An Implicit Registration Framework Integrating Kolmogorov–Arnold Networks with Velocity Regularization for Image-Guided Radiation Therapy
Abstract
1. Introduction
- We propose the first KAN-based implicit neural representation of the velocity field for CT–CBCT diffeomorphism registration.
- The KAN network is use to encode velocity information into a compact representation, followed by inverse principal component analysis reconstruction, significantly improving registration efficiency.
- The proposed approach is validated on a paired CT–CBCT public dataset from 19 pelvic patients, excellent registration accuracy was achieved.
2. Method
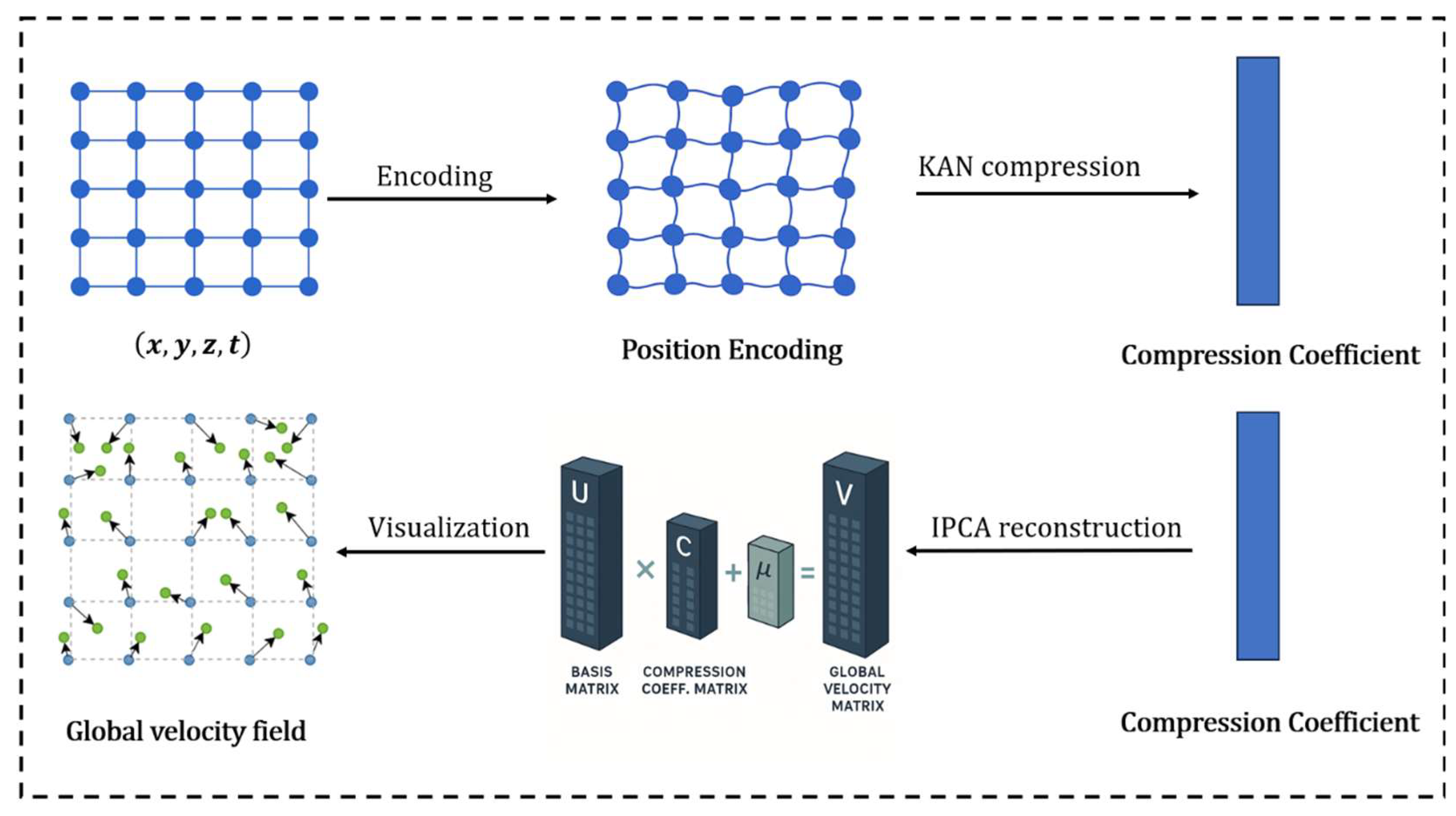
2.1. Method Overview
2.2. Deformation Based on Implicit Representation
2.3. KAN Network Modeling
2.4. Inverse PCA for Reconstructing the Velocity Field
2.4.1. Motivation
2.4.2. Mathematical Derivation
2.5. Coarse-to-Fine Strategy
3. Experiment
3.1. Dataset
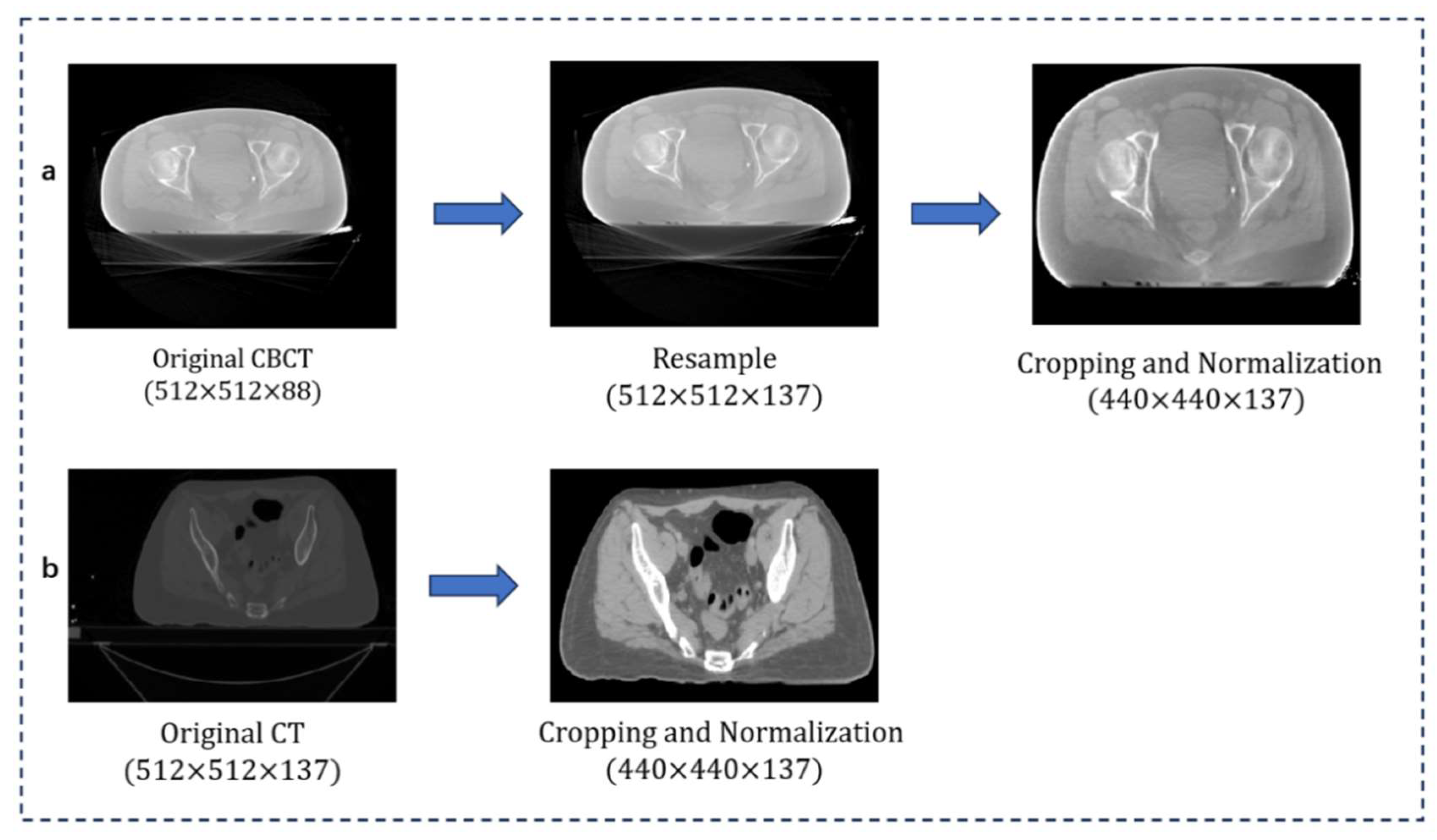
3.2. Image Preprocessing
3.3. Organ Contour Delineation
3.4. Evaluation
3.4.1. Efficiency of Inverse PCA
