A Collaborative Data Sharing Platform to Accelerate Translation of Biomedical Innovations
Abstract
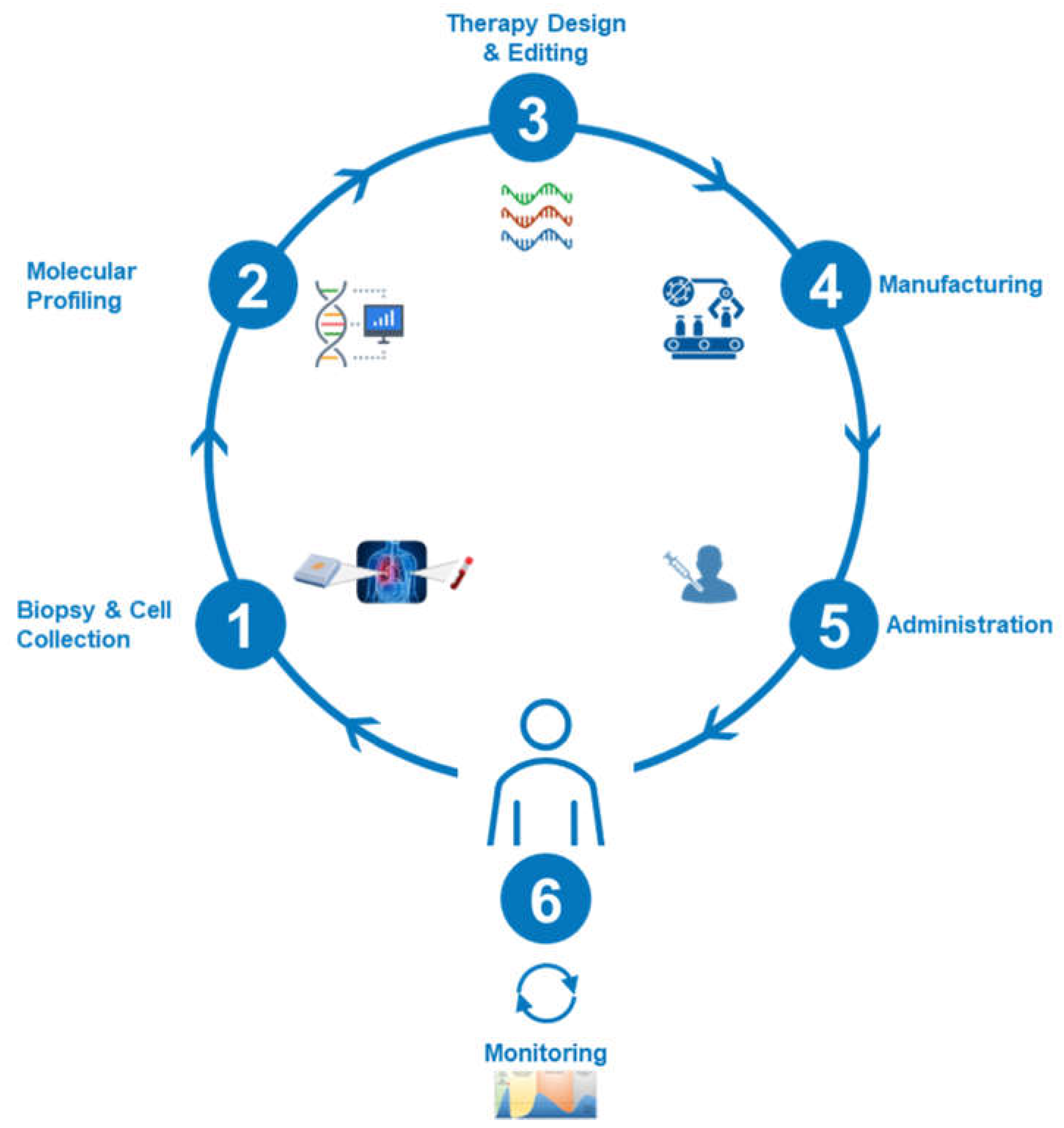
1. Introduction
2. Data Sharing Challenges in the Translation of Biomedical Products
3. Data Sharing Technology Solutions for Biomedical Innovations
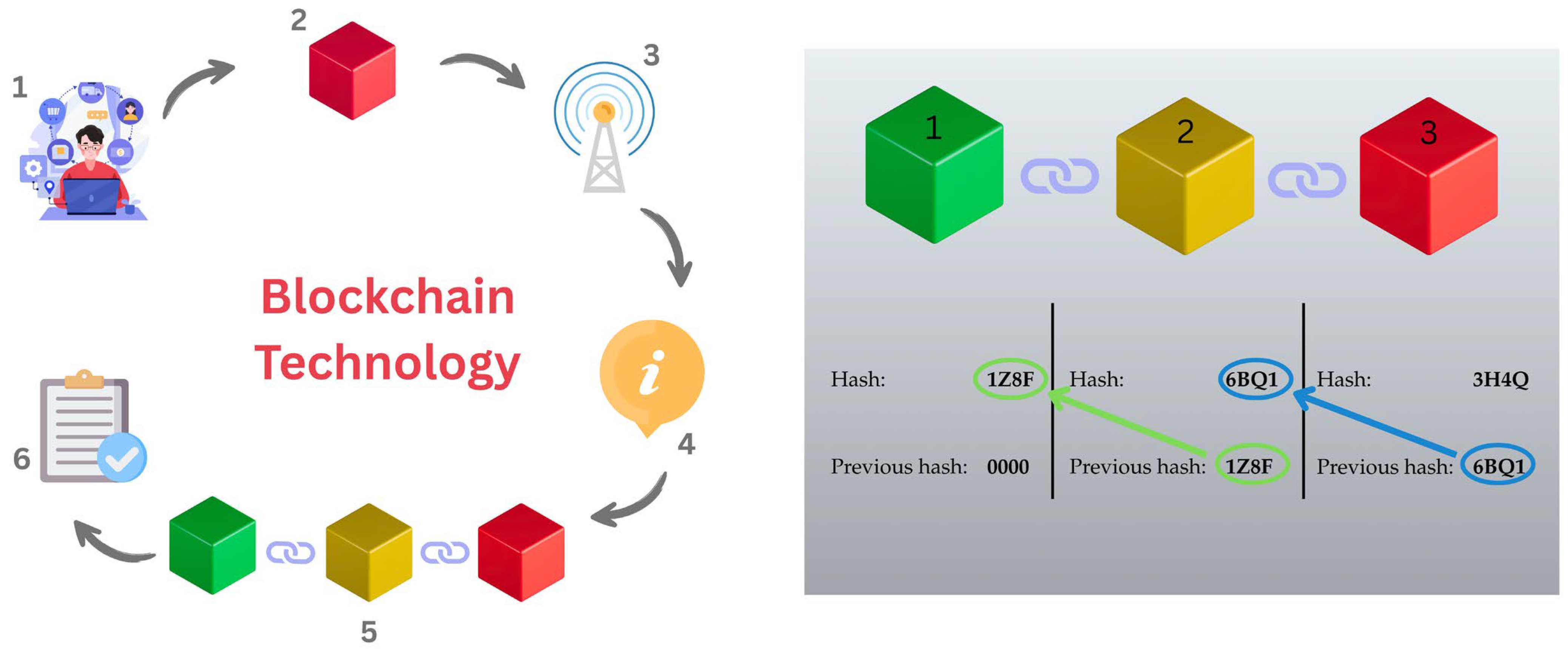
3.1. Blockchain as Foundational Infrastucture for the Knowledge Hub (KH)
- Connection Establishment: Two blockchains establish a connection, which involves agreeing on a common set of rules for communication. This set of rules is established when the blockchain is initially set up and helps to provide a framework for a high level of security and integrity.
