Ro(a)d to New Functional Materials: Sustainable Isolation of High-Aspect-Ratio β-Chitin Microrods from Marine Algae
Abstract
1. Introduction
2. Materials and Methods
2.1. Thalassiosira Rotula Cell Culture
2.2. Chitin Microrod Isolation
2.3. Light Microscopy
2.4. Electron Microscopy
2.5. HAADF-STEM Microscopy
2.6. Imaging and Statistical Analysis of the Chitin Microrods
3. Results and Discussion
3.1. Native Chitin Rod Formation in Thalassiosira rotula
3.2. Thalassiosira rotula Chitin Rod Isolation
3.3. Electron Microscopical Analysis of the Isolated Chitin Rods
3.4. Comparison with Nanochitins from Other Sources
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AWI | Alfred Wegener Institut |
| DP | Degree of polymerization |
| DA | Degree of acetylation |
| ddH2O | Double distilled water |
| ESAW | Enriched artificial sea water |
| FP | Fultoportulae |
| GlcNAc | N-acetyl glucosamine |
| HAADF-STEM | High-angle annular dark-field scanning transmission electron microscopy |
| PA | Patterns of acetylation |
| RT | Room Temperature |
| SEM | Scanning electron microscopy |
References
- Dhillon, G.S.; Kaur, S.; Brar, S.K.; Verma, M. Green Synthesis Approach: Extraction of Chitosan from Fungus Mycelia. Crit. Rev. Biotechnol. 2013, 33, 379–403. [Google Scholar] [CrossRef] [PubMed]
- Novis, P.M.; Sales, R.E.; Gordon, K.; Manning, N.; Duleba, M.; Ács, É.; Dressler, M.; Schallenberg, M. Lindavia Intermedia (Bacillariophyceae) and Nuisance Lake Snow in New Zealand: Chitin Content and Quantitative PCR Methods to Estimate Cell Concentrations and Expression of Chitin Synthase1. J. Phycol. 2020, 56, 1232–1244. [Google Scholar] [CrossRef]
- Hou, J.; Aydemir, B.E.; Dumanli, A.G. Understanding the Structural Diversity of Chitins as a Versatile Biomaterial. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2021, 379, 20200331. [Google Scholar] [CrossRef]
- Yan, N.; Chen, X. Sustainability: Don’t Waste Seafood Waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef]
- Ozkan, A. Screening Diatom Strains Belonging to Cyclotella Genus for Chitin Nanofiber Production under Photobioreactor Conditions: Chitin Productivity and Characterization of Physicochemical Properties. Algal Res. 2023, 70, 103015. [Google Scholar] [CrossRef]
- Aldila, H.; Asmar; Fabiani, V.A.; Dalimunthe, D.Y.; Irwanto, R. The Effect of Deproteinization Temperature and NaOH Concentration on Deacetylation Step in Optimizing Extraction of Chitosan from Shrimp Shells Waste. IOP Conf. Ser. Earth Environ. Sci. 2020, 599, 012003. [Google Scholar] [CrossRef]
- Kozma, M.; Acharya, B.; Bissessur, R. Chitin, Chitosan, and Nanochitin: Extraction, Synthesis, and Applications. Polymers 2022, 14, 3989. [Google Scholar] [CrossRef]
- Mohan, K.; Ganesan, A.R.; Ezhilarasi, P.N.; Kondamareddy, K.K.; Rajan, D.K.; Sathishkumar, P.; Rajarajeswaran, J.; Conterno, L. Green and Eco-Friendly Approaches for the Extraction of Chitin and Chitosan: A Review. Carbohydr. Polym. 2022, 287, 119349. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.-C.; Liu, C.; Liu, X.; Lu, Y.; Zhou, G.; Liu, C.; Mei, C. Microwave-Assisted Deep Eutectic Solvent Extraction of Chitin from Crayfish Shell Wastes for 3D Printable Inks. Ind. Crops Prod. 2023, 194, 116325. [Google Scholar] [CrossRef]
- Saravana, P.S.; Ho, T.C.; Chae, S.-J.; Cho, Y.-J.; Park, J.-S.; Lee, H.-J.; Chun, B.-S. Deep Eutectic Solvent-Based Extraction and Fabrication of Chitin Films from Crustacean Waste. Carbohydr. Polym. 2018, 195, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Shi, L.; Ma, S.; Chen, X.; Cao, S.; Li, W.; Zhao, Z.; Chen, C.; Deng, H. Chitin/Chitosan Nanofibers Toward a Sustainable Future: From Hierarchical Structural Regulation to Functionalization Applications. Nano Lett. 2024, 24, 12014–12026. [Google Scholar] [CrossRef]
