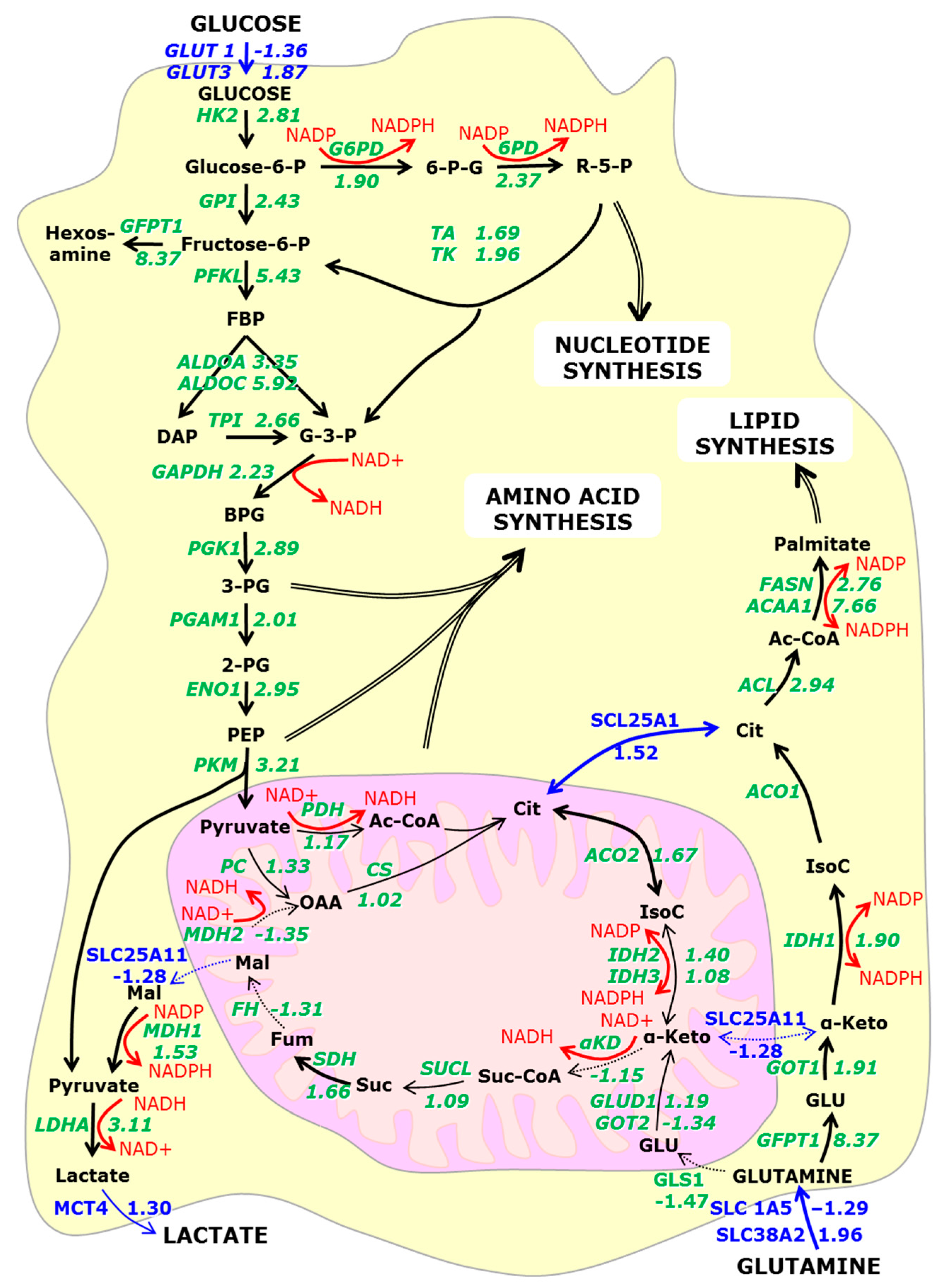
3.2. Is Metabolic Reprogramming Driven by Oxygen or Glucose Insufficiency?
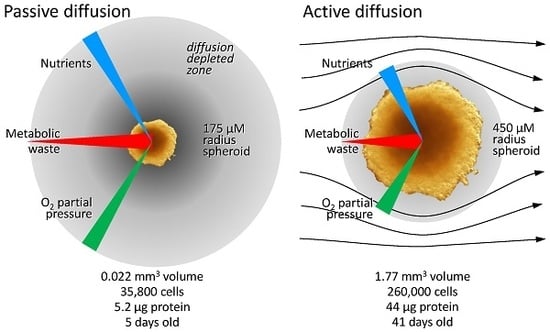
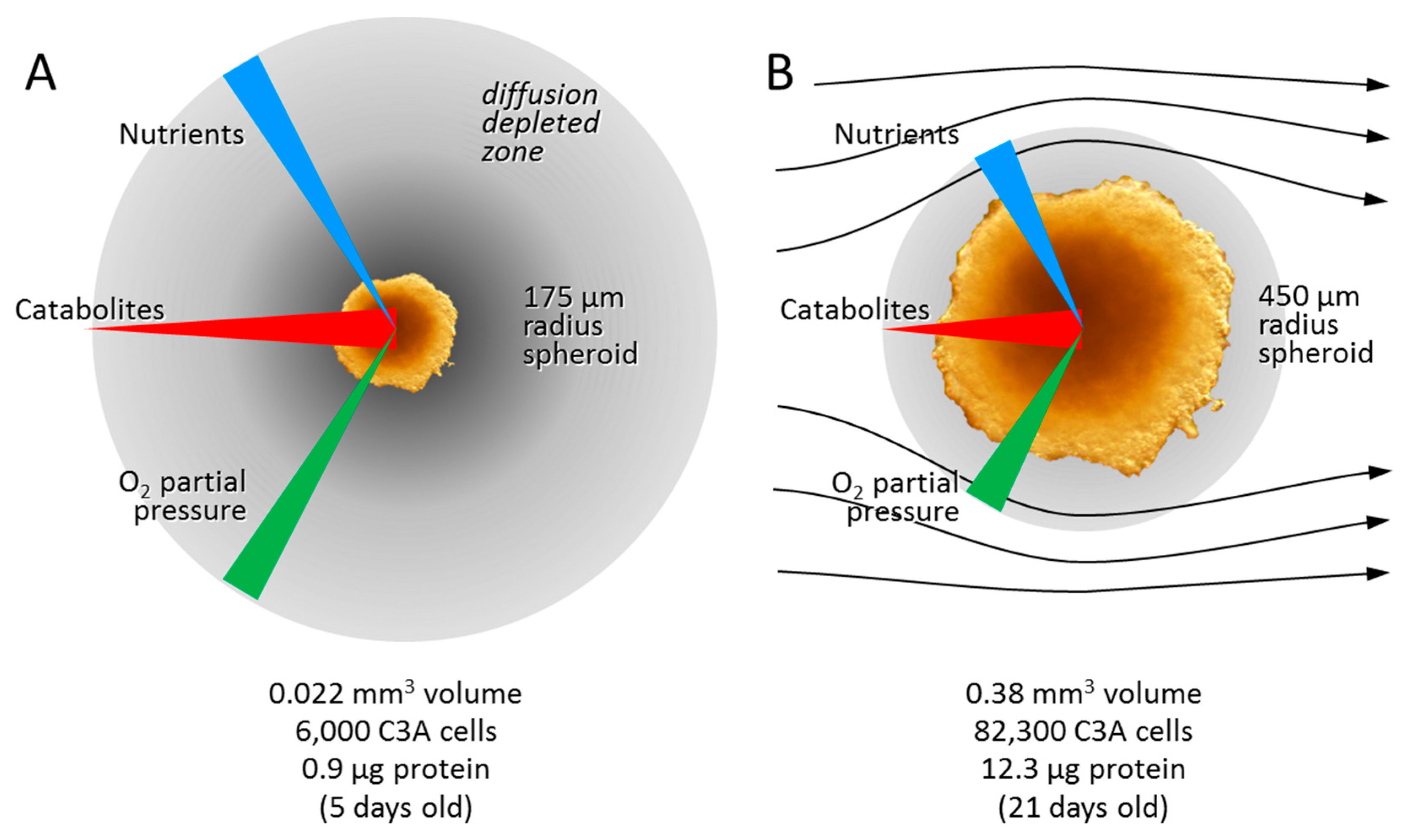
3.2.1. Diffusion Gradients and the Importance of Irrigation
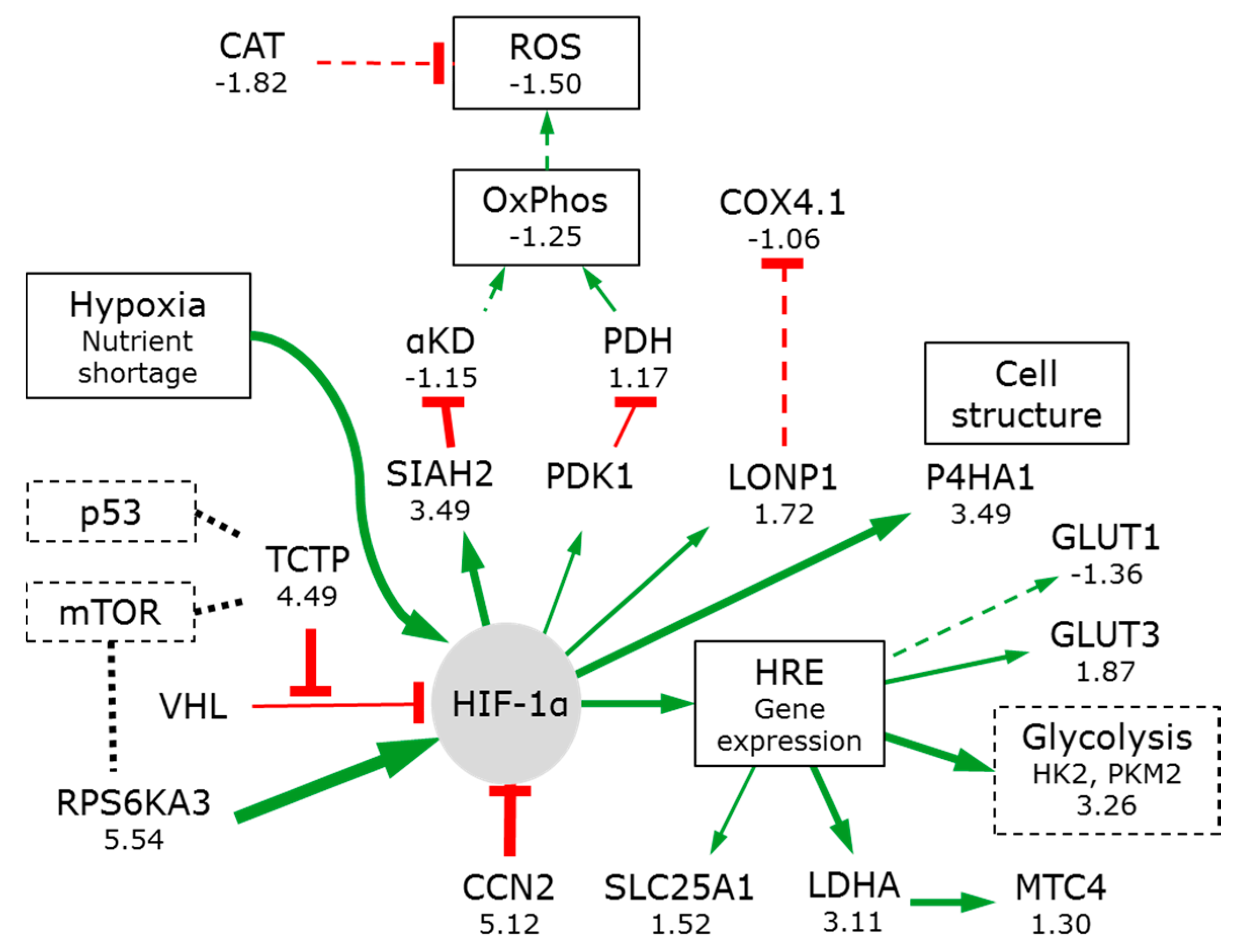
3.2.2. Hypoxia Affects Glycolysis and Oxidative Phosphorylation
3.3. Glucose Starvation Has Little Effect on Metabolic Reprogramming
3.4. Metabolic Reprogramming ‘Links’ Glutamine Metabolism to the Hexosamine Pathway
3.4.1. Conversion of Glutamine to Glutamate
3.4.2. α-Ketoglutarate
3.4.3. NADH
3.4.4. Citrate
3.5. Metabolic Reprogramming Is Associated with Chromatin Remodelling
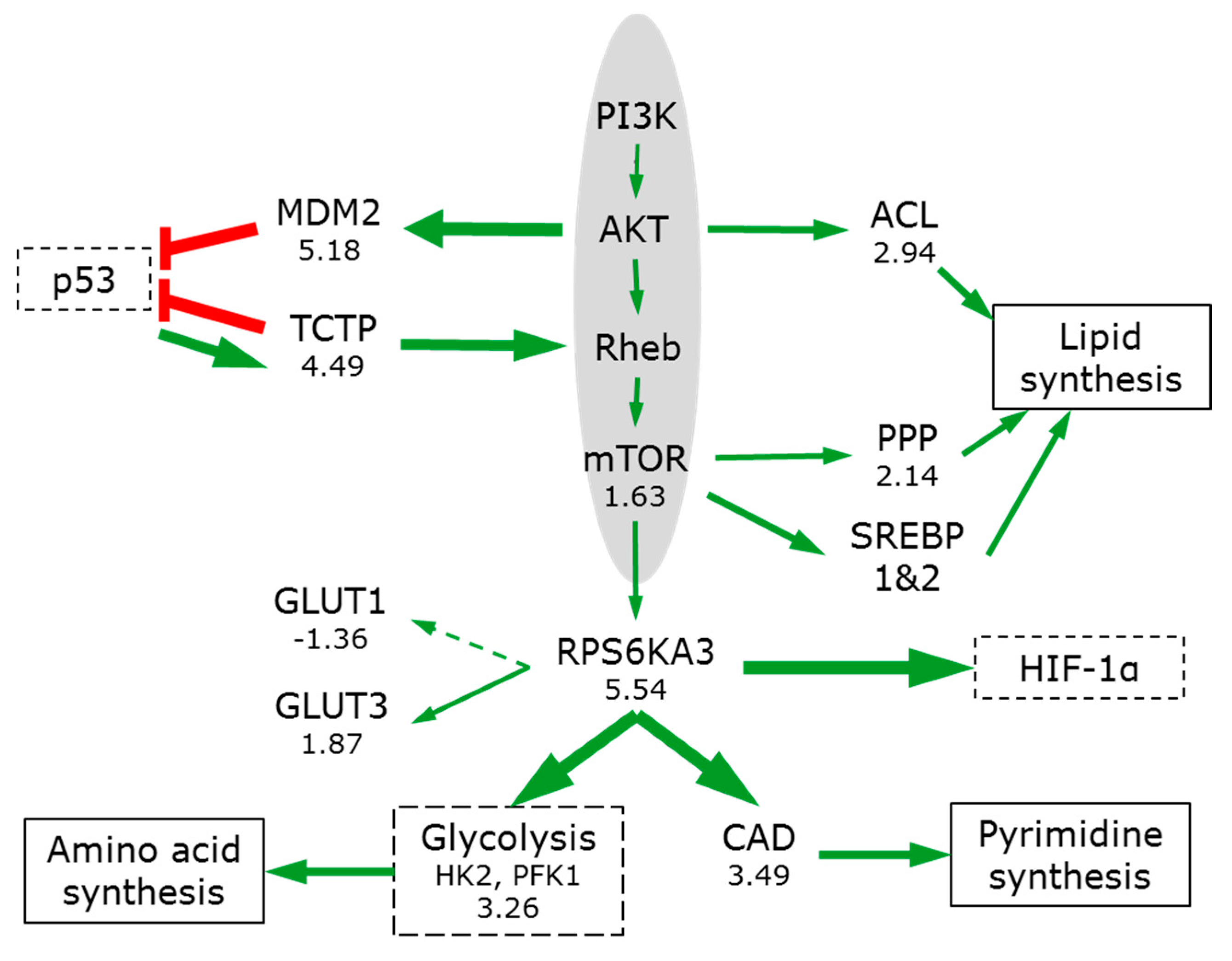
3.6. The Switch to Anabolic Metabolism
3.7. Signal Pathways Involved in Orchestrating Metabolic Reprogramming
3.7.1. PIK3/AKT/mTOR
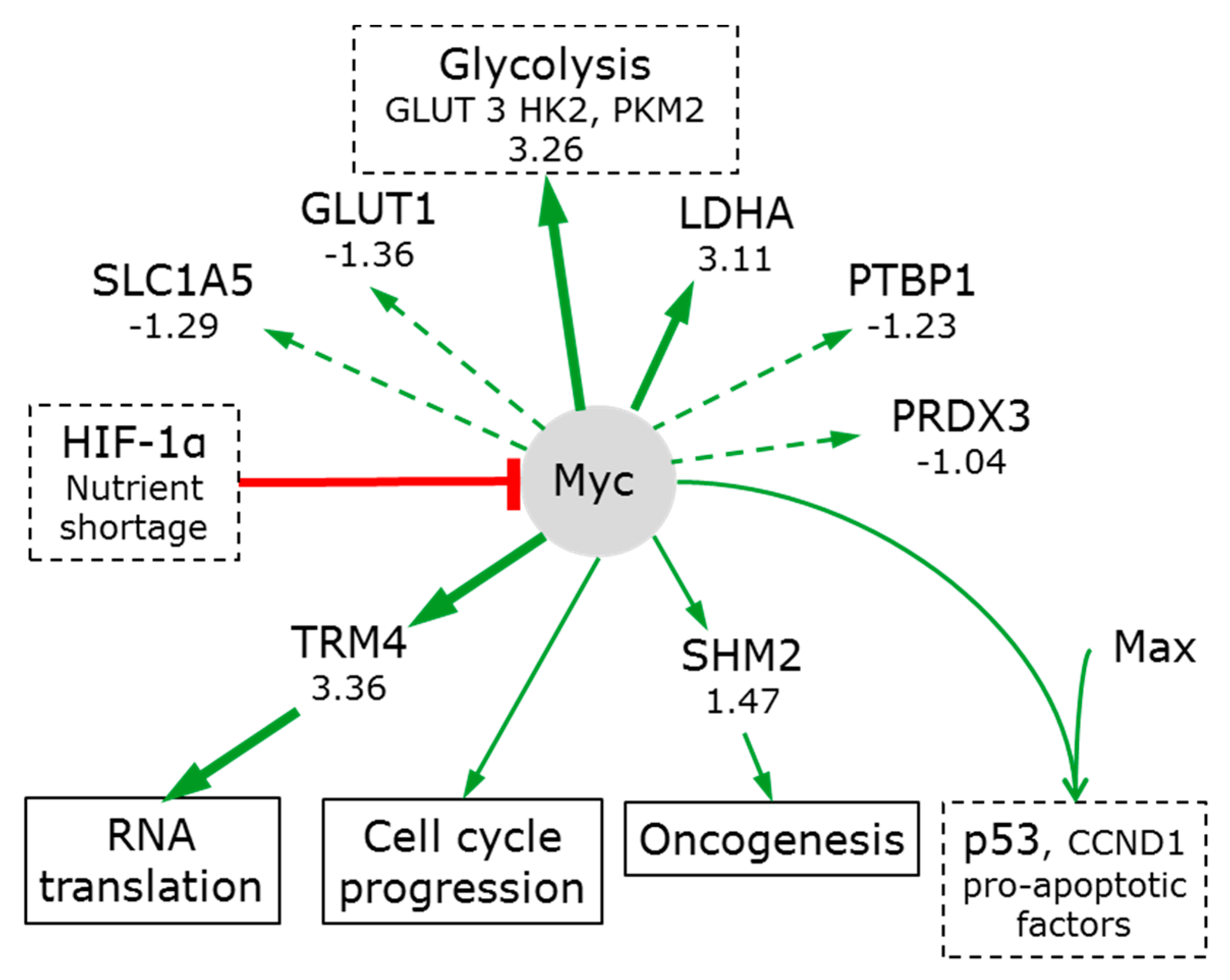
3.7.2. Myc
3.7.3. p53
3.7.4. Wnt GSK-3β/β-Catenin
3.7.5. NF-κB
3.7.6. Cell Death
4. Conclusions
- Oxygen limitations (and to a less extent glucose) induce metabolic reprogramming from oxidative phosphorylation to aerobic glycolysis and result in a strong anabolic phenotype.
- The metabolic reprogramming includes an activation of glutaminolysis (via extra-mitochondrial pathways) (consistent with physiological increases in lipid and cholesterol synthesis).
- Glutamine conversion to the lipid ‘precursor’ glutamate is linked to the hexosamine pathway activation. This correlates to increased glycogen production and protein glycosylation.
- The additional NADPH needed for citrate and lipid synthesis is mainly generated by pentose phosphate pathway activation. Increases in acetyl-CoA also provide precursors for the observed histone acetylation.
- Signalling pathway activities (activation of mTOR and p53, repression of NF-κB and canonical Wnt) are consistent with significant retardation of proliferation and the accumulation of cells in G1/G0, (resulting in a rate resembling that seen in both healthy and transformed cells in tissues and tumours).
- The reduction in proliferation rate allows the cell to achieve higher ATP levels.
- Activation of the non-canonical Wnt signalling pathway orchestrates the significant ultrastructural changes.
- The rate of proliferation is not coupled to aerobic glycolysis.
- Metabolic reprogramming underpins the recovery of traits mimicking in vivo physiology.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Balasubramanian, S.; Packard, J.A.; Leach, J.B.; Powell, E.M. Three-dimensional environment sustains morphological heterogeneity and promotes phenotypic progression during astrocyte development. Tissue Eng. Part A 2016, 22, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016, 11, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Bersini, S.; Moretti, M. 3D functional and perfusable microvascular networks for organotypic microfluidic models. J. Mater. Sci. Mater. Med. 2015, 26, 180. [Google Scholar] [CrossRef] [PubMed]
- Ashley, N.; Jones, M.; Ouaret, D.; Wilding, J.; Bodmer, W.F. Rapidly derived colorectal cancer cultures recapitulate parental cancer characteristics and enable personalized therapeutic assays. J. Pathol. 2014, 234, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hong, J.H.; Park, H.K.; Park, J.S.; Kim, B.K.; Lee, J.Y.; Jeong, J.Y.; Yoon, G.S.; Inoue, M.; Choi, G.S.; et al. Colorectal cancer-derived tumor spheroids retain the characteristics of original tumors. Cancer Lett. 2015, 367, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ruppen, J.; Wildhaber, F.D.; Strub, C.; Hall, S.R.; Schmid, R.A.; Geiser, T.; Guenat, O.T. Towards personalized medicine: Chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip 2015, 15, 3076–3085. [Google Scholar] [CrossRef] [PubMed]