- Channel Creation: Within the established connection a channel is created for sending the data. Note: Specific channels can be created for different types of messages or transactions.
- Message Exchange: Blockchains can send packets of data (messages) to each other over the established channels. Each message can carry information about transactions, state changes, or other relevant data.
- Verification: Messages are verified using cryptographic proofs to ensure that they are legitimate and have been sent from an authenticated source. These authenticated sources are established by the developers and the stakeholders.
- State Updates: The receiving blockchain is updated based on the information contained in the incoming message.
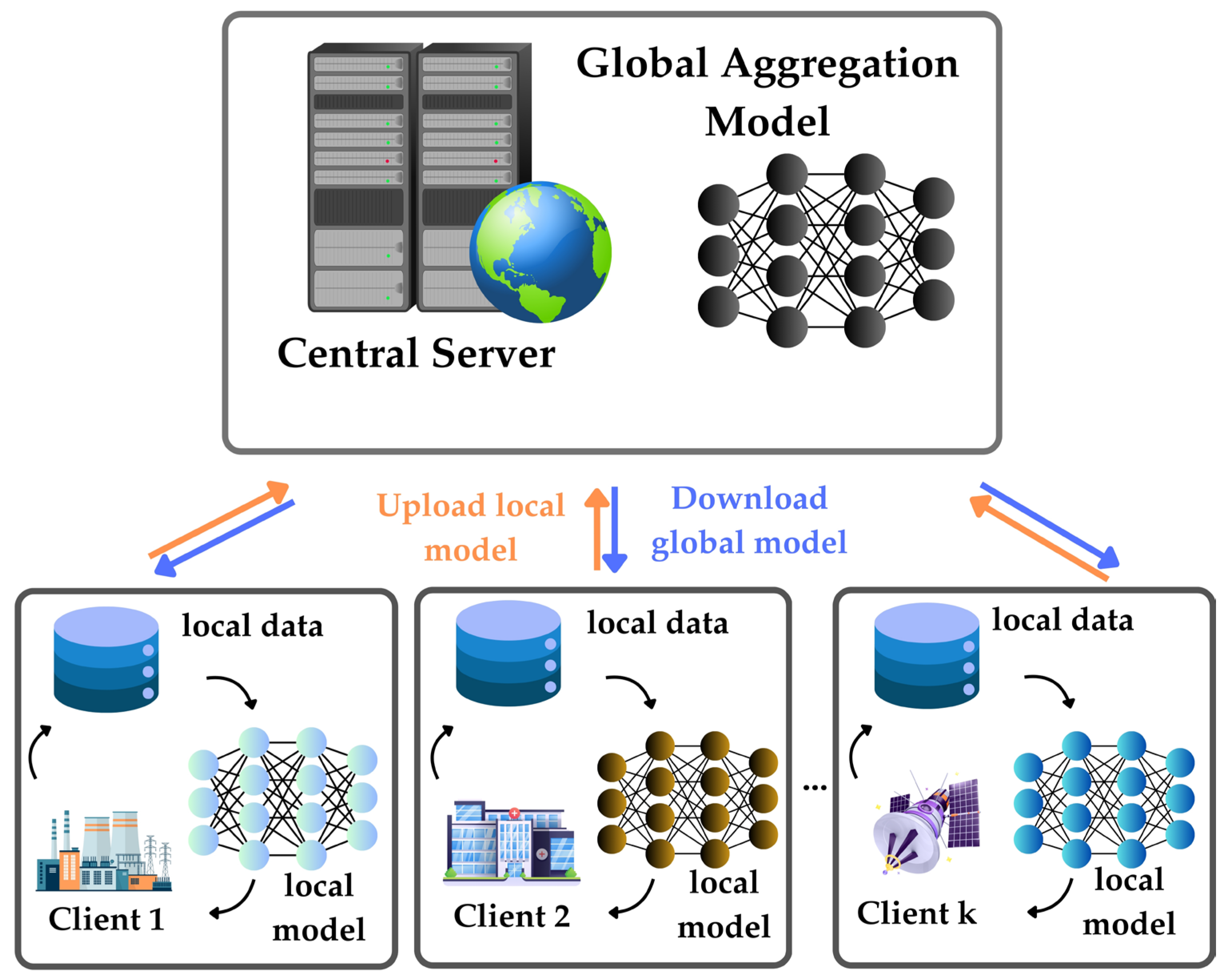
3.2. Federated Learning
- Decentralized Model Training: FL enables model training to occur on local devices or servers where the data resides, rather than centralizing all data on a single server.
- Model Distribution: The initial model or its parameters are distributed to participating devices. Each device trains the model locally using its data and then sends only the model updates (gradients) back to the central server.
- Aggregation: The central server aggregates the received updates from multiple devices to update the global model. Common aggregation methods include simple averaging or more sophisticated methods like weighted aggregation.
- Privacy-Preserving: Raw data never leaves the local device, and only the model updates are shared, helping address privacy concerns associated with centralized models.
- Iterative Process: The process of distributing the model, training locally, and aggregating updates is performed iteratively. The global model improves over time as each local model contributes information.
- Communication Efficiency: FL reduces the need for large-scale data transmission since only model updates are exchanged. This can be beneficial in scenarios where bandwidth or communication costs are significant concerns.