- Kovaleva, V.V.; Kuznetsov, N.M.; Istomina, A.P.; Bogdanova, O.I.; Vdovichenko, A.Y.; Streltsov, D.R.; Malakhov, S.N.; Kamyshinsky, R.A.; Chvalun, S.N. Low-Filled Suspensions of α-Chitin Nanorods for Electrorheological Applications. Carbohydr. Polym. 2022, 277, 118792. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Ji, C.; Wang, T.; Wang, Y.; Yang, H. Hairy Chitin Nanocrystals: Sustainable Adsorbents for Efficient Removal of Organic Dyes. Int. J. Biol. Macromol. 2025, 298, 139948. [Google Scholar] [CrossRef]
- Dickinson, E. Biopolymer-Based Particles as Stabilizing Agents for Emulsions and Foams. Food Hydrocoll. 2017, 68, 219–231. [Google Scholar] [CrossRef]
- Holzwarth, M.; Ludwig, J.; Bernz, A.; Claasen, B.; Majoul, A.; Reuter, J.; Zens, A.; Pawletta, B.; Bilitewski, U.; Weiss, I.M.; et al. Modulating Chitin Synthesis in Marine Algae with Iminosugars Obtained by SmI2 and FeCl3-Mediated Diastereoselective Carbonyl Ene Reaction. Org. Biomol. Chem. 2022, 20, 6606–6618. [Google Scholar] [CrossRef]
- LeDuff, P.; Rorrer, G.L. Formation of Extracellular β-Chitin Nanofibers during Batch Cultivation of Marine Diatom Cyclotella Sp. at Silicon Limitation. J. Appl. Phycol. 2019, 31, 3479–3490. [Google Scholar] [CrossRef]
- Herth, W.; Zugenmaier, P. Ultrastructure of the Chitin Fibrils of the Centric Diatom Cyclotella Cryptica. J. Ultrastruct. Res. 1977, 61, 230–239. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Noishiki, Y.; Wada, M. X-Ray Structure of Anhydrous β-Chitin at 1 Å Resolution. Macromolecules 2011, 44, 950–957. [Google Scholar] [CrossRef]
- Lee, S.; Hao, L.T.; Park, J.; Oh, D.X.; Hwang, D.S. Nanochitin and Nanochitosan: Chitin Nanostructure Engineering with Multiscale Properties for Biomedical and Environmental Applications. Adv. Mater. 2023, 35, 2203325. [Google Scholar] [CrossRef]
- Bai, L.; Liu, L.; Esquivel, M.; Tardy, B.L.; Huan, S.; Niu, X.; Liu, S.; Yang, G.; Fan, Y.; Rojas, O.J. Nanochitin: Chemistry, Structure, Assembly, and Applications. Chem. Rev. 2022, 122, 11604–11674. [Google Scholar] [CrossRef] [PubMed]
- Pakizeh, M.; Moradi, A.; Ghassemi, T. Chemical Extraction and Modification of Chitin and Chitosan from Shrimp Shells. Eur. Polym. J. 2021, 159, 110709. [Google Scholar] [CrossRef]
- Sreekumar, S.; Wattjes, J.; Niehues, A.; Mengoni, T.; Mendes, A.C.; Morris, E.R.; Goycoolea, F.M.; Moerschbacher, B.M. Biotechnologically Produced Chitosans with Nonrandom Acetylation Patterns Differ from Conventional Chitosans in Properties and Activities. Nat. Commun. 2022, 13, 7125. [Google Scholar] [CrossRef] [PubMed]
- Panackal Shibu, R.; Jafari, M.; Sagala, S.L.; Shamshina, J.L. Chitin Nanowhiskers: A Review of Manufacturing, Processing, and the Influence of Content on Composite Reinforcement and Property Enhancement. RSC Appl. Polym. 2025. [Google Scholar] [CrossRef]
- Cheng, H.; Bowler, C.; Xing, X.; Bulone, V.; Shao, Z.; Duan, D. Full-Length Transcriptome of Thalassiosira Weissflogii as a Reference Resource and Mining of Chitin-Related Genes. Mar. Drugs 2021, 19, 392. [Google Scholar] [CrossRef] [PubMed]
- Di Dato, V.; Di Costanzo, F.; Barbarinaldi, R.; Perna, A.; Ianora, A.; Romano, G. Unveiling the Presence of Biosynthetic Pathways for Bioactive Compounds in the Thalassiosira Rotula Transcriptome. Sci. Rep. 2019, 9, 9893. [Google Scholar] [CrossRef]
- Sumper, M.; Weiss, I.M.; Eichner, N. Chitin Synthase 1 [Thalassiosira Rotula]. Available online: https://www.ncbi.nlm.nih.gov/protein/ACL00587.1 (accessed on 10 August 2025).