- Rajcevic, U.; Knol, J.C.; Piersma, S.; Bougnaud, S.; Fack, F.; Sundlisaeter, E.; Sondenaa, K.; Myklebust, R.; Pham, T.V.; Niclou, S.P.; et al. Colorectal cancer derived organotypic spheroids maintain essential tissue characteristics but adapt their metabolism in culture. Proteome Sci. 2014, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Horning, J.L.; Sahoo, S.K.; Vijayaraghavalu, S.; Dimitrijevic, S.; Vasir, J.K.; Jain, T.K.; Panda, A.K.; Labhasetwar, V. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol. Pharm. 2008, 5, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Vantangoli, M.M.; Wilson, S.; Madnick, S.J.; Huse, S.M.; Boekelheide, K. Morphologic effects of estrogen stimulation on 3D MCF-7 microtissues. Toxicol. Lett. 2016, 248, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, L.; Gerber, D.A. Improved function and growth of pancreatic cells in a three-dimensional bioreactor environment. Tissue Eng. Part C Methods 2013, 19, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Joo, D.J.; Kim, J.Y.; Lee, J.I.; Jeong, J.H.; Cho, Y.; Ju, M.K.; Huh, K.H.; Kim, M.S.; Kim, Y.S. Manufacturing of insulin-secreting spheroids with the RIN-5F cell line using a shaking culture method. Transplant. Proc. 2010, 42, 4225–4227. [Google Scholar] [CrossRef] [PubMed]
- Morabito, C.; Steimberg, N.; Mazzoleni, G.; Guarnieri, S.; Fano-Illic, G.; Mariggio, M.A. RCCS bioreactor-based modelled microgravity induces significant changes on in vitro 3D neuroglial cell cultures. BioMed Res. Int. 2015, 2015, 754283. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.; Hohn, H.P.; Mareel, M.M.; Denker, H.W. Adhesion and invasion of three human choriocarcinoma cell lines into human endometrium in a three-dimensional organ culture system. Placenta 1994, 15, 411–429. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.; van de Water, B.; Price, L.S. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, T.; Tsuboi, S.; Fukaya, K.; Pu, H.; Ohno, T.; Tsuji, T.; Miyazaki, M.; Namba, M. Spheroid cultures of human hepatoblastoma cells (HuH-6 line) and their application for cytotoxicity assay of alcohols. Acta Med. Okayama 1996, 50, 61–66. [Google Scholar] [PubMed]
- Fey, S.J.; Wrzesinski, K. Determination of acute lethal and chronic lethal dose thresholds of valproic acid using 3D spheroids constructed from the immortal human hepatocyte cell line HepG2/C3A. In Valproic Acid; Boucher, A., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2013; pp. 141–165. [Google Scholar]
- Wrzesinski, K.; Fey, S.J. After trypsinisation, 3D spheroids of C3A hepatocytes need 18 days to re-establish similar levels of key physiological functions to those seen in the liver. Toxicol. Res. 2013, 2, 123–135. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Magnone, M.C.; Visby Hansen, L.; Kruse, M.E.; Bergauer, T.; Bobadilla, M.; Gubler, M.; Mizrahi, J.; Zhang, K.; Andreasen, C.M.; et al. HepG2/C3a 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation. Toxicol. Res. 2013, 2, 163–172. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCS. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Greggio, C.; De Franceschi, F.; Figueiredo-Larsen, M.; Gobaa, S.; Ranga, A.; Semb, H.; Lutolf, M.; Grapin-Botton, A. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 2013, 140, 4452–4462. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Cata, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Caiazzo, M.; Okawa, Y.; Ranga, A.; Piersigilli, A.; Tabata, Y.; Lutolf, M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 2016, 15, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Peloso, A.; Dhal, A.; Zambon, J.P.; Li, P.; Orlando, G.; Atala, A.; Soker, S. Current achievements and future perspectives in whole-organ bioengineering. Stem Cell Res. Ther. 2015, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Croughan, M.S.; Wang, D.I. Hydrodynamic effects on animal cells in microcarrier bioreactors. Biotechnology 1991, 17, 213–249. [Google Scholar] [PubMed]