- Personalization: FL allows for personalized model updates based on local data. This is especially useful in applications where individual user preferences or characteristics are important.
- Edge Computing Integration: FL is well-suited for edge computing environments where devices at the edge of the network (e.g., smartphones, IoT devices) can participate in model training without relying heavily on centralized cloud servers.
- Robustness: The distributed nature of FL can enhance model robustness. Local models may adapt to diverse data distributions, leading to a more robust global model.
- Data Discovery. There is a primary graphical user interface (GUI) that allows data users to discover data schematic information and perform Exploratory Data Analysis (EDA) tasks.
- Data Management. Each data owner has complete Digital Rights Management (DRM) capability for their data. Each owner can determine who uses their data and for what purpose.
- Data Usage Management and Audit. TripleBlind ensures that data is only used for authorized purposes (meaning no unauthorized secondary usage of data is allowed).
- Data User Functionality. A wide range of functionalities are available such as:
- Logical data aggregation across data nodes, both horizontally (same data at multiple locations) and vertically (same subject at multiple locations). In the case of vertically arranged data, the system supports sophisticated private record linkage functionality.
- Data harmonization tools. Disparate data can be “pre-processed” into the appropriate format for a given analytical purpose.
- Analytical tools. The system supports everything from simple queries to sophisticated dashboards.
- Machine Learning/Artificial Intelligence (ML/AI) tools. The system supports multiple python data science libraries such as SciKit Learn, PyTorch and Pandas.
- Model delivery. ML/AI models frequently need to be delivered to data in organizations other than the organization in which they were trained, on data that is private to the owning organization (i.e., a diagnostic at a hospital). TripleBlind ensures the appropriate privacy for both the data owner and the model owner.
- Audit Trail. TripleBlind is a secure, closed system (versus an open-source project) that insists that all assets (datasets, models, and algorithms) have cryptographically enforced identifiers. All users have access to credentials. These immutable identifiers are used to record a complete audit trail for every interaction on the system.
- Multi-modal data capability. The system operationalizes any type of data that can be stored electronically—tabular, text, image (i.e., dicom, etc.), video, etc.
- Data Location flexibility. TripleBlind software can run on any server capable of running Linux. Therefore, any data location is accessible—in the cloud, across cloud providers, on-premise, or in space—as long as there is an internet connection available.
- True Data Mesh. The TripleBlind router makes any data useful to any user in the collaboration, effortlessly. Users use the dataset names available via the data discovery tools; the TripleBlind router takes care of the server-to-server (peer-to-peer) connectivity necessary to accomplish the required computation.
3.3. Homomorphic Encryption and Other Approaches to Data and Data Model Security
3.4. New Strategies for Intellectual Property
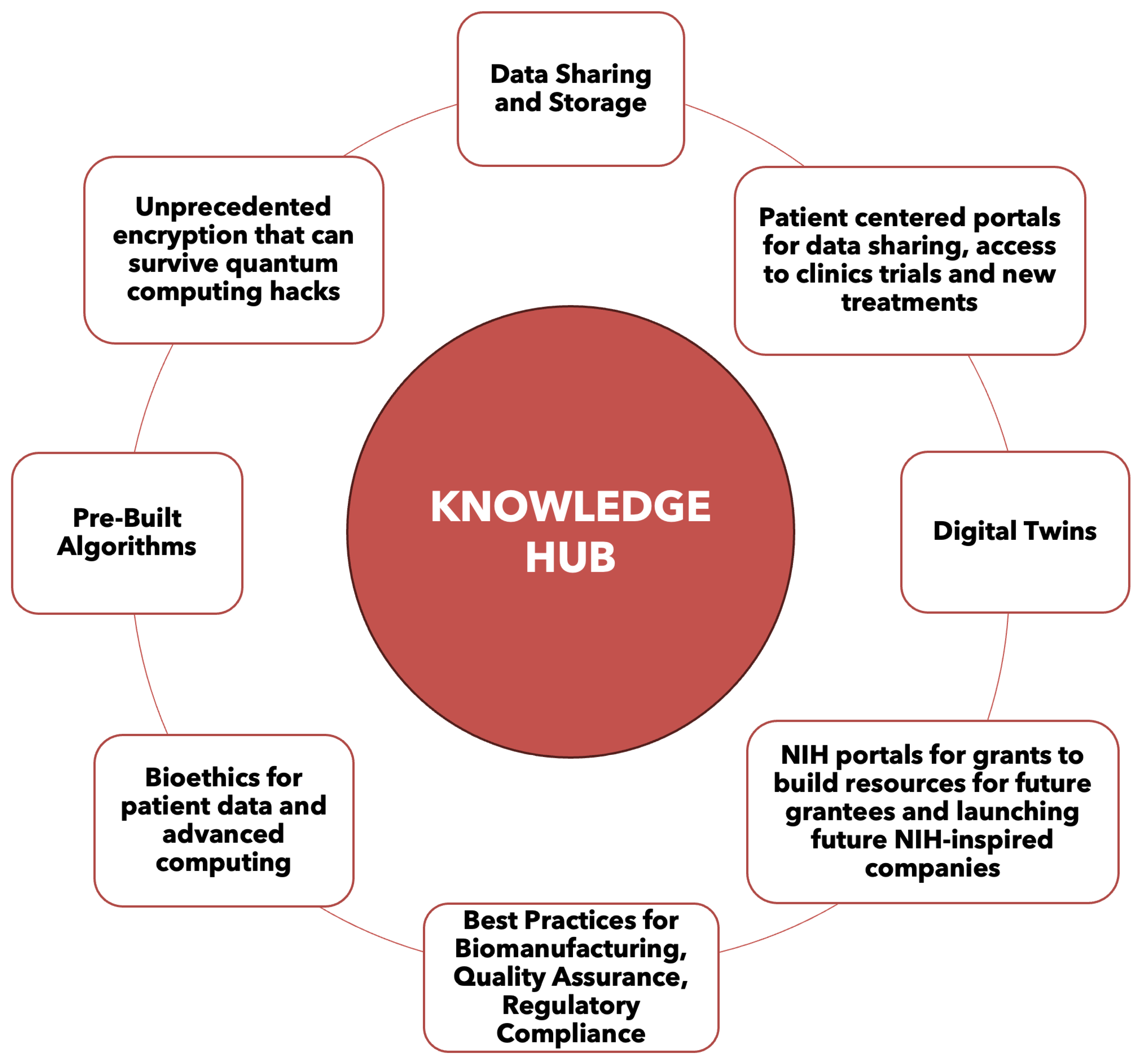
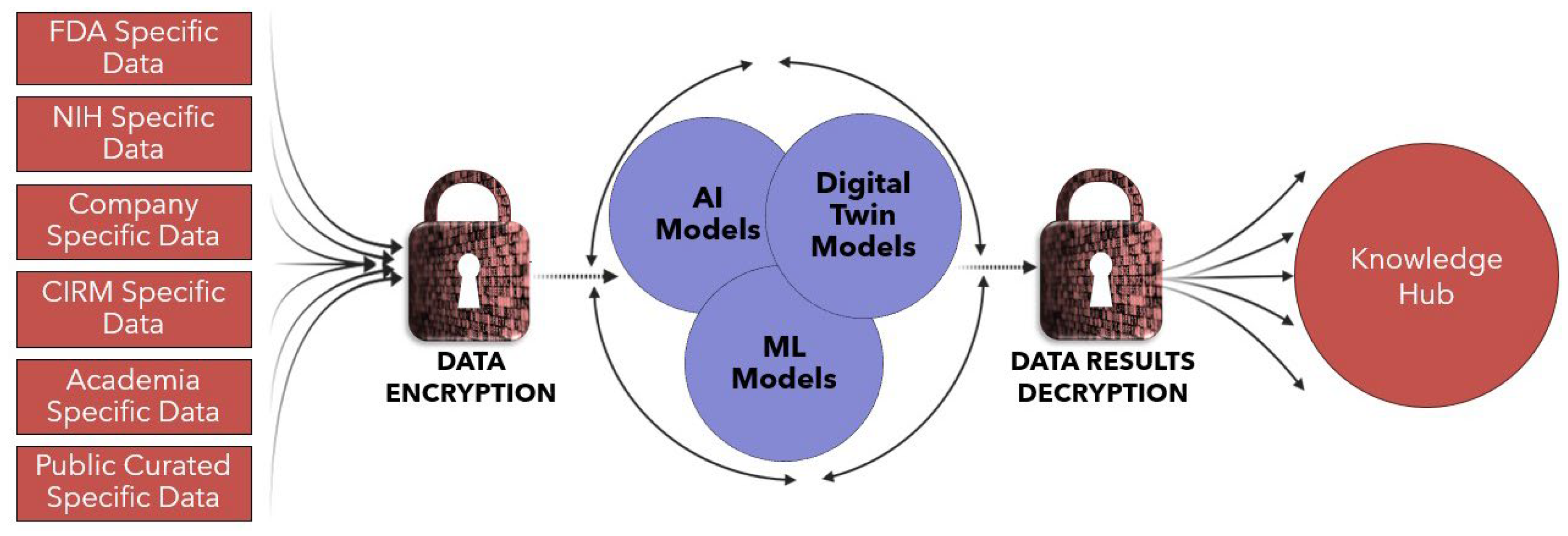
4. Knowledge Hub Capabilities for Advancing Biomanufacturing
5. The Emergent Importance of Data Provenance for Management and Control of Chain of Identity (COI) in TERM Biomanufacturing
- High complexity operations: These End-to-End (E2E) operations are highly discontinuous and complex due to the extensive myriad of interdependent sub-processes, each with the potential to generate extensive and highly variable datasets.
- Multi-institutional: Multiple disparate institutions and entities are required to seamlessly integrate their subprocess across the E2E operation.