- Harrison, P.J.; Waters, R.E.; Taylor, F.J.R. A Broad Spectrum Artificial Sea Water Medium for Coastal and Open Ocean Phytoplankton1. J. Phycol. 1980, 16, 28–35. [Google Scholar] [CrossRef]
- Berges, J.A.; Franklin, D.J.; Harrison, P.J. Evolution of an Artificial Seawater Medium: Improvements in Enriched Seawater, Artificial Water Over the Last Two Decades. J. Phycol. 2001, 37, 1138–1145. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Scrucca, L.; Fraley, C.; Murphy, T.B.; Raftery, A.E. Model-Based Clustering, Classification, and Density Estimation Using Mclust in R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2023; ISBN 978-1-032-23495-3. [Google Scholar]
- Khan, M.J.; Singh, R.; Shewani, K.; Shukla, P.; Bhaskar, P.V.; Joshi, K.B.; Vinayak, V. Exopolysaccharides Directed Embellishment of Diatoms Triggered on Plastics and Other Marine Litter. Sci. Rep. 2020, 10, 18448. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-Q.; Carter, B.R.; Feely, R.A.; Lauvset, S.K.; Olsen, A. Surface Ocean pH and Buffer Capacity: Past, Present and Future. Sci. Rep. 2019, 9, 18624. [Google Scholar] [CrossRef]
- Montroni, D.; Marzec, B.; Valle, F.; Nudelman, F.; Falini, G. β-Chitin Nanofibril Self-Assembly in Aqueous Environments. Biomacromolecules 2019, 20, 2421–2429. [Google Scholar] [CrossRef]
- McLachlan, J.; McInnes, A.G.; Falk, M. Studies on the Chitan (Chitin: Poly-n-Acetylglucosamine) Fibers of the Diatom Thalassiosira Fluviatilis Hustedt: I Production and Isolation of Chitan Fibers. Can. J. Bot. 1965, 43, 707–713. [Google Scholar] [CrossRef]