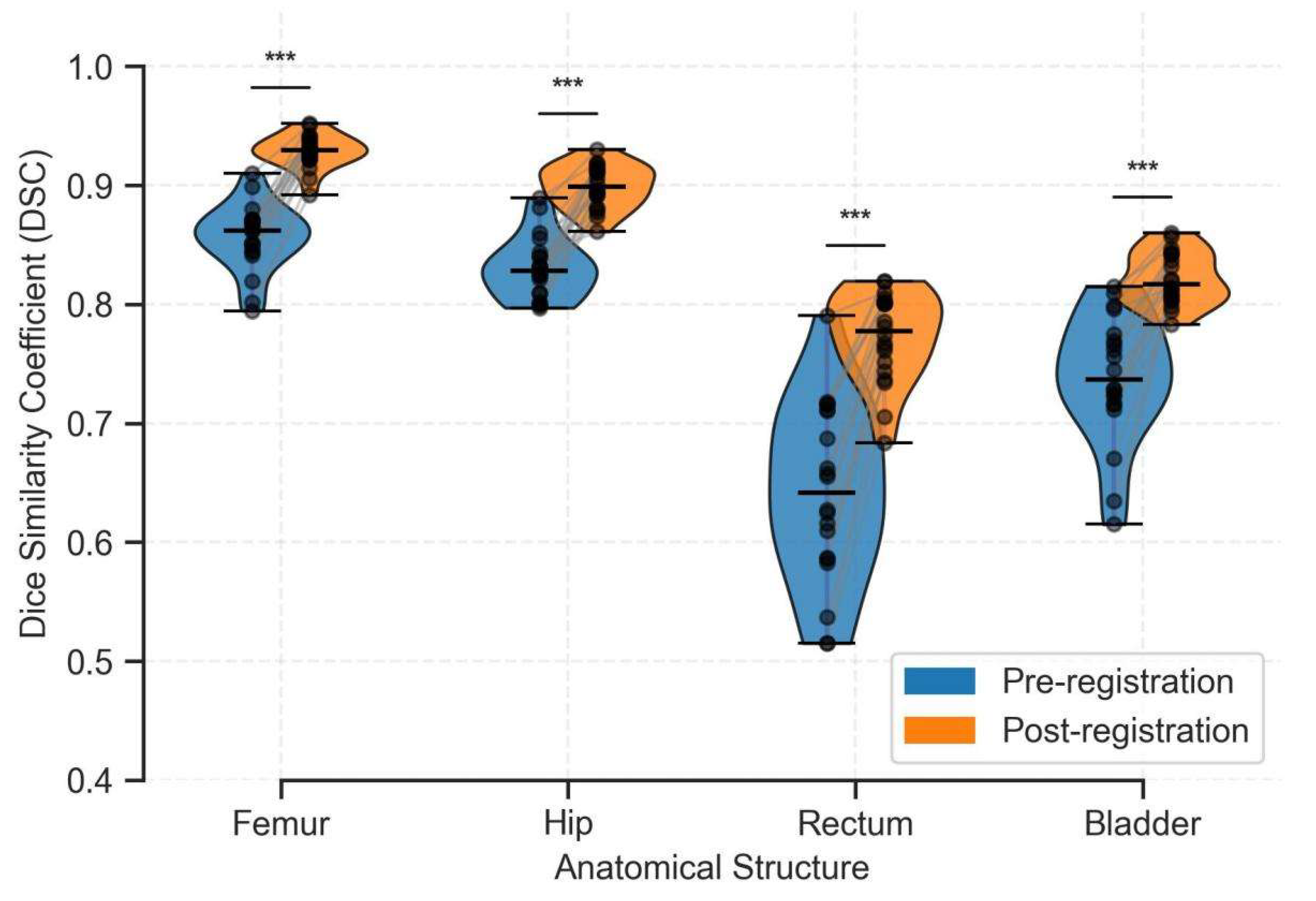
3.4.2. Accuracy Evaluation of Registration
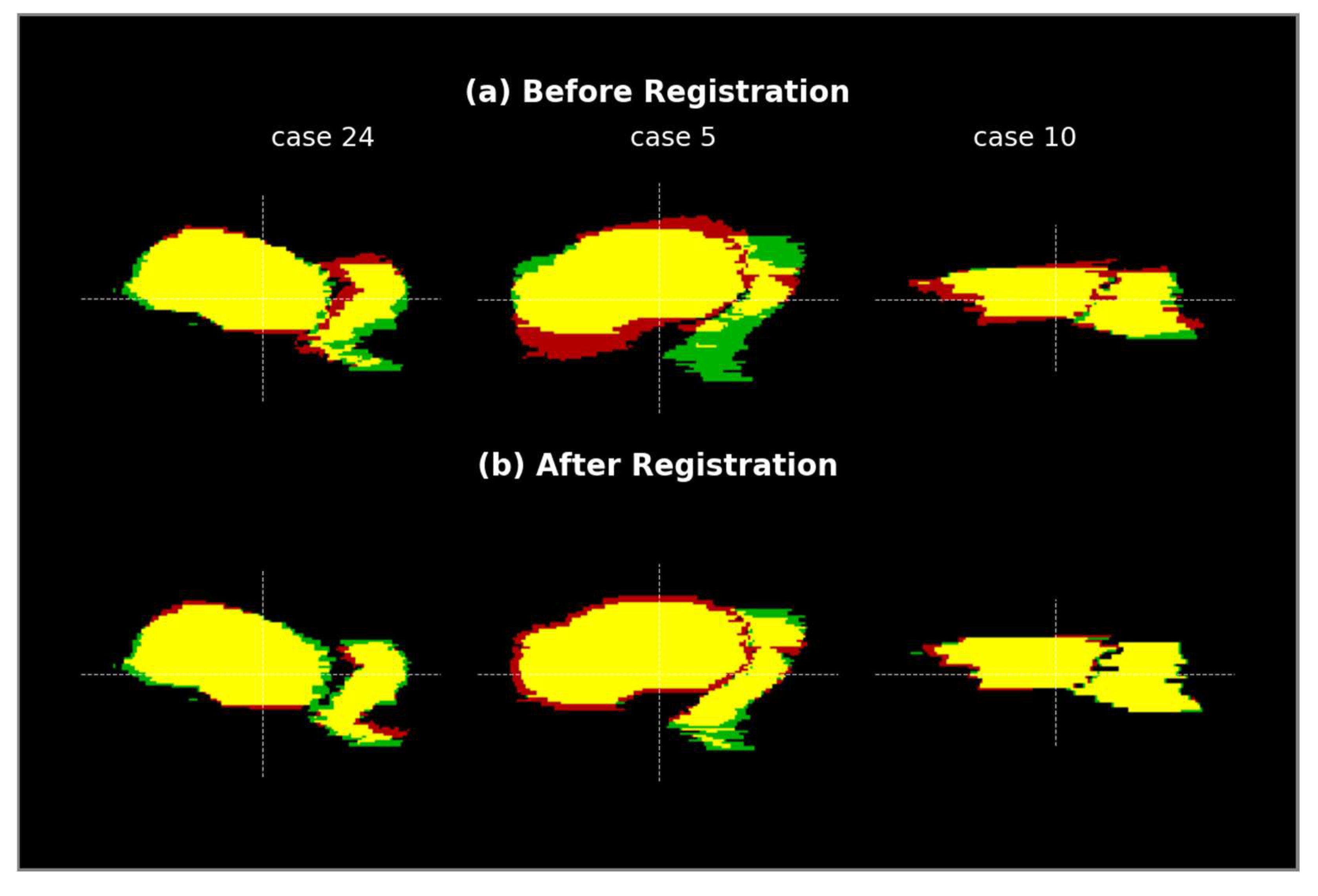
3.4.3. Quality of Deformation Field
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | computed tomography |
| CBCT | cone beam computed tomography |
| KAN | Kolmogorov–Arnold Network |
| PCA | principal component analysis |
| pCT | planning computed tomography |
| IGRT | image-guided radiation therapy |
| INR | implicit neural representation |
| 3D | three-dimensional |
| MLP | multilayer perceptron |
| DSC | Dice Similarity Coefficient |
| HD | Hausdorff Distance |
References
- Olanloye, E.E.; Aarthi Ramlaul Ntekim, A.I.; Adeyemi, S.S. FDG-PET/CT and MR imaging for target volume delineation in rectal cancer radiotherapy treatment planning: A systematic review. J. Radiother. Pract. 2021, 21, 529–539. [Google Scholar] [CrossRef]
- Ding, G.X.; Duggan, D.M.; Coffey, C.W. Characteristics of kilovoltage x-ray beams used for cone-beam computed tomography in radiation therapy. Phys. Med. Biol. 2007, 52, 1595–1615. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Liu, Y.; Wang, T.; Curran, W.J.; Liu, T.; Patel, P.; Yang, X. Cone-beam Computed Tomography (CBCT) and CT image registration aided by CBCT-based synthetic CT. In Medical Imaging 2022: Image Processing; SPIE: Bellingham, WA, USA, 2020; Volume 11313, pp. 721–727. [Google Scholar] [CrossRef]
- Liang, X.; Chen, L.; Nguyen, D.; Zhou, Z.; Gu, X.; Yang, M.; Wang, J.; Jiang, S. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys. Med. Biol. 2019, 64, 125002. [Google Scholar] [CrossRef]
- Thirion, J.P. Image matching as a diffusion process: An analogy with Maxwell’s demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef]
- Hellier, P.; Barillot, C.; Mémin, E.; Pérez, P. Hierarchical estimation of a dense deformation field for 3-D robust registration. IEEE Trans. Med. Imaging 2001, 20, 388–402. [Google Scholar] [CrossRef]
- Bajcsy, R.; Kovačič, S. Multiresolution elastic matching. Comput. Vis. Graph. Image Process. 1989, 46, 1–21. [Google Scholar] [CrossRef]
- Modat, M.; Ridgway, G.R.; Taylor, Z.A.; Lehmann, M.; Barnes, J.; Hawkes, D.J.; Fox, N.C.; Ourselin, S. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 2010, 98, 278–284. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 2013, 7, 39. [Google Scholar] [CrossRef]