- Data-dependent efficacy: The efficacy of these therapies is dependent on error-free digital process control, traceability, and data provenance to ensure authenticated COI.
- Process variability: The processes across clinical, laboratory and bioproduction operations are highly variable due to the diversity of materials, cell systems, devices and formulations used in these processes
- Control & Ownership of COI Risk: The operational implementation and deployment of the E2E process requires capabilities for entity(ies) to have access to a myriad of data and controls that enable mitigation and ownership of the COI risk.
- Unique identifiers linking multiple products to a single therapy event
- Handling and transport controls, barcoding, tracking and unique IDs
- Mandates for labeling, tracking and identity documentation for human cellular and tissue-Based Products (HCT/Ps)
- Validated systems for managing COI and chain of custody (COC)
- Audit trails, data integrity and electronic controls
- Visually readable barcodes on consumables can be corrupted, misread and cloned.
- Adhesive labels on labware and clinical consumables typically do not survive harsh laboratory conditions and chemicals, such as organic solvents and extreme temperatures.
- The small form factor of many consumables has limited surface availability to allow for multiple ID labels to be added by multiple institutions.
- Tamper-proof identification that indelibly links patient samples and subsequent derivatives to their identity.
- Immutable tracking to ensure that every operation subprocess is recorded in an unchangeable ledger
- Interoperability across stakeholders that enable multi-institutional entities across manufacturers, hospitals, diagnostics laboratories, logistics providers, patient services centers and pharmaceutical companies to seamlessly verify the authenticity of the therapy and the E2E process.
- Durability and size to enable tolerance for harsh environmental exposures and sufficient microscopic dimensions to be integrated with the small size of labware and clinical consumables.
6. Knowledge Hub Case Studies
6.1. Case 1: Personalized Stem Cell Therapy KH—A BT
- Patient Consultation and Initial Assessment
- -
- Patients consult with healthcare providers to assess their conditions and determine the need for stem cell therapy.
- -
- Patients opt into the Clinical Trial Advocacy Portal of the KH through their electronic health record profile (e.g., EPIC and CERNER).
- -
- Relevant medical history and conditions are recorded in a newly created block.
- Stem Cell Source Identification
- -
- The source of stem cells (e.g., autologous, allogeneic) is determined. This is based on the patient’s condition, and specific therapeutic goals.
- Informed Consent and Data Collection
- -
- Patients provide informed consent for data collection and treatment through EPIC or CERNER.
- -
- Patient specific data including genetic information, health records, and treatment preferences are added to the block.
- Data Encryption and Hashing
- -
- All collected data is encrypted and hashed to ensure patient privacy and security.
- -
- This step maintains data integrity and confidentiality.
- Personalized Treatment Plan Development
- -
- Based on the collected data and available therapeutic options, personalized treatment plans are developed for each patient and added to the block.
- -
- Treatment plans may include specific stem cell therapies tailored to the patient’s condition.
- Treatment Preparation and Administration
- -
- Information on the used materials, technical protocols, and procedures for preparation of each patient’s therapeutic product is documented and assigned with COI tags and linked to the patient profile in the blockchain.
- -
- Stem cell therapy is administered to the patient based on the personalized treatment plan.
- -
- The administration procedure is documented on the blockchain, maintaining a complete history of treatments.