- Noishiki, Y.; Nishiyama, Y.; Wada, M.; Okada, S.; Kuga, S. Inclusion Complex of Beta-Chitin and Aliphatic Amines. Biomacromolecules 2003, 4, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Brunner, E.; Richthammer, P.; Ehrlich, H.; Paasch, S.; Simon, P.; Ueberlein, S.; van Pée, K.-H. Chitin-Based Organic Networks: An Integral Part of Cell Wall Biosilica in the Diatom Thalassiosira Pseudonana. Angew. Chem. Int. Ed. Engl. 2009, 48, 9724–9727. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Kimura, S.; Wada, M. Electron Diffraction and High-Resolution Imaging on Highly-Crystalline β-Chitin Microfibril. J. Struct. Biol. 2011, 176, 83–90. [Google Scholar] [CrossRef]
- Cheng, M.; Shao, Z.; Wang, X.; Lu, C.; Li, S.; Duan, D. Novel Chitin Deacetylase from Thalassiosira Weissflogii Highlights the Potential for Chitin Derivative Production. Metabolites 2023, 13, 429. [Google Scholar] [CrossRef]
- Wu, Q.; Jungstedt, E.; Šoltésová, M.; Mushi, N.E.; Berglund, L.A. High Strength Nanostructured Films Based on Well-Preserved β-Chitin Nanofibrils. Nanoscale 2019, 11, 11001–11011. [Google Scholar] [CrossRef]
- Fan, Y.; Saito, T.; Isogai, A. Preparation of Chitin Nanofibers from Squid Pen β-Chitin by Simple Mechanical Treatment under Acid Conditions. Biomacromolecules 2008, 9, 1919–1923. [Google Scholar] [CrossRef] [PubMed]
- Bamba, Y.; Ogawa, Y.; Saito, T.; Berglund, L.A.; Isogai, A. Estimating the Strength of Single Chitin Nanofibrils via Sonication-Induced Fragmentation. Biomacromolecules 2017, 18, 4405–4410. [Google Scholar] [CrossRef]
- Tsai, W.-C.; Wang, S.-T.; Chang, K.-L.B.; Tsai, M.-L. Enhancing Saltiness Perception Using Chitin Nanomaterials. Polymers 2019, 11, 719. [Google Scholar] [CrossRef]
