Deep Ensemble Learning for Multiclass Skin Lesion Classification
Abstract
1. Introduction
- Class imbalance: Many studies highlight the uneven distribution of skin lesion categories in datasets, which can lead to biased model performance.
- Lack of standardized evaluation metrics: There is a notable inconsistency in the metrics used across studies, making it difficult to compare results and assess model reliability.
- Use of ensemble techniques: Ensemble learning has gained popularity for improving classification accuracy and robustness, especially in multiclass tasks.
- Need for explainable AI: The black-box nature of deep learning models limits clinical adoption, prompting calls for interpretable and transparent systems.
- Data diversity and augmentation: To address data scarcity and improve generalization, researchers emphasize the importance of diverse datasets and augmentation strategies.
- Integration of clinical metadata: Future directions suggest combining imaging data with patient history to enhance diagnostic precision and clinical relevance.
- Examine the impact of removal of the background image on the performance of skin lesion classification.
- Explore strategies for addressing class imbalance in dermatological image datasets.
- Investigate the design of a multi-architecture ensemble model for the classification of multiple skin lesion types.
| Reference | Years | Papers | Conclusions | Future Directions |
|---|---|---|---|---|
| [25] | 2000~2022 | 91 |
|
|
| [26] | 2015~2023 | 153 |
|
|
| [27] | 2017~2023 | 89 |
|
|
| [28] | 2007~2023 | 37 |
|
|
| [29] | 2001~2023 | 632 |
|
|
| [30] | 1997~2024 | 74 |
|
|
| [31] | 2017~2024 | 143 |
|
|
2. Materials and Methods
2.1. CSMUH Dataset
2.2. Research Framework
- Accuracy: The proportion of correctly classified samples among all samples, as defined in Equation (1).
- Sensitivity (Recall): The ability of the model to correctly identify positive cases, as defined in Equation (2).
- Specificity: The ability of the model to correctly identify negative cases, as defined in Equation (3).
- Precision (PPV): The proportion of true positives among all predicted positives, as defined in Equation (4).
- Negative predictive value (NPV): The proportion of predicted negatives that are truly negative, as defined in Equation (5).
- False positive rate (FPR): The rates of incorrect predictions in positive classes, as defined in Equation (6).
- False negative rate (FNR): The rates of incorrect predictions in negative classes, as defined in Equation (7).
- F1 Score: The harmonic mean of precision and recall, providing a balanced measure of model performance, as defined in Equation (8).
2.3. Data Preprocessing
2.4. Transfer Learning
2.5. Ensemble Architecture
2.6. Model Architecture
3. Results
3.1. Effect of Removing a Dermoscopic Scale
3.2. Effect of Different Pretrained Models
3.3. Effect of Hard Voting and Soft Voting Ensemble Methods
3.4. Effect of Different Ensemble Weights in Soft Voting Methods
3.5. Performance Indices of the Best Model
4. Discussion
- Multi-architecture ensemble model design: This research introduces a deep learning ensemble framework that integrates Swin Transformer, Vision Transformer (ViT), and EfficientNetB4. By employing a soft voting strategy, the model effectively combines the strengths of these three architectures, achieving a test accuracy of 98.5% in multiclass skin lesion classification—significantly outperforming individual models.
- Optimization for multiclass classification tasks: While most prior studies have focused on binary classification of melanoma, this study addresses the classification of seven distinct skin lesion types, thereby enhancing the breadth and practical relevance of clinical applications.
- Strong generalization capability: The proposed architecture is not tailored to a specific dataset, demonstrating robust generalizability across different datasets and clinical environments.
- Class imbalance handling strategy: To address the uneven class distribution in the CSMUH dataset, the study employs both static and dynamic data augmentation techniques, effectively improving the model’s ability to recognize minority classes.
- Comprehensive performance evaluation: Beyond accuracy, the study reports multiple evaluation metrics including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and F1 score, underscoring the model’s stability and reliability in clinical settings.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Foudation, S.C. Skin Cancer Facts & Statistics. Available online: https://www.skincancer.org/skin-cancer-information/skin-cancer-facts/ (accessed on 1 January 2024).