- Post-Treatment Monitoring and Data Collection
- -
- Patients undergo monitoring to assess the effectiveness of the treatment.
- -
- Outcomes and side effects are recorded and encrypted on the blockchain for future reference.
- Smart Contract Creation
- -
- Smart contracts are created for the block to govern the usage and sharing of patient data and the treatment outcomes.
- -
- Contracts specify conditions for data access and treatment protocols.
- -
- Smart contracts in the blockchain are executed automatically when the specified conditions are met, ensuring that all parties fulfill their obligations without manual intervention. If obligations are not met, depending on how the blockchain is administered, an “error” message is created, or a no execution command will be initiated meaning the process will not proceed to the next step or the whole process can revert to the beginning.
- Stem Cell Processing and Quality Control
- -
- Stem cells are processed in a controlled environment, and in some cases by multiple commercial and academic stakeholders. COI data from the blockchain provides documentation of this complex process for every personalized therapy.
- -
- Quality control measures are implemented, and results are recorded on the blockchain for transparency.
- -
- Biomanufacturing environmental data can also be included on the blockchain to ensure proper handling guidelines are met. As with smart contracts, deviations in environmental conditions or product preparation conditions, recorded on the blockchain, can send alerts and a series of appropriate protocols can be implemented.
- -
- Lab errors that could cause unique patient biological materials to be mixed up are also eliminated.
- Data Storage on Blockchain
- -
- Encrypted patient data, treatment protocols, and quality control results are stored on the blockchain.
- -
- This creates an immutable record that can be accessed by authorized parties.
- Research and Development
- -
- Authorized researchers can access and analyze this immutable record to study the anonymized data with the goal of improving stem cell therapies.
- -
- Insights from research are fed back into the treatment protocols.
- Feedback Loop for Continuous Improvement
- -
- Continuous collection of data from treated patients helps refine stem cell therapies and treatment protocols.
- -
- This feedback loop allows for ongoing research and development based on real-world outcomes.
- Patient Control and Data Access
- -
- Patients can control access to their data and choose to share their treatment outcomes with researchers or providers.
- -
- The use of smart contracts ensures that data sharing is secure and compliant with regulations.
- -
- Patient outcomes could also be linked with manufacturing processes.
- Regulatory Compliance
- -
- Throughout the workflow, compliance with healthcare regulations (like HIPAA, GDPR) is maintained.
- -
- Blockchain provides a transparent and traceable record of data handling.
6.2. Case 2: KidsightsTM Pediatric Database—A FL and TripleBlind Technology
7. Vision for Knowledge Hub and Next Steps
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| API | Application Programming Interface |
| CIRM | California Institute of Regenerative Medicine |
| COC | Chain of Custody |
| COI | Chain of Identity |
| CT | Computer Tomography |
| DeFi | Decentralized Finance |
| DHDM | Digital Health Data Marketplace |
| DLT | Distributed Ledger Technology |
| DRM | Digital Rights Management |
| ECG | Electrocardiogram |
| EDA | Exploratory Data Analysis |
| EMR | Electronic Medical Record |
| EU | European Union |
| E2E | End-to-End |
| FRAND | Fair, Reasonable, and Non-Discriminatory |
| FDA | Food and Drug Administration |
| FL | Federated Learning |
| FHE | Fully Homomorphic Encryption |
| GDPR | General Data Protection Regulation |
| GMP | Good Manufacturing Practices |
| GUI | Graphical User Interface |
| HCT/Ps | Human Cellular Tissue-based Products |
| HIPAA | Health Insurance Portability and Accountability Act |
| IBC | Inter-Blockchain Communication |
| IETF | Internet Engineering Task Force |
| IIoT | Industrial-internet-of-things |
| IP | Intellectual Property |
| KH | Knowledge Hub |
| LEO | Low Earth Orbit |
| MPC | Multi-party computation |
| ML | Machine Learning |
| NAM | New Approach Methodologies |
| NIH | National Institutes of Health |
| NIIMB | National Institute for Innovation in Manufacturing Biopharmaceuticals |
| ODC | Orbital Data Center |
| PSCT | Personalized Stem Cell Therapy |
| RFID | Radio Frequency Identification |
| RMMS | Regenerative Medicine Manufacturing Society |
| R&D | Research and Development |
| TERM | Tissue Engineering and Regenerative Medicine |
| TEE | Trusted Execution Environments |
| UNCTAD | United Nations Trade and Development |
References
- Steeves, J.D. Chapter 11—Bench to bedside: Challenges of clinical translation. Prog. Brain Res. 2015, 218, 227–239. [Google Scholar]
- Wehling, M. Principles of Translational Science in Medicine: From Bench to Bedside; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Buxbaum, J.D.; Cohen, A.J.; Fendrick, A.M. Measures of the burden of medical expenses. JAMA 2018, 319, 1621. [Google Scholar] [CrossRef]
- de Kanter, A.J.; Jongsma, K.R.; Verhaar, M.C.; Bredenoord, A.L. The ethical implications of tissue engineering for regenerative purposes: A systematic review. Tissue Eng. Part B 2023, 29, 167–187. [Google Scholar] [CrossRef] [PubMed]
- Data Protection and Privacy Legislation Worldwide. Available online: https://unctad.org/page/data-protection-and-privacy-legislation-worldwide (accessed on 21 March 2025).
- Regulation, P. Regulation (EU) 2016/679 of the European Parliament and of the Council. Regulation (EU) 2016, 679, 2016. [Google Scholar]
- European Commission Directive (EU). 2022/2555 of the European Parliament and of the Council of 14 December 2022 on measures for a high common level of cybersecurity across the Union, amending Regulation (EU) No 910/2014 and Directive (EU) 2018/1972, and repealing Directive (EU) 2016/1148 (NIS 2 Directive). Off. J. Eur. Union 2022, 50, 80. [Google Scholar]
- Summary of the HIPAA Security Rule. Available online: https://www.hhs.gov/hipaa/for-professionals/security/laws-regulations/index.html (accessed on 18 March 2025).
- Data Protection Laws in Canada. Available online: https://www.dlapiperdataprotection.com/?c=CA (accessed on 11 August 2025).