- Fan, Y.; Saito, T.; Isogai, A. Individual Chitin Nano-Whiskers Prepared from Partially Deacetylated α-Chitin by Fibril Surface Cationization. Carbohydr. Polym. 2010, 79, 1046–1051. [Google Scholar] [CrossRef]
- Mushi, N.E.; Butchosa, N.; Salajkova, M.; Zhou, Q.; Berglund, L.A. Nanostructured Membranes Based on Native Chitin Nanofibers Prepared by Mild Process. Carbohydr. Polym. 2014, 112, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Salaberria, A.M.; Fernandes, S.C.M.; Diaz, R.H.; Labidi, J. Processing of α-Chitin Nanofibers by Dynamic High Pressure Homogenization: Characterization and Antifungal Activity against A. Niger. Carbohydr. Polym. 2015, 116, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-C.; Wu, Q.; Song, K.; Cheng, H.N.; Suzuki, S.; Lei, T. Chitin Nanofibers as Reinforcing and Antimicrobial Agents in Carboxymethyl Cellulose Films: Influence of Partial Deacetylation. ACS Sustain. Chem. Eng. 2016, 4, 4385–4395. [Google Scholar] [CrossRef]
- Algae, U.C.C. Enriched Seawater Medium. Available online: https://utex.org/products/enriched-seawater-medium (accessed on 29 August 2025).
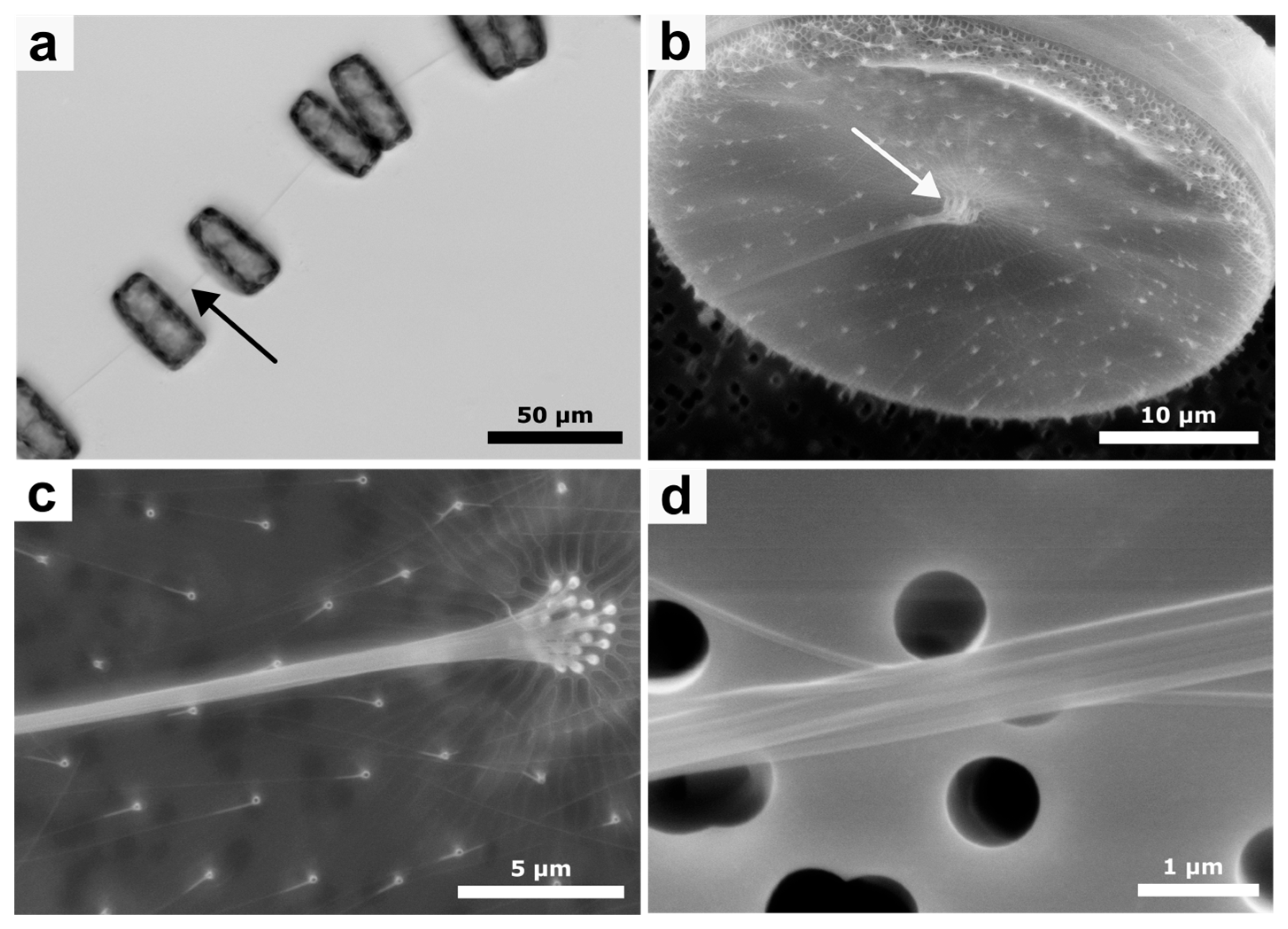
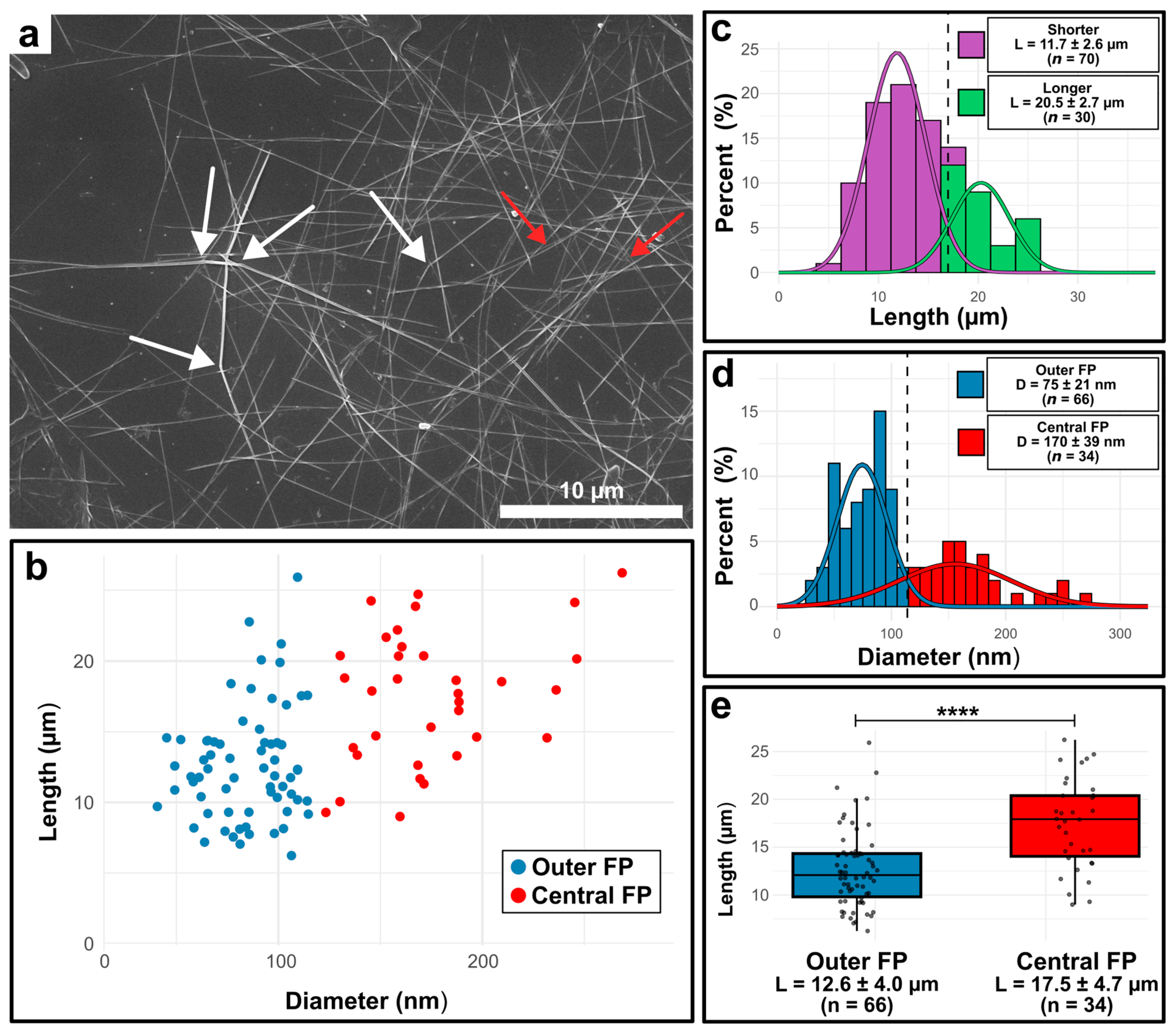
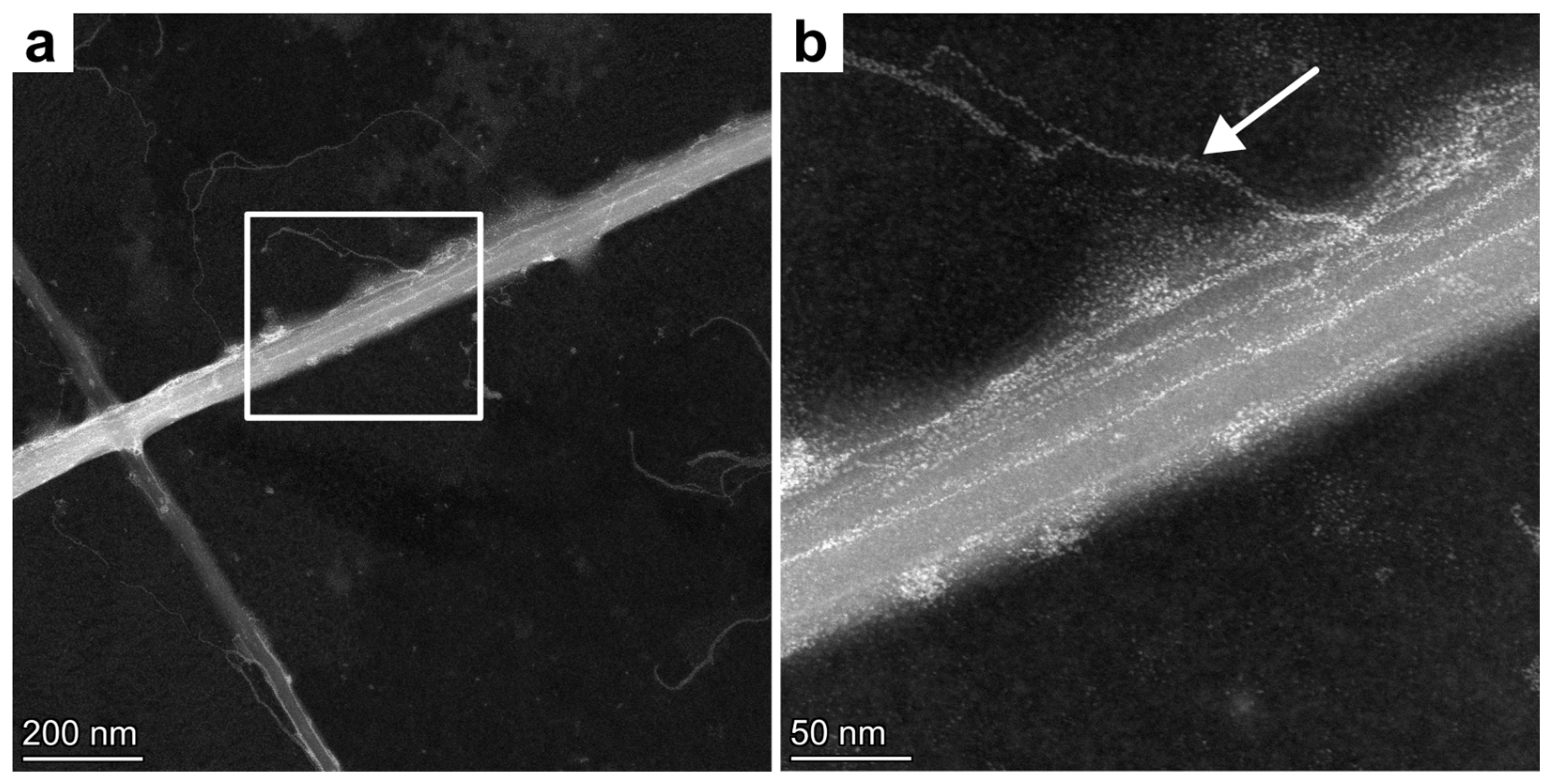
| Central Position | Outer Position | |
|---|---|---|
| Number of fultoportulae per valve (#) | 16.1 ± 1.2 (n = 12) | 109.0 ± 8.5 (n = 4) |
| Fultoportula diameter (nm) | 216 ± 44 (n = 35) | 165 ± 33 (n = 68) |
| Chitin rod diameter (nm) | 111 ± 33 (n = 34) | 69 ± 18 (n = 50) |
| Algal Chitin Source | Method Used to Extract Chitin Rods | Year of Publication | Average Rod Measurements | Reference | |