- Majtner, T.; Yildirim-Yayilgan, S.; Hardeberg, J.Y. Combining deep learning and hand-crafted features for skin lesion classification. In Proceedings of the 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA), Oulu, Finland, 12–15 December 2016; pp. 1–6. [Google Scholar]
- Nasr-Esfahani, E.; Samavi, S.; Karimi, N.; Soroushmehr, S.M.; Jafari, M.H.; Ward, K.; Najarian, K. Melanoma detection by analysis of clinical images using convolutional neural network. In Proceedings of the 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1373–1376. [Google Scholar]
- Çevik, E.; Zengin, K. Classification of Skin Lesions in Dermatoscopic Images with Deep Convolution Network. Eur. J. Sci. Technol. 2019, 309–318. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef]
- Mobiny, A.; Singh, A.; Van Nguyen, H. Risk-aware machine learning classifier for skin lesion diagnosis. J. Clin. Med. 2019, 8, 1241. [Google Scholar] [CrossRef] [PubMed]
- Lucius, M.; De All, J.; De All, J.A.; Belvisi, M.; Radizza, L.; Lanfranconi, M.; Lorenzatti, V.; Galmarini, C.M. Deep neural frameworks improve the accuracy of general practitioners in the classification of pigmented skin lesions. Diagnostics 2020, 10, 969. [Google Scholar] [CrossRef] [PubMed]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Hoang, L.; Lee, S.-H.; Lee, E.-J.; Kwon, K.-R. Multiclass skin lesion classification using a novel lightweight deep learning framework for smart healthcare. Appl. Sci. 2022, 12, 2677. [Google Scholar] [CrossRef]
- Gururaj, H.L.; Manju, N.; Nagarjun, A.; Aradhya, V.M.; Flammini, F. DeepSkin: A deep learning approach for skin cancer classification. IEEE Access 2023, 11, 50205–50214. [Google Scholar] [CrossRef]
- Alwakid, G.; Gouda, W.; Humayun, M.; Jhanjhi, N. Diagnosing melanomas in Dermoscopy images using deep learning. Diagnostics 2023, 13, 1815. [Google Scholar] [CrossRef]
- Chatterjee, S.; Gil, J.-M.; Byun, Y.-C. Early Detection of Multiclass Skin Lesions Using Transfer Learning-based IncepX-Ensemble Model. IEEE Access 2024, 12, 113677–113693. [Google Scholar] [CrossRef]
- Musthafa, M.M.; Mahesh, T.R.; Vinoth Kumar, V.; Suresh, G. Enhanced skin cancer diagnosis using optimized CNN architecture and checkpoints for automated dermatological lesion classification. BMC Med. Imaging 2024, 24, 201. [Google Scholar] [CrossRef]
- Kumar Lilhore, U.; Simaiya, S.; Sharma, Y.K.; Kaswan, K.S.; Rao, K.B.; Rao, V.M.; Baliyan, A.; Bijalwan, A.; Alroobaea, R. A precise model for skin cancer diagnosis using hybrid U-Net and improved MobileNet-V3 with hyperparameters optimization. Sci. Rep. 2024, 14, 4299. [Google Scholar] [CrossRef]
- Asaduzzaman, A.; Thompson, C.C.; Uddin, M.J. Machine Learning Approaches for Skin Neoplasm Diagnosis. ACS Omega 2024, 9, 32853–32863. [Google Scholar] [CrossRef]
- Islam, M.S.; Panta, S. Skin cancer images classification using transfer learning techniques. arXiv 2024, arXiv:2406.12954. [Google Scholar] [CrossRef]