- Beg, M.F.; Miller, M.I.; Trouvé, A.; Younes, L. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. Int. J. Comput. Vis. 2005, 61, 139–157. [Google Scholar] [CrossRef]
- Arsigny, V.; Olivier Commowick Pennec, X.; Ayache, N. A Log-Euclidean Framework for Statistics on Diffeomorphisms. Lect. Notes Comput. Sci. 2006, 9, 924–931. [Google Scholar] [CrossRef]
- Iqbal, I.; Shahzad, G.; Rafiq, N.; Mustafa, G.; Ma, J. Deep learning based automated detection of human knee joint’s synovial fluid from magnetic resonance images with transfer learning. IET Image Process. 2020, 14, 1990–1998. [Google Scholar] [CrossRef]
- Iqbal, I.; Ullah, I.; Peng, T.; Wang, W.; Ma, N. An end-to-end deep convolutional neural network-based data-driven fusion framework for identification of human induced pluripotent stem cell-derived endothelial cells in photomicrographs. Eng. Appl. Artif. Intell. 2024, 139, 109573. [Google Scholar] [CrossRef]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans. Med. Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef]
- Chen, J.; Frey, E.C.; He, Y.; Segars, W.P.; Li, Y.; Du, Y. TransMorph: Transformer for unsupervised medical image registration. Med. Image Anal. 2022, 82, 102615. [Google Scholar] [CrossRef]
- Kim, B.; Han, I.; Ye, J.C. DiffuseMorph: Unsupervised Deformable Image Registration Using Diffusion Model. In Lecture Notes in Computer Science; Springer Nature: Cham, Switzerland, 2022; pp. 347–364. [Google Scholar] [CrossRef]
- Hoffmann, M.; Billot, B.; Greve, D.N.; Iglesias, J.E.; Fischl, B.; Dalca, A.V. SynthMorph: Learning Contrast-Invariant Registration Without Acquired Images. IEEE Trans. Med. Imaging 2022, 41, 543–558. [Google Scholar] [CrossRef]
- Zhang, C.; Liang, X. INR-LDDMM: Fluid-based Medical Image Registration Integrating Implicit Neural Representation and Large Deformation Diffeomorphic Metric Mapping. arXiv 2025, arXiv:2308.09473. [Google Scholar]
- van Harten, L.D.; Stoker, J.; Isgum, I. Robust Deformable Image Registration Using Cycle-Consistent Implicit Representations. IEEE Trans. Med. Imaging 2024, 43, 784–793. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Meng, D. Continuous Representation Methods, Theories, and Applications: An Overview and Perspectives. arXiv 2025, arXiv:2505.15222. [Google Scholar] [CrossRef]
- Rahaman, N.; Baratin, A.; Arpit, D.; Draxler, F.; Lin, M.; Hamprecht, F.; Bengio, Y.; Courville, A. On the Spectral Bias of Neural Networks. PMLR 2019, 97, 5301–5310. Available online: https://proceedings.mlr.press/v97/rahaman19a.html (accessed on 8 August 2025).
- Sitzmann, V.; Martel, J.N.P.; Bergman, A.W.; Lindell, D.B.; Wetzstein, G. Implicit Neural Representations with Periodic Activation Functions. arXiv 2006, arXiv:2006.09661. [Google Scholar] [CrossRef]
- Tancik, M.; Srinivasan, P.; Mildenhall, B.; Fridovich-Keil, S.; Raghavan, N.; Singhal, U.; Ramamoorthi, R.; Barron, J.; Ng, R. Fourier Features Let Networks Learn High Frequency Functions in Low Dimensional Domains. Neural Inf. Process. Syst. 2020, 33, 7537–7547. Available online: https://proceedings.neurips.cc/paper_files/paper/2020/hash/55053683268957697aa39fba6f231c68-Abstract.html (accessed on 20 May 2024).