- Cinbiz, M.N.; Tigli, R.S.; Beskardes, I.G.; Gumusderelioglu, M.; Colak, U. Computational fluid dynamics modeling of momentum transport in rotating wall perfused bioreactor for cartilage tissue engineering. J. Biotechnol. 2010, 150, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.C.; Liu, Y.; Yuan, X.; Chella, R.; Ma, T. Aggregation kinetics of human mesenchymal stem cells under wave motion. Biotechnol. J. 2017, 12, 1600448. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, A.; Bordas, R.; Oncul, A.A.; Thevenin, D.; Genzel, Y.; Reichl, U. Experimental characterization of flow conditions in 2- and 20-L bioreactors with wave-induced motion. Biotechnol. Prog. 2011, 27, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Prot, J.M.; Wang, Y.I.; Miller, P.; Llamas-Vidales, J.R.; Naughton, B.A.; Applegate, D.R.; Shuler, M.L. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip 2015, 15, 2269–2277. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.F.; Silva, M.M.; Giroux, D.; Hashimura, Y.; Wesselschmidt, R.; Lee, B.; Roldao, A.; Carrondo, M.J.; Alves, P.M.; Serra, M. Production of oncolytic adenovirus and human mesenchymal stem cells in a single-use, vertical-wheel bioreactor system: Impact of bioreactor design on performance of microcarrier-based cell culture processes. Biotechnol. Prog. 2015, 31, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Ismadi, M.Z.; Gupta, P.; Fouras, A.; Verma, P.; Jadhav, S.; Bellare, J.; Hourigan, K. Flow characterization of a spinner flask for induced pluripotent stem cell culture application. PLoS ONE 2014, 9, e106493. [Google Scholar] [CrossRef] [PubMed]
- Dardik, A.; Chen, L.; Frattini, J.; Asada, H.; Aziz, F.; Kudo, F.A.; Sumpio, B.E. Differential effects of orbital and laminar shear stress on endothelial cells. J. Vasc. Surg. 2005, 41, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Gareau, T.; Lara, G.G.; Shepherd, R.D.; Krawetz, R.; Rancourt, D.E.; Rinker, K.D.; Kallos, M.S. Shear stress influences the pluripotency of murine embryonic stem cells in stirred suspension bioreactors. J. Tissue Eng. Regen. Med. 2014, 8, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Bee, J.S.; Biddlecombe, J.G.; Chen, Q.; Leach, W.T. Computational fluid dynamics (CFD) insights into agitation stress methods in biopharmaceutical development. Int. J. Pharm. 2012, 423, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Filipovic, N.; Ghimire, K.; Saveljic, I.; Milosevic, Z.; Ruegg, C. Computational modeling of shear forces and experimental validation of endothelial cell responses in an orbital well shaker system. Comput. Methods Biomech. Biomed. Eng. 2016, 19, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sanchez, R.; Rodriguez-Enriquez, S.; Marin-Hernandez, A.; Saavedra, E. Energy metabolism in tumor cells. FEBS J. 2007, 274, 1393–1418. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Day, S.E.; Kettunen, M.I.; Gallagher, F.A.; Hu, D.E.; Lerche, M.; Wolber, J.; Golman, K.; Ardenkjaer-Larsen, J.H.; Brindle, K.M. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 2007, 13, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Albers, M.J.; Bok, R.; Chen, A.P.; Cunningham, C.H.; Zierhut, M.L.; Zhang, V.Y.; Kohler, S.J.; Tropp, J.; Hurd, R.E.; Yen, Y.F.; et al. Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008, 68, 8607–8615. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Serrao, E.M.; Kennedy, B.W.; Hu, D.E.; Kettunen, M.I.; Brindle, K.M. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat. Med. 2014, 20, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Shannon, B.J.; Vaishnavi, S.N.; Vlassenko, A.G.; Shimony, J.S.; Rutlin, J.; Raichle, M.E. Brain aerobic glycolysis and motor adaptation learning. Proc. Natl. Acad. Sci. USA 2016, 113, E3782–E3791. [Google Scholar] [CrossRef] [PubMed]