|---|---|---|---|---|---|
| Length (µm) | Diameter (nm) | ||||
| Thalassiosira fluviatilis Hustedt (extruded chitin) | Dislodging chitin with Waring blender for 1–2 s, removing the cells with Sharples continuous flow centrifugation of supernatant at 2/3 maximum speed, filtration of chitin-rich supernatant through 1.2 µm membrane filter; chitin formed a “mesh” on the filter, which was water-washed, air- or oven-dried at 40 °C, separated from the filter by scraping off the mesh, water-washed, treated with MeOH or ether, and dried under vacuum over P2O5 | 1965 | 60–80 | 100–200 | [36] |
| Cyclotella cryptica (extruded chitin) | Collection of algal cell pellet by centrifugation, vacuum-assisted filtration of the supernatant to collect chitin “mesh” on membrane, 5×water wash on the filter, scraping off chitin “mesh” and wash 2× with ethanol, collection by centrifugation and drying (15 min at 100 °C or 200 °C) | 1977 | 50 | 5–30 | [17] |
| Thalassiosira weissflogii (cell wall and extruded chitin) | Cell collection via centrifugation, treatment of the pellet with 5% KOH (overnight, RT), methanol (80 °C, 2 h), 0.34% NaClO2 (pH 4, 70 °C, 6 h), 0.1 N HCl (boiling, 1 h), 1% HF (RT, overnight) rinsing with water after each step, lyophilization, storage under vacuum | 2003 | Not provided | Not provided | [37] |
| Thalassiosira pseudonana (cell wall chitin) | Cell walls harvested by high-speed centrifugation in a Westfalia separator or filtration on nylon filters, twice boiling of the pellet in 0.1 M EDTA and 2% SDS, centrifugation and water wash until supernatant was colorless, lyophilization overnight. Dissolution of silica frustules using 8M NH4F/2M HF (RT, pH 4–5, 20 min), centrifugation, 4× water wash, lyophilization overnight. Treatment with 2.5 M NaOH (37 °C, 2 h), centrifugation, 4× water wash, lyophilization overnight | 2009 | Not provided | 25 | [38] |