- Beheshtizadeh, N.; Gharibshahian, M.; Pazhouhnia, Z.; Rostami, M.; Zangi, A.R.; Maleki, R.; Azar, H.K.; Zalouli, V.; Rajavand, H.; Farzin, A.; et al. Commercialization and regulation of regenerative medicine products: Promises, advances and challenges. Biomed. Pharmacother. 2022, 153, 113431. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; Zhang, Y. A Blockchain-Based Framework for Data Sharing with Fine-Grained Access Control in Decentralized Storage Systems. IEEE Access 2018, 6, 38437–38450. [Google Scholar] [CrossRef]
- Isichei, J.C.; Khorsandroo, S.; Desai, S. Cybersecurity and privacy in smart bioprinting. Bioprinting 2023, 36, e00321. [Google Scholar] [CrossRef]
- Hoenig, A.; Roy, K.; Acquaah, Y.T.; Yi, S.; Desai, S.S. Explainable AI for cyber-physical systems: Issues and challenges. IEEE Access 2024, 12, 73113–73140. [Google Scholar] [CrossRef]
- Tettey, F.; Parupelli, S.K.; Desai, S. A review of biomedical devices: Classification, regulatory guidelines, human factors, software as a medical device, and cybersecurity. Biomed. Mater. Devices 2024, 2, 316–341. [Google Scholar] [CrossRef]
- Ogunsanya, M.; Desai, S. Physics-based and data-driven modeling for biomanufacturing 4.0. Manuf. Lett. 2023, 36, 91–95. [Google Scholar] [CrossRef]
- Jarman, H.; Rozenblum, S.; Huang, T.J. Neither protective nor harmonized: The crossborder regulation of medical devices in the EU. Health Econ. Policy Law 2021, 16, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, S. Bitcoin: A Peer-to-Peer Electronic Cash System. Bitcoin 2008, 4, 15. [Google Scholar]
- Haber, S.; Stornetta, W.S. How to Time-Stamp a Digital Document. In Proceedings of the Conference on the Theory and Application of Cryptography, Fujiyoshida, Japan, 11–14 November 1991; Springer: Berlin/Heidelberg, Germany, 1991. [Google Scholar]
- Buterin, V. Ethereum whitepaper: A next-generation smart contract and decentralized application platform. White Pap. 2013, 3, 1–36. [Google Scholar]
- Petcu, A.; Pahontu, B.; Frunzete, M.; Stoichescu, D.A. A secure and decentralized authentication mechanism based on web 3.0 and ethereum blockchain technology. Appl. Sci. 2023, 13, 2231. [Google Scholar] [CrossRef]
- Tariq, U.U.; Sabrina, F.; Rashid, M.M.; Gordon, S.; Lin, Y.; Wang, Z.; Azad, S. Blockchain-Based Secured Data Sharing in Healthcare: A Systematic Literature Review. IEEE Access 2025, 13, 45415–45435. [Google Scholar] [CrossRef]
- Kshetri, N. The Rise of Blockchains: Disrupting Economies and Transforming Societies; Edward Elgar Publishing: Northampton, MA, USA, 2022. [Google Scholar]
- Jin, Y.; Zhu, H.; Xu, J.; Chen, Y. Federated Learning Fundamentals and Advances; 1 online resource; Springer: Singapore, 2023. [Google Scholar]
- Qi, P.; Chiaro, D.; Guzzo, A.; Ianni, M.; Fortino, G.; Piccialli, F. Model aggregation techniques in federated learning: A comprehensive survey. Future Gener. Comput. Syst. 2024, 150, 272–293. [Google Scholar] [CrossRef]
- Accelerating Pediatric Innovation. Available online: https://www.kidsights.com/ (accessed on 31 March 2025).
- Exploring a Federated Approach to Data Management. Available online: https://www.mayoclinicplatform.org/2023/09/14/exploring-a-federated-approach-to-data-management/ (accessed on 24 March 2025).
- Seh, A.H.; Zarour, M.; Alenezi, M.; Sarkar, A.K.; Agrawal, A.; Kumar, R.; Khan, R.A. Healthcare Data Breaches: Insights and Implications. Healthcare 2020, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Truhn, D.; Tayebi Arasteh, S.; Saldanha, O.L.; Müller-Franzes, G.; Khader, F.; Quirke, P.; West, N.P.; Gray, R.; Hutchins, G.G.A.; James, J.A.; et al. Encrypted federated learning for secure decentralized collaboration in cancer image analysis. Med. Image Anal. 2024, 92, 103059. [Google Scholar] [CrossRef]
- Su, Y.; Huang, C.; Zhu, W.; Lyu, X.; Ji, F. Multi-party Diabetes Mellitus risk prediction based on secure federated learning. Biomed. Signal Process. Control. 2023, 85, 104881. [Google Scholar] [CrossRef]
- Gharibi, G.; Gilkalaye, B.P.; Patel, R.; Rademacher, A.; Wagner, D.; Fay, J.; Moore, G.; Penrod, S.; Storm, G.; Das, R. Tripleblind: A Privacy-Preserving Framework for Decentralized Data and Algorithms. In Proceedings of the NeurIPS 2021 Competitions and Demonstrations Track, Online, 6–14 December 2021. [Google Scholar]
- Sun, X.; Zhang, P.; Sookhak, M.; Yu, J.; Xie, W. Utilizing fully homomorphic encryption to implement secure medical computation in smart cities. Pers. Ubiquitous Comput. 2017, 21, 831–839. [Google Scholar] [CrossRef]
- AMalik, N.; Ratha, B.; Yalavarthi, T.; Sharma, A.; Kaushik, C. Jutla; Confidential and Protected Disease Classifier using Fully Homomorphic Encryption. In Proceedings of the 2024 IEEE Conference on Artificial Intelligence (CAI), Singapore, 25–27 June 2024. [Google Scholar]
- Shaikh, M.U.; Adnan, W.A.W.; Ahmad, S.A. Sensitivity and Positive Prediction of Secured Electrocardiograph (ECG) Transmission using Fully Homomorphic Encryption Technique (FHE). In Proceedings of the 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Langkawi Island, Malaysia, 1–3 March 2021. [Google Scholar]
- Du, W.; Atallah, M.J. Secure Multi-Party Computation Problems and their Applications: A Review and Open Problems. In Proceedings of the 2001 Workshop on New Security Paradigms, Cloudcroft, NM, USA, 10–13 September 2001. [Google Scholar]
- Lindell, Y. Secure multiparty computation. Commun ACM 2020, 64, 86–96. [Google Scholar] [CrossRef]
- Ficek, J.; Wang, W.; Chen, H.; Dagne, G.; Daley, E. Differential privacy in health research: A scoping review. J. Am. Med. Inform. Assoc. 2021, 28, 2269–2276. [Google Scholar] [CrossRef]
- Joshi, M.; Pal, A.; Sankarasubbu, M. Federated learning for healthcare domain-pipeline, applications and challenges. ACM Trans. Comput. Healthc. 2022, 3, 1–38. [Google Scholar] [CrossRef]
- Lee, P. Reconceptualizing the role of intellectual property rights in shaping industry structure. Vand. L. Rev. 2019, 72, 1197. [Google Scholar]
- Gottschalk, U.; Brorson, K.; Shukla, A.A. The need for innovation in biomanufacturing. Nat. Biotechnol. 2012, 30, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Love, J.C.; Reynolds, E.B.; Goldston, D.; Frye, H.E. Biomanufacturing in the US: A MIT Policy Brief. 2025. Available online: https://dspace.mit.edu/bitstream/handle/1721.1/158134/Biomanufacturing%20in%20US%20MIT%20Brief.pdf (accessed on 2 June 2025).