- Lumata, L.; Yang, C.; Ragavan, M.; Carpenter, N.; DeBerardinis, R.J.; Merritt, M.E. Hyperpolarized (13)C magnetic resonance and its use in metabolic assessment of cultured cells and perfused organs. Methods Enzymol. 2015, 561, 73–106. [Google Scholar] [PubMed]
- Fan, T.W.; Kucia, M.; Jankowski, K.; Higashi, R.M.; Ratajczak, J.; Ratajczak, M.Z.; Lane, A.N. Rhabdomyosarcoma cells show an energy producing anabolic metabolic phenotype compared with primary myocytes. Mol. Cancer 2008, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Fantin, V.R.; St-Pierre, J.; Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006, 9, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Hatzivassiliou, G.; Zhao, F.; Bauer, D.E.; Andreadis, C.; Shaw, A.N.; Dhanak, D.; Hingorani, S.R.; Tuveson, D.A.; Thompson, C.B. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 2005, 8, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, F.; Hamanaka, R.; Wheaton, W.W.; Weinberg, S.; Joseph, J.; Lopez, M.; Kalyanaraman, B.; Mutlu, G.M.; Budinger, G.R.; Chandel, N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA 2010, 107, 8788–8793. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Moriguchi, M.; Mitsumoto, Y.; Katagishi, T.; Kimura, H.; Shintani, H.; Deguchi, T.; Okanoue, T.; Kagawa, K.; Ashihara, T. Simple tumor profile chart based on cell kinetic parameters and histologic grade is useful for estimating the natural growth rate of hepatocellular carcinoma. Hum. Pathol. 2002, 33, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Zu, X.L.; Guppy, M. Cancer metabolism: Facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 2004, 313, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. Selection and adaptation during metastatic cancer progression. Nature 2013, 501, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Sabo, A.; Kress, T.R.; Pelizzola, M.; de Pretis, S.; Gorski, M.M.; Tesi, A.; Morelli, M.J.; Bora, P.; Doni, M.; Verrecchia, A.; et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014, 511, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Wrzesinski, K.; Rogowska-Wrzesinska, A.; Kanlaya, R.; Borkowski, K.; Schwammle, V.; Dai, J.; Joensen, K.E.; Wojdyla, K.; Carvalho, V.B.; Fey, S.J. The cultural divide: Exponential growth in classical 2d and metabolic equilibrium in 3D environments. PLoS ONE 2014, 9, e106973. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shestov, A.A.; Swain, P.; Yang, C.; Parker, S.J.; Wang, Q.A.; Terada, L.S.; Adams, N.D.; McCabe, M.T.; Pietrak, B.; et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016, 532, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Wrzesinski, K.; Fey, S.J. From 2D to 3D—A new dimension for modelling the effect of natural products on human tissue. Curr. Pharm. Des. 2015, 21, 5605–5616. [Google Scholar] [CrossRef] [PubMed]