- Suleiman, T.A.; Anyimadu, D.T.; Permana, A.D.; Ngim, H.A.A.; Scotto di Freca, A. Two-step hierarchical binary classification of cancerous skin lesions using transfer learning and the random forest algorithm. Vis. Comput. Ind. Biomed. Art 2024, 7, 15. [Google Scholar] [CrossRef]
- Chiu, T.-M.; Li, Y.-C.; Chi, I.-C.; Tseng, M.-H. AI-Driven Enhancement of Skin Cancer Diagnosis: A Two-Stage Voting Ensemble Approach Using Dermoscopic Data. Cancers 2025, 17, 137. [Google Scholar] [CrossRef]
- Ray, S. Disease Classification within Dermascopic Images Using Features Extracted by ResNet50 and Classification through Deep Forest. arXiv 2018, arXiv:1807.05711. [Google Scholar] [CrossRef]
- Milton, M.A.A. Automated Skin Lesion Classification Using Ensemble of Deep Neural Networks in ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection Challenge. arXiv 2019, arXiv:1901.10802. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef]
- Thurnhofer-Hemsi, K.; López-Rubio, E.; Dominguez, E.; Elizondo, D.A. Skin lesion classification by ensembles of deep convolutional networks and regularly spaced shifting. IEEE Access 2021, 9, 112193–112205. [Google Scholar] [CrossRef]
- Alshawi, S.A.; Musawi, G.F.K.A. Skin cancer image detection and classification by cnn based ensemble learning. Int. J. Adv. Comput. Sci. Appl. 2023, 14. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Jeong, H.K.; Park, C.; Henao, R.; Kheterpal, M. Deep learning in dermatology: A systematic review of current approaches, outcomes, and limitations. JID Innov. 2023, 3, 100150. [Google Scholar] [CrossRef]
- Zhang, J.; Zhong, F.; He, K.; Ji, M.; Li, S.; Li, C. Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review. Diagnostics 2023, 13, 3506. [Google Scholar] [CrossRef]
- Debelee, T.G. Skin Lesion Classification and Detection Using Machine Learning Techniques: A Systematic Review. Diagnostics 2023, 13, 3147. [Google Scholar] [CrossRef]
- Choy, S.P.; Kim, B.J.; Paolino, A.; Tan, W.R.; Lim, S.M.L.; Seo, J.; Tan, S.P.; Francis, L.; Tsakok, T.; Simpson, M. Systematic review of deep learning image analyses for the diagnosis and monitoring of skin disease. npj Digit. Med. 2023, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.K.; Ahamad, M.A.; Yap, C.H.; Yang, G. A survey, review, and future trends of skin lesion segmentation and classification. Comput. Biol. Med. 2023, 155, 106624. [Google Scholar] [CrossRef] [PubMed]
- Vayadande, K. Innovative approaches for skin disease identification in machine learning: A comprehensive study. Oral Oncol. Rep. 2024, 10, 100365. [Google Scholar] [CrossRef]
- Meedeniya, D.; De Silva, S.; Gamage, L.; Isuranga, U. Skin cancer identification utilizing deep learning: A survey. IET Image Process. 2024, 18, 3731–3749. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Zhou, Z.-H. Ensemble Methods: Foundations and Algorithms; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Taşar, B. SkinCancerNet: Automated Classification of Skin Lesion Using Deep Transfer Learning Method. Trait. Du Signal 2023, 40, 285. [Google Scholar] [CrossRef]