- Zhou, Y.; Sun, F. Creating knowledge assets under biocapitalism: Analyzing China’s biomedical industry and its patent networks. Econ. Geogr. 2022, 98, 411–437. [Google Scholar] [CrossRef]
- Joshi, A.M.; Nerkar, A. When do strategic alliances inhibit innovation by firms? Evidence from patent pools in the global optical disc industry. Strateg. Manag. J. 2011, 32, 1139–1160. [Google Scholar] [CrossRef]
- Clark, J.; Piccolo, J.; Stanton, B.; Tyson, K. Patent pools: A solution to the problem of access in biotechnology patents? Biotechnol. Law Rep. 2001, 20, 607–622. [Google Scholar] [CrossRef]
- Wang, L.X. Global drug diffusion and innovation with the medicines patent pool. J. Health Econ. 2022, 85, 102671. [Google Scholar] [CrossRef]
- Galasso, A.; Schankerman, M. Licensing life-saving drugs for developing countries: Evidence from the medicines patent pool. Rev. Econ. Stat. 2024, 106, 1529–1541. [Google Scholar] [CrossRef]
- Shadlen, K.C. Accelerating pooled licensing of medicines to enhance global production and equitable access. Lancet 2022, 400, 632–634. [Google Scholar] [CrossRef]
- Panetta, J. A tale in three parts: The success of california’s life science clusters. J. Commer. Biotechnol. 2021, 26, 40–42. [Google Scholar] [CrossRef]
- Reynolds, E.B. Institutions, Public Policy and the Product Life Cycle: The Globalization of Biomanufacturing and Implications for Massachusetts. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 2010. [Google Scholar]
- Stower, C. Strengthening the US Biomanufacturing Sector Through Standardization. Ind. Biotechnol. 2023, 19, 201–203. [Google Scholar]
- Raab, R.; Küderle, A.; Zakreuskaya, A.; Stern, A.D.; Klucken, J.; Kaissis, G.; Rueckert, D.; Boll, S.; Eils, R.; Wagener, H.; et al. Federated electronic health records for the European Health Data Space. Lancet Digit. Health 2023, 5, e840–e847. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Humayun, M.S.; Huang, S.S. Future Vision Forum Faculty Gaps and future of human-centered artificial intelligence in ophthalmology: Future Vision Forum consensus statement. Curr. Opin. Ophthalmol. 2023, 34, 431–436. [Google Scholar] [CrossRef]
- Wang, Y.; Maes, E.M.; Heinle, L.; Ruterbories, K.; Doktor, S.; Larsen, M.; Olson, A.; Lee, A.; Van Handel, C.; Ji, Q.C.; et al. Integrity and efficiency: AbbVie’s journey of building an integrated nonregulated bioanalytical laboratory. Bioanalysis 2023, 15, 161–176. [Google Scholar] [CrossRef]
- Liu, J.C.; Goetz, J.; Sen, S.; Tewari, A. Learning From Others Without Sacrificing Privacy: Simulation Comparing Centralized and Federated Machine Learning on Mobile Health Data. JMIR Mhealth Uhealth 2021, 9, e23728. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.X.; Ran, A.R.; Hu, X.; Yang, D.; Jiang, M.; Dou, Q.; Cheung, C.Y.; Palm, C. Federated Learning in Ocular Imaging: Current Progress and Future Direction. Diagnostics 2022, 12, 2835. [Google Scholar] [CrossRef]
- Final NIH Policy for Data Management and Sharing. Available online: https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html (accessed on 18 March 2025).
- Roadmap to Reducing Animal Testing in Preclinical Safety Studies. Available online: https://www.fda.gov/media/186092/download?attachment (accessed on 18 June 2025).
- ICE: Integrated Chemical Environment. Available online: https://ntp.niehs.nih.gov/whatwestudy/niceatm/comptox/ct-ice/ice (accessed on 18 June 2025).
- Badwan, B.A.; Liaropoulos, G.; Kyrodimos, E.; Skaltsas, D.; Tsirigos, A.; Gorgoulis, V.G. Machine learning approaches to predict drug efficacy and toxicity in oncology. Cells Rep. Methods 2023, 3, 100413. [Google Scholar] [CrossRef]
- Huang, R. A quantitative high-throughput screening data analysis pipeline for activity profiling. In High-Throughput Screening Assays in Toxicology; Springer: New York, NY, USA, 2016. [Google Scholar]
- Hunsberger, J.G.; Pandya, P.; Mulligan, M.K.; Marotta, D.; Moroni, L.; Shusteff, M.; Brogan, G.; Brovold, M.; Yoo, J.; Koffler, J. Review of Disruptive Technologies in 3D Bioprinting. Curr. Stem Cell Rep. 2025, 11, 5. [Google Scholar] [CrossRef]
- Adarkwa, E.; Roy, A.; Ohodnicki, J.; Lee, B.; Kumta, P.N.; Desai, S. 3D printing of drug-eluting bioactive multifunctional coatings for orthopedic applications. Int. J. Bioprint. 2023, 9, 661. [Google Scholar] [CrossRef] [PubMed]
- Nandipati, M.; Fatoki, O.; Desai, S. Bridging nanomanufacturing and artificial intelligence—A comprehensive review. Materials 2024, 17, 1621. [Google Scholar] [CrossRef] [PubMed]
- Tettey-Engmann, F.; Parupelli, S.K.; Bauer, S.R.; Bhattarai, N.; Desai, S. Advances in Artificial Intelligence-Based Medical Devices for Healthcare Applications. Biomed. Mater. Devices 2025, 1–21. [Google Scholar] [CrossRef]
- Considerations for the Development of Chimeric Antigen Receptor (CAR) T Cell Products. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/considerations-development-chimeric-antigen-receptor-car-t-cell-products (accessed on 18 August 2025).