- Rogowska-Wrzesinska, A.; Wrzesinski, K.; Fey, S.J. Heteromer score-using internal standards to assess the quality of proteomic data. Proteomics 2014, 14, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012, 481, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, S.; Boschek, C.B.; Hugo, F.; Eigenbrodt, E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 2005, 15, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, B.; Gottlieb, E. Rocking cell metabolism: Revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem. Sci. 2012, 37, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Swinnen, J.V.; Smans, K. ATP-citrate lyase: A key player in cancer metabolism. Cancer Res. 2012, 72, 3709–3714. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sai, K.; Gong, F.; Yang, Q.; Chen, F.; Lin, J. Mutation of isocitrate dehydrogenase 1 induces glioma cell proliferation via nuclear factor-kappaB activation in a hypoxia-inducible factor 1-alpha dependent manner. Mol. Med. Rep. 2014, 9, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; O’Sullivan, D.; Klein Geltink, R.I.; Curtis, J.D.; Chang, C.H.; Sanin, D.E.; Qiu, J.; Kretz, O.; Braas, D.; van der Windt, G.J.; et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 2016, 166, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Harrison, C.; Jin, E.S.; Chuang, D.T.; Sherry, A.D.; Malloy, C.R.; Merritt, M.E.; DeBerardinis, R.J. Simultaneous steady-state and dynamic 13C NMR can differentiate alternative routes of pyruvate metabolism in living cancer cells. J. Biol. Chem. 2014, 289, 6212–6224. [Google Scholar] [CrossRef] [PubMed]
- Keshari, K.R.; Kurhanewicz, J.; Jeffries, R.E.; Wilson, D.M.; Dewar, B.J.; Van Criekinge, M.; Zierhut, M.; Vigneron, D.B.; Macdonald, J.M. Hyperpolarized (13)C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism. Magn. Reson. Med. 2010, 63, 322–329. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.R.; Kelly, C.; Bloch, K.; Partridge, M. A method for estimating the oxygen consumption rate in multicellular tumour spheroids. J. R. Soc. Interface 2014, 11, 20131124. [Google Scholar] [CrossRef] [PubMed]
- Carreau, A.; El Hafny-Rahbi, B.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Lenihan, C.R.; Taylor, C.T. The impact of hypoxia on cell death pathways. Biochem. Soc. Trans. 2013, 41, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, I.; Krishna, C.; Kaper, F.; Cai, D.; Giaccia, A.J.; Denko, N.C. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 2005, 65, 3171–3178. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, R.I.; Zhdanov, A.V.; Nolan, Y.M.; Papkovsky, D.B. Imaging of neurosphere oxygenation with phosphorescent probes. Biomaterials 2013, 34, 9307–9317. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.M. Cell and environment interactions in tumor microregions: The multicell spheroid model. Science 1988, 240, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Klieser, W.F.; Sutherland, R.M. Influence of convection in the growth medium on oxygen tensions in multicellular tumor spheroids. Cancer Res. 1982, 42, 237–242. [Google Scholar] [PubMed]
- Hulikova, A.; Swietach, P. Rapid CO2 permeation across biological membranes: Implications for CO2 venting from tissue. FASEB J. 2014, 28, 2762–2774. [Google Scholar] [CrossRef] [PubMed]
- Hulikova, A.; Vaughan-Jones, R.D.; Swietach, P. Dual role of CO2/HCO3(-) buffer in the regulation of intracellular pH of three-dimensional tumor growths. J. Biol. Chem. 2011, 286, 13815–13826. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Kawana, K.; Adachi, K.; Fujimoto, A.; Yoshida, M.; Nakamura, H.; Nishida, H.; Inoue, T.; Taguchi, A.; Takahashi, J.; et al. Spheroid cancer stem cells display reprogrammed metabolism and obtain energy by actively running the tricarboxylic acid (TCA) cycle. Oncotarget 2016, 7, 33297–33305. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, R.; Zhang, H.; Kim, J.W.; Shimoda, L.; Dang, C.V.; Semenza, G.L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007, 129, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, S.; Huang, C.; Cheng, H.; Zhou, R. TCTP increases stability of hypoxia-inducible factor 1alpha by interaction with and degradation of the tumour suppressor VHL. Biol. Cell 2013, 105, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.J.; Bui, T.; Gruber, M.; Gordan, J.D.; DeBerardinis, R.J.; Covello, K.L.; Simon, M.C.; Thompson, C.B. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007, 21, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, D.M.; Bajpai, S.; Chaturvedi, P.; Wirtz, D.; Semenza, G.L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013, 288, 10819–10829. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.M.; Espanol, Y.; Acin-Perez, R.; Rodriguez, F.; Barcena, C.; Watanabe, K.; Calvo, E.; Loureiro, M.; Fernandez-Garcia, M.S.; Fueyo, A.; et al. ATP-dependent lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014, 8, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Wojdyla, K.; Wrzesinski, K.; Williamson, J.; Fey, S.J.; Rogowska-Wrzesinska, A. Acetaminophen-induced S-nitrosylation and S-sulfenylation changes in 3D cultured hepatocarcinoma cell spheroids. Toxicol. Res. 2016, 5, 905–920. [Google Scholar] [CrossRef]
- Lee, A.S. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat. Rev. Cancer 2014, 14, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Endoplasmic reticulum and the unfolded protein response: Dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 2013, 301, 215–290. [Google Scholar] [PubMed]
- Korennykh, A.; Walter, P. Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2012, 28, 251–277. [Google Scholar] [CrossRef] [PubMed]
- Behnke, J.; Feige, M.J.; Hendershot, L.M. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J. Mol. Biol. 2015, 427, 1589–1608. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, Y.D.; Ganapathy, V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta 2016, 1863, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Khelifa, S.; Ratnikov, B.; Scott, D.A.; Feng, Y.; Parisi, F.; Ruller, C.; Lau, E.; Kim, H.; Brill, L.M.; et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell 2015, 27, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Broer, A.; Rahimi, F.; Broer, S. Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 2016, 291, 13194–13205. [Google Scholar] [CrossRef] [PubMed]
- Guillaumond, F.; Leca, J.; Olivares, O.; Lavaut, M.N.; Vidal, N.; Berthezene, P.; Dusetti, N.J.; Loncle, C.; Calvo, E.; Turrini, O.; et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. USA 2013, 110, 3919–3924. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Hypoxia-inducible histone lysine demethylases: Impact on the aging process and age-related diseases. Aging Dis. 2016, 7, 180–200. [Google Scholar] [PubMed]
- Tvardovskiy, A.; Schwammle, V.; Kempf, S.J.; Rogowska-Wrzesinska, A.; Jensen, O.N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017, 45, 9272–9289. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.K.; Jeong, Y.W.; Kim, D.I.; Park, M.J.; Choi, J.H.; Kim, S.U.; Kang, S.S.; Han, H.J.; Park, S.H. Activation of PRMT1 and PRMT5 mediates hypoxia- and ischemia-induced apoptosis in human lung epithelial cells and the lung of miniature pigs: The role of p38 and JNK mitogen-activated protein kinases. Biochem. Biophys. Res. Commun. 2013, 440, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Reintjes, A.; Fuchs, J.E.; Kremser, L.; Lindner, H.H.; Liedl, K.R.; Huber, L.A.; Valovka, T. Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFalpha/NF-kappaB response. Proc. Natl. Acad. Sci. USA 2016, 113, 4326–4331. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Wolfe, L.A.; Ichikawa, M.; Markello, T.; He, M.; Tifft, C.J.; Gahl, W.A.; Freeze, H.H. Biallelic mutations in cad, impair de novo pyrimidine biosynthesis and decrease glycosylation precursors. Hum. Mol. Genet. 2015, 24, 3050–3057. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Duvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 2010, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, A.M.; Christen, S.; Shimobayashi, M.; Cornu, M.; Fava, L.L.; Moes, S.; Prescianotto-Baschong, C.; Sauer, U.; Jenoe, P.; Hall, M.N. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 2013, 339, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. Myc, metabolism, and cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Gao, P.; Liu, Y.C.; Semenza, G.L.; Dang, C.V. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 2007, 27, 7381–7393. [Google Scholar] [CrossRef] [PubMed]
- Koshiji, M.; Kageyama, Y.; Pete, E.A.; Horikawa, I.; Barrett, J.C.; Huang, L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004, 23, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Wonsey, D.R.; Zeller, K.I.; Dang, C.V. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. USA 2002, 99, 6649–6654. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.J.; Redman, K.L. Trm4 and Nsun2 RNA:m5c methyltransferases form metabolite-dependent, covalent adducts with previously methylated RNA. Biochemistry 2014, 53, 7132–7144. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, T.W.; Ko, K.S.; Xia, M.; Lu, S.C. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology 2009, 49, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, S.; Afzal, S.; Tomasini, R.; Guillaumond, F.; Tardivel-Lacombe, J.; Mak, T.W.; Iovanna, J.L. Consequences of DJ-1 upregulation following p53 loss and cell transformation. Oncogene 2012, 31, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Moscovitz, O.; Ben-Nissan, G.; Fainer, I.; Pollack, D.; Mizrachi, L.; Sharon, M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015, 6, 6609. [Google Scholar] [CrossRef] [PubMed]
- Mak, G.W.; Lai, W.L.; Zhou, Y.; Li, M.; Ng, I.O.; Ching, Y.P. CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells. PLoS ONE 2012, 7, e42210. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Ellsworth, B.A.; Nirschl, A.A.; McCann, P.J.; Patel, M.; Girotra, R.N.; Wu, G.; Sher, P.M.; Morrison, E.P.; Biller, S.A.; et al. Discovery of dapagliflozin: A potent, selective renal sodium-dependent glucose cotransporter 2 (SGLT2) inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 2008, 51, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Amson, R.; Pece, S.; Marine, J.C.; Di Fiore, P.P.; Telerman, A. TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol. 2013, 23, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, H.; Tao, S.; Zheng, Y.; Wu, W.; Lian, F.; Jaramillo, M.; Fang, D.; Zhang, D.D. Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle 2013, 12, 2321–2328. [Google Scholar] [CrossRef] [PubMed]
- Amson, R.; Pece, S.; Lespagnol, A.; Vyas, R.; Mazzarol, G.; Tosoni, D.; Colaluca, I.; Viale, G.; Rodrigues-Ferreira, S.; Wynendaele, J.; et al. Reciprocal repression between p53 and TCTP. Nat. Med. 2012, 18, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Vizlin-Hodzic, D.; Simonsson, T.; Simonsson, S. Translationally controlled tumor protein interacts with nucleophosmin during mitosis in ES cells. Cell Cycle 2010, 9, 2160–2169. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.; Metukuri, M.; Scott, D.; Rothermund, K.; Prochownik, E.V. Regulation of reactive oxygen species homeostasis by peroxiredoxins and c-myc. J. Biol. Chem. 2009, 284, 6520–6529. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, L.; Fish, M.; Logan, C.Y.; Nusse, R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 2015, 524, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Wlodarchak, N.; Xing, Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 162–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Zhang, Y.; Santella, R.M.; Weinstein, I.B. HINT1 inhibits beta-catenin/TCF4, USF2 and NFkappab activity in human hepatoma cells. Int. J. Cancer 2009, 124, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Weiske, J.; Huber, O. The histidine triad protein hint1 triggers apoptosis independent of its enzymatic activity. J. Biol. Chem. 2006, 281, 27356–27366. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, P.; Zhao, L.; Huang, L.; Zhang, Z.; Zhao, S.; Huang, J. NF-kappaB expression and outcomes in solid tumors: A systematic review and meta-analysis. Medicine 2015, 94, e1687. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.R.; Robinson, J.Y.; Sanchez, N.S.; Townsend, T.A.; Arrieta, J.A.; Merryman, W.D.; Trykall, D.Z.; Olivey, H.E.; Hong, C.C.; Barnett, J.V. Common pathways regulate type III TGFbeta receptor-dependent cell invasion in epicardial and endocardial cells. Cell. Signal. 2016, 28, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Blomme, A.; Debois, D.; Somja, J.; Delvaux, D.; Patsos, G.; Di Valentin, E.; Peulen, O.; Mutijima, E.N.; De Pauw, E.; et al. Organized proteomic heterogeneity in colorectal cancer liver metastases and implications for therapies. Hepatology 2014, 59, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, H.; Li, X.; Ding, Q.; Wei, P.; Zhou, J. Transforming growth factor beta-induced is essential for endotoxin tolerance induced by a low dose of lipopolysaccharide in human peripheral blood mononuclear cells. Iran. J. Allergy Asthma Immunol. 2015, 14, 321–330. [Google Scholar] [PubMed]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Kepp, O.; Kroemer, G. Regulated cell death and adaptive stress responses. Cell. Mol. Life Sci. CMLS 2016, 73, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, A.V.; Marchenko, N.D.; Ji, K.; Tsirka, S.E.; Holzmann, S.; Moll, U.M. P53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 2012, 149, 1536–1548. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Kroemer, G. Organelle-specific initiation of cell death. Nat. Cell Biol. 2014, 16, 728–736. [Google Scholar] [CrossRef] [PubMed]


| Pathway | Gene | Fold Change |
|---|---|---|
| Glucose phosphorylation | HK2 | 2.81 |
| Glycogenolysis | PYGB | 4.06 |
| Glycolysis | PFKL | 5.43 |
| Glycolysis | PKM | 3.21 |
| Pentose Phosphate | G6PD | 1.90 |
| Hexose | GFPT1 | 8.37 |
| TCA Cycle | IDH2 & 3 | 1.40 & 1.08 |
| Pyrimidine synthesis | CAD | 3.49 |
| Purine synthesis | PRPS1 | 3.74 |
| Fatty acid synthesis | FASN | 2.76 |
| Fatty acid synthesis | ACAA1 | 7.66 |
| Fatty acid oxidation | CRAT | 1.17 |
| Alanine synthesis | ALT | n.d. |
| Asparagine synthesis | ASNS | 5.58 |
| Aspartate synthesis | GOT1 | 1.91 |
| Cysteine synthesis | MAT1 | 5.62 |
| Glutamine-glutamate conversion | GLUD | 1.19 |
| GFPT1 | 8.37 | |
| Glycine synthesis | SHMT | 1.47 |
| Methionine synthesis | MTR | n.d. |
| Proline synthesis | PYCR1 & 2 | 1.06 & 1.03 |
| Serine synthesis | PHGDH | 7.67 |
| Tyrosine synthesis | PAH | 3.80 |
| Urea synthesis | CPS | 3.49 |
| Folate synthesis | MDHFD1 | 2.49 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wrzesinski, K.; Fey, S.J. Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors. Bioengineering 2018, 5, 22. https://doi.org/10.3390/bioengineering5010022
Wrzesinski K, Fey SJ. Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors. Bioengineering. 2018; 5(1):22. https://doi.org/10.3390/bioengineering5010022
Chicago/Turabian StyleWrzesinski, Krzysztof, and Stephen J. Fey. 2018. "Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors" Bioengineering 5, no. 1: 22. https://doi.org/10.3390/bioengineering5010022
APA StyleWrzesinski, K., & Fey, S. J. (2018). Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors. Bioengineering, 5(1), 22. https://doi.org/10.3390/bioengineering5010022