- Liu, X.; Yu, Z.; Tan, L.; Yan, Y.; Shi, G. Enhancing skin lesion diagnosis with ensemble learning. In Proceedings of the 2024 4th International Conference on Computer Science and Blockchain (CCSB), Shenzhen, China, 6–8 September 2024; pp. 94–98. [Google Scholar]
- Thwin, S.M.; Park, H.-S. Skin Lesion Classification Using a Deep Ensemble Model. Appl. Sci. 2024, 14, 5599. [Google Scholar] [CrossRef]

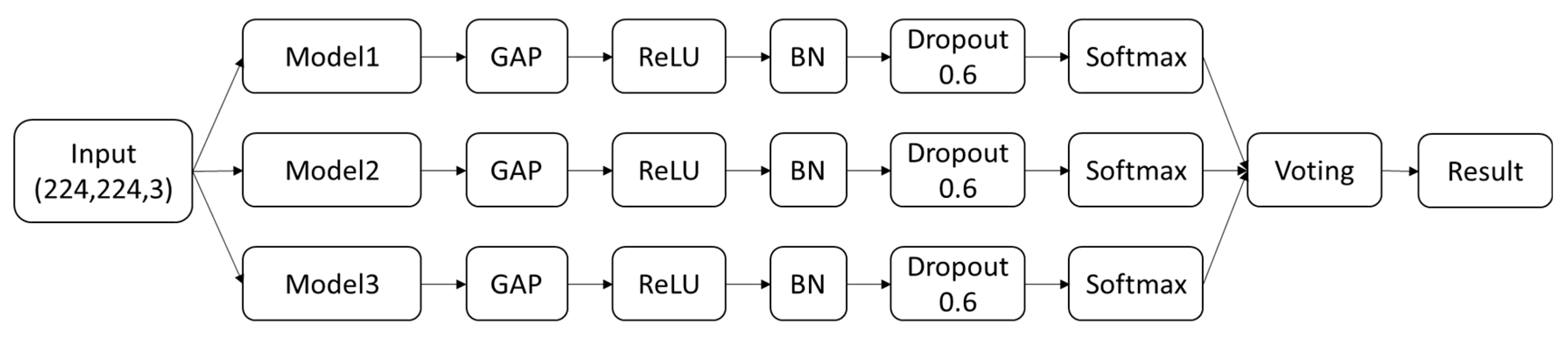
| Class | Num | Label | Image |
|---|---|---|---|
| Malignant Melanoma | 111 | 0 |  |
| Melanocytic Nevus | 199 | 1 | 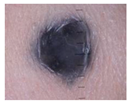 |
| Basal cell carcinoma | 45 | 2 |  |
| Actinic keratosis | 82 | 3 | 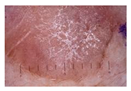 |
| Benign keratosis | 134 | 4 | 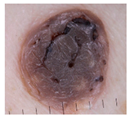 |
| Dermatofibroma | 42 | 5 | 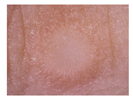 |
| Vascular | 53 | 6 | 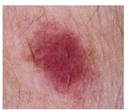 |
| Dataset | Example | Model | Train ACC | Test ACC |
|---|---|---|---|---|
| Original | 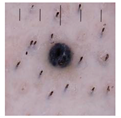 | Swin base Transformer | 0.991 ± 0.005 | 0.818 ± 0.046 |
| cropped | 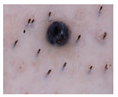 | 0.992 ± 0.005 | 0.845 ± 0.027 |
| Architecture | No. of Models | Model | Train ACC | Test ACC |
|---|---|---|---|---|
| CNN | 18 | EfficientNetB1 | 0.993 ± 0.004 | 0.793 ± 0.050 |
| EfficientNetB2 | 0.960 ± 0.009 | 0.748 ± 0.015 | ||
| EfficientNetB3 | 0.989 ± 0.007 | 0.815 ± 0.027 | ||
| EfficientNetB4 | 0.993 ± 0.004 | 0.816 ± 0.016 | ||
| EfficientNetB5 | 0.991 ± 0.005 | 0.800 ± 0.021 | ||
| EfficientNetV2B2 | 0.965 ± 0.028 | 0.797 ± 0.036 | ||
| EfficientNetV2B3 | 0.985 ± 0.007 | 0.806 ± 0.015 | ||
| MobileNetV2 | 0.947 ± 0.020 | 0.749 ± 0.033 | ||
| DenseNet121 | 0.945 ± 0.038 | 0.736 ± 0.069 | ||
| DenseNet169 | 0.941 ± 0.058 | 0.748 ± 0.038 | ||
| DenseNet201 | 0.979 ± 0.016 | 0.785 ± 0.026 | ||
| VGGNet16 | 0.917 ± 0.038 | 0.685 ± 0.055 | ||
| Xception | 0.981 ± 0.007 | 0.778 ± 0.034 | ||
| ResNet50 | 0.964 ± 0.017 | 0.787 ± 0.025 | ||
| InceptionV3 | 0.918 ± 0.049 | 0.733 ± 0.049 | ||
| InceptionResnetV2 | 0.973 ± 0.022 | 0.769 ± 0.051 | ||
| ConvNeXtSmall | 0.947 ± 0.024 | 0.785 ± 0.042 | ||
| ConvNeXtBase | 0.981 ± 0.023 | 0.806 ± 0.038 | ||
| ViT | 7 | Vision b16 Transformer | 0.995 ± 0.003 | 0.823 ± 0.022 |
| Vision b32 Transformer | 0.996 ± 0.004 | 0.824 ± 0.032 | ||
| Vision l16 Transformer | 0.995 ± 0.003 | 0.840 ± 0.023 | ||
| Vision l32 Transformer | 0.997 ± 0.005 | 0.840 ± 0.012 | ||
| Swin small Transformer | 0.987 ± 0.006 | 0.824 ± 0.023 | ||
| Swin base Transformer | 0.992 ± 0.005 | 0.845 ± 0.027 | ||
| Swin large Transformer | 0.991 ± 0.006 | 0.849 ± 0.022 |
| Voting Method | Architecture | Model | Train ACC | Test ACC |
|---|---|---|---|---|
| Hard voting | CNN | EfficientNetB3-EfficientNetB4-EfficientNetV2B3 | 0.999 ± 0.001 | 0.948 ± 0.017 |
| ViT | ViTb16-ViTl16-ViTl32 | 1.000 ± 0.000 | 0.954 ± 0.013 | |
| SwinLarge-SwinBase-SwinSmall | 0.995 ± 0.005 | 0.903 ± 0.022 | ||
| Ensemble | Swin-ViT-EfficientNetB4 | 0.999 ± 0.001 | 0.954 ± 0.020 | |
| Soft voting | CNN | EfficientNetB3-EfficientNetB4-EfficientNetV2B3 | 0.999 ± 0.001 | 0.952 ± 0.010 |