| Thalassiosira weissflogii (extruded chitin) | Dislodging fibers from algal cells by blending in a kitchen mixer for several seconds, low-speed centrifugation and collection of chitin-rich supernatant, high-speed centrifugation to obtain chitin-rich pellet, treatment with 1 N KOH overnight at RT, 0.3% NaClO2 (pH 4.8, 80 °C, 3 h) repeated three times with water washing between each step | 2011 | Not provided | 29.8 | [39] |
| Cyclotella sp. (extruded chitin) | Dislodging of chitin using a Waring blender, low-speed centrifugation and collection of supernatant, high-speed centrifugation to obtain chitin-rich pellet, HPLC-grade water wash, treatment with 1M HCl at 70 °C (30 min), 0.5% (m/m) SDS, 95% EtOH (RT), air-drying at 45 °C for 4 h | 2019 | 60 | 56 | [16] |
| Thalassiosira weissflogii (cell wall and extruded chitin) | Cell collection via centrifugation, treatment of cell pellets and supernatant with methanol (65 °C, 2 h), 5% KOH (RT, overnight), 0.34% NaClO2 (70 °C, 6h), 0.1 N HCl (boiling, 1 h), 1% HF (RT, overnight) with high-speed centrifugation steps and removal of supernatant in between. Sample was dried at 80 °C, stored at −80 °C | 2023 | Not provided | Not provided | [40] |
| Chitin Source | Chitin Polymorph | Length [µm] | Diameter [nm] | Aspect Ratio (L/d) | Reference |
|---|---|---|---|---|---|
| Squid pen (Illex argentinus) | β | 1–3 | 14 ± 7 | ~143 (up to 750) | [41] |