- What Is ISBT 128? Available online: https://www.isbt128.org/isbt-128-basics (accessed on 18 August 2025).
- EudraLex The Rules Governing Medicinal Products in the European Union. Available online: https://health.ec.europa.eu/system/files/2016-11/annex11_01-2011_en_0.pdf (accessed on 18 August 2025).
- ISPE. GAMP® 5: A Risk-Based Approach to Compliant GxP Computerized Systems, 2nd ed.; International Society for Pharmaceutical Engineering: North Bethesda, MD, USA, 2022. [Google Scholar]
- PART 1271—Human Cells, Tissues, and Cellular and Tissue-Based Products. Available online: https://www.ecfr.gov/current/title-21/chapter-I/subchapter-L/part-1271 (accessed on 18 August 2025).
- Chemistry, Manufacturing, and Control (CMC) Information for Human Gene Therapy Investigational New Drug Applications (INDs). Available online: https://www.fda.gov/media/113760/download (accessed on 18 August 2025).
- Vivian, J.C. Medication Errors and Liability Issues. pp. 43–46. Available online: https://www.uspharmacist.com/article/medication-errors-and-liability-issues#:~:text=In%20the%20U.S.%2C%20medication%20errors,result%20of%20a%20medication%20error.&text=Reporting%20of%20medication%20errors%20in,annually%20related%20to%20medication%20errors.&text=Forty%2Done%20percent%20of%20Americans,hospital%2C%20and%20other%20pharmacy%20settings (accessed on 27 June 2025).
- Axiom Space to Launch Orbital Data Center Nodes to Support National Security, Commercial, International Customers. Available online: https://www.axiomspace.com/release/axiom-space-to-launch-orbital-data-center-nodes-to-support-national-security-commercial-international-customers (accessed on 11 April 2025).
- Zhang, E.; Chai, J.; Ye, R.; Wang, Y.; Chen, S. Incentivizing Inclusive Contributions in Model Sharing Markets. arXiv 2025, arXiv:2505.02462. [Google Scholar] [CrossRef]
- Moonen, J.; Zomer, T.; Van der Veen, A.; Ploeg, S. Sustainable Revenue Models for Data Sharing Initiatives; Jheronimus Academy of Data Science: DA’s-Hertogenbosch, The Netherlands, 2025. Available online: https://www.government.nl/documents/reports/2025/04/22/sustainable-revenue-models-for-data-sharing-initiatives (accessed on 27 June 2025).
- Semiconductor Manufacturing in Low-Earth Orbit for Terrestrial Use. Available online: https://osf.io/preprints/osf/d6ar4_v1 (accessed on 8 August 2025).
- In Space Production: Applications Within Reach. Available online: https://www.nasa.gov/missions/station/applications-within-reach/ (accessed on 8 August 2023).
- High-Performance Data Monetization. Available online: https://cisr.mit.edu/publication/2024_1101_HighPerformanceDataMonetization_WixomBeathDuane (accessed on 29 June 2025).
- de Laage, R.; Yuhala, P.; Wicht, F.; Felber, P.; Cachin, C.; Schiavoni, V. Practical Secure Aggregation by Combining Cryptography and Trusted Execution Environments. In Proceedings of the 19th ACM International Conference on Distributed and Event-Based Systems, Gothenburg, Sweden, 10–13 June 2025. [Google Scholar]
- Li, X.; Zhao, B.; Yang, G.; Xiang, T.; Weng, J.; Deng, R.H. A survey of secure computation using trusted execution environments. arXiv 2023, arXiv:2302.12150. [Google Scholar] [CrossRef]

| Region | Regulations | Focus Areas |
|---|---|---|
| European Union | GDPR (General Data Protection Regulation) [6] | Classifies healthcare data as sensitive data with additional protection rules |
| NIS2 Directive (effective since 2024) [7] | Aims to reduce cybersecurity risks for healthcare institutions | |
| United States | HIPAA (Health Insurance Portability and Accountability Act of 1996) [8] | Regulates use and disclosure of patients’ health information |
| HITECH Act (2009) [8] | Promotes the adoption and meaningful use of health information technology | |
| HIPAA Final Omnibus Rule (2013) [8] | Strengthens privacy and security protections for health information | |
| Canada | PIPEDA [9] | Complex, multi-jurisdictional framework with 29 + privacy statutes Governs federal and interprovincial data privacy |
| Feature | Current Status |
|---|---|
| Consortium Membership | Gillette Children’s (U.S.) and Holland Bloorview (Canada) |
| Scale of Use | Two leading pediatric specialty institutions |
| Outputs & Publications | No publicly disclosed publications or validated outputs yet |
| Data Access & Use | Aggregated de-identified EMR data available to researchers and innovators |
| Privacy & Security | Compliance with PHIPA, PIPEDA, HIPAA; advanced de-identification |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izadifar, Z.; Storm, G.; Joshi, A.M.; Hochberg, A.; Hadjisavas, M.; Rodrigue, G.; Bauer, S.R.; Schmidt, J.B.; Somara, S.; Atala, A.; et al. A Collaborative Data Sharing Platform to Accelerate Translation of Biomedical Innovations. Bioengineering 2025, 12, 938. https://doi.org/10.3390/bioengineering12090938
Izadifar Z, Storm G, Joshi AM, Hochberg A, Hadjisavas M, Rodrigue G, Bauer SR, Schmidt JB, Somara S, Atala A, et al. A Collaborative Data Sharing Platform to Accelerate Translation of Biomedical Innovations. Bioengineering. 2025; 12(9):938. https://doi.org/10.3390/bioengineering12090938
Chicago/Turabian StyleIzadifar, Zohreh, Greg Storm, Amol M. Joshi, Anna Hochberg, Michael Hadjisavas, Gary Rodrigue, Steven R. Bauer, James B. Schmidt, Sita Somara, Anthony Atala, and et al. 2025. "A Collaborative Data Sharing Platform to Accelerate Translation of Biomedical Innovations" Bioengineering 12, no. 9: 938. https://doi.org/10.3390/bioengineering12090938
APA StyleIzadifar, Z., Storm, G., Joshi, A. M., Hochberg, A., Hadjisavas, M., Rodrigue, G., Bauer, S. R., Schmidt, J. B., Somara, S., Atala, A., Heyward, I., Desai, S., & Hunsberger, J. (2025). A Collaborative Data Sharing Platform to Accelerate Translation of Biomedical Innovations. Bioengineering, 12(9), 938. https://doi.org/10.3390/bioengineering12090938