| ViT | ViTb16-ViTl16-ViTl32 | 1.000 ± 0.000 | 0.955 ± 0.013 | |
| SwinLarge-SwinBase-SwinSmall | 0.995 ± 0.005 | 0.921 ± 0.026 | ||
| Ensemble | Swin-ViT-EfficientNetB4 | 0.999 ± 0.001 | 0.961 ± 0.020 |
| Ensemble Weight (EfficientNetB4, ViT, Swin Base) | Train ACC | Test ACC |
|---|---|---|
| (1, 0, 0) | 0.993 ± 0.004 | 0.816 ± 0.016 |
| (0, 1, 0) | 0.995 ± 0.003 | 0.840 ± 0.022 |
| (0, 0, 1) | 0.982 ± 0.018 | 0.824 ± 0.048 |
| (1, 1, 0) | 0.999 ± 0.002 | 0.952 ± 0.020 |
| (1, 0, 1) | 0.995 ± 0.009 | 0.937 ± 0.030 |
| (0, 1, 1) | 0.997 ± 0.004 | 0.954 ± 0.020 |
| (1, 1, 1) | 0.999 ± 0.001 | 0.961 ± 0.020 |
| Predict | ||||||||
|---|---|---|---|---|---|---|---|---|
| True | label | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| 0 | 89 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 0 | 159 | 0 | 0 | 0 | 0 | 0 | |
| 2 | 0 | 0 | 36 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 107 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 0 | 0 | 34 | 0 | |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 42 | |
| Predict | ||||||||
|---|---|---|---|---|---|---|---|---|
| True | label | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | |
| 2 | 0 | 1 | 8 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 26 | 0 | 0 | |
| 5 | 0 | 1 | 0 | 0 | 0 | 7 | 0 | |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | |
| Class | Sensitivity | Specificity | NPV | PPV | FPR | FNR | F1 Score |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| 1 | 1 | 0.978 | 1 | 0.952 | 0.022 | 0 | 0.976 |
| 2 | 0.889 | 1 | 0.992 | 1 | 0 | 0.111 | 0.941 |
| 3 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| 4 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| 5 | 0.875 | 1 | 0.992 | 1 | 0 | 0.125 | 0.933 |
| 6 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| Author, Year | Method | Validation | Dataset | Class | Test ACC |
|---|---|---|---|---|---|
| [37], 2023 | DenseNet121 | Holdout (8:2) full: 666 | CSMUH | 7 | 0.851 |
| VGGNet16 | 0.731 | ||||
| ResNet50 | 0.784 | ||||
| MobileNet | 0.813 | ||||
| Xception | 0.801 | ||||
| this study, 2025 | Ensemble (vote) (Swin, ViT, EfficientNetB4) | Holdout (8:2) full: 666 | CSMUH | 7 | 0.985 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, T.-M.; Chi, I.-C.; Li, Y.-C.; Tseng, M.-H. Deep Ensemble Learning for Multiclass Skin Lesion Classification. Bioengineering 2025, 12, 934. https://doi.org/10.3390/bioengineering12090934
Chiu T-M, Chi I-C, Li Y-C, Tseng M-H. Deep Ensemble Learning for Multiclass Skin Lesion Classification. Bioengineering. 2025; 12(9):934. https://doi.org/10.3390/bioengineering12090934
Chicago/Turabian StyleChiu, Tsu-Man, I-Chun Chi, Yun-Chang Li, and Ming-Hseng Tseng. 2025. "Deep Ensemble Learning for Multiclass Skin Lesion Classification" Bioengineering 12, no. 9: 934. https://doi.org/10.3390/bioengineering12090934
APA StyleChiu, T.-M., Chi, I.-C., Li, Y.-C., & Tseng, M.-H. (2025). Deep Ensemble Learning for Multiclass Skin Lesion Classification. Bioengineering, 12(9), 934. https://doi.org/10.3390/bioengineering12090934