| Squid pen (Todarodes pacificus) | β | >1 | 3–4 | >250 | [42] |
| Squid pen (Loligo bleekeri) | β | 0.48 | 4.1 | ~117 | [43] |
| Squid pen (Illex argentinus) | β | 1.73 ± 0.59 | 17.24 ± 2.02 | ~100 | [44] |
| Algae (Phaeocystis globosa) | α | 3 | 37 ± 8 | ~81 | [43] |
| Crab | α | 0.25 ± 0.14 | 6.2 ± 1.1 | ~40 | [45] |
| Lobster (Homarus americanus) | α | 0.697–1.167 | 3.1–3.5 | 199–376 | [46] |
| Lobster (Cervimunida johni) | α | 5 | 80–100 | >50 | [47] |
| Fresh speckled swimming crabs (Arenaeus cribrarius) | α | 5–10 | 80–100 | >50 | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludwig, J.; Kauffmann, F.; Laschat, S.; Weiss, I.M. Ro(a)d to New Functional Materials: Sustainable Isolation of High-Aspect-Ratio β-Chitin Microrods from Marine Algae. Bioengineering 2025, 12, 969. https://doi.org/10.3390/bioengineering12090969
Ludwig J, Kauffmann F, Laschat S, Weiss IM. Ro(a)d to New Functional Materials: Sustainable Isolation of High-Aspect-Ratio β-Chitin Microrods from Marine Algae. Bioengineering. 2025; 12(9):969. https://doi.org/10.3390/bioengineering12090969
Chicago/Turabian StyleLudwig, Jan, Florian Kauffmann, Sabine Laschat, and Ingrid M. Weiss. 2025. "Ro(a)d to New Functional Materials: Sustainable Isolation of High-Aspect-Ratio β-Chitin Microrods from Marine Algae" Bioengineering 12, no. 9: 969. https://doi.org/10.3390/bioengineering12090969
APA StyleLudwig, J., Kauffmann, F., Laschat, S., & Weiss, I. M. (2025). Ro(a)d to New Functional Materials: Sustainable Isolation of High-Aspect-Ratio β-Chitin Microrods from Marine Algae. Bioengineering, 12(9), 969. https://doi.org/10.3390/bioengineering12090969