- Liu, Z.; Wang, Y.; Vaidya, S.; Ruehle, F.; Halverson, J.; Soljacic, M.; Hou, T.Y.; Tegmark, M. KAN: Kolmogorov–Arnold Networks. Int. Conf. Represent. Learn. 2025, 2025, 70367–70413. Available online: https://proceedings.iclr.cc/paper_files/paper/2025/hash/afaed89642ea100935e39d39a4da602c-Abstract-Conference.html (accessed on 22 July 2025).
- Ziad Al-Haj, H.; Vogt, N.; Quillien, L.; Weihsbach, C.; Heinrich, M.P.; Oster, J. CineJENSE: Simultaneous Cine MRI Image Reconstruction and Sensitivity Map Estimation Using Neural Representations. Lect. Notes Comput. Sci. 2024, 14507, 467–478. [Google Scholar] [CrossRef]
- Ruyi Zha Zhang, Y.; Li, H. NAF: Neural Attenuation Fields for Sparse-View CBCT Reconstruction. In Lecture Notes in Computer Science; Springer Nature: Cham, Switzerland, 2022; pp. 442–452. [Google Scholar] [CrossRef]
- Du, M.; Zheng, Z.; Wang, W.; Quan, G.; Shi, W.; Shen, L.; Zhang, L.; Li, L.; Liu, Y.; Xing, Y. Nonperiodic dynamic CT reconstruction using backward-warping INR with regularization of diffeomorphism (BIRD). arXiv 2025, arXiv:2505.03463. [Google Scholar] [CrossRef]
- Shen, X.; Yang, J.; Wei, C.; Deng, B.; Huang, J.; Hua, X.; Cheng, X.; Liang, K. DCT-Mask: Discrete Cosine Transform Mask Representation for Instance Segmentation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Virtually, Nashville, TN, USA, 19–25 June 2021; pp. 8716–8725. [Google Scholar] [CrossRef]
- Staub, D.; Docef, A.; Brock, R.S.; Vaman, C.; Murphy, M.J. 4D Cone-beam CT reconstruction using a motion model based on principal component analysis. Med. Phys. 2011, 38, 6697–6709. [Google Scholar] [CrossRef]
- Argota-Perez, R.; Robbins, J.; Green, A.; Herk Mvan Korreman, S.; Vásquez-Osorio, E. Evaluating principal component analysis models for representing anatomical changes in head and neck radiotherapy. Phys. Imaging Radiat. Oncol. 2022, 22, 13–19. [Google Scholar] [CrossRef]
- The Pelvic-Reference-Data. The Cancer Imaging Archive (TCIA). Available online: https://www.cancerimagingarchive.net/collection/pelvic-reference-data/ (accessed on 18 March 2025).
- Wasserthal, J.; Breit, H.C.; Meyer, M.T.; Pradella, M.; Hinck, D.; Sauter, A.W.; Heye, T.; Boll, D.T.; Cyriac, J.; Yang, S.; et al. TotalSegmentator: Robust Segmentation of 104 Anatomic Structures in CT Images. Radiology 2023, 5, e230024. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Huttenlocher, D.P.; Klanderman, G.A.; Rucklidge, W.J. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 850–863. [Google Scholar] [CrossRef]




| Type | Method | Femur | Hip | Bladder | Rectum | Average |
|---|---|---|---|---|---|---|
| (a) DSC (unit: %) | ||||||
| Iteration-based | Demons | 87.89 ± 2.24 | 86.65 ± 2.91 | 74.11 ± 4.65 | 70.33 ± 5.79 | 79.75 |
| Elastix | 88.09 ± 3.01 | 88.87 ± 2.77 | 75.13 ± 3.16 | 71.91 ± 3.65 | 81.00 | |
| INR-based | MLP only | 90.91 ± 4.06 | 90.11 ± 2.65 | 79.44 ± 4.14 | 74.03 ± 4.17 | 83.62 |
| KAN only | 91.18 ± 3.34 | 90.37 ± 2.30 | 80.15 ± 3.05 | 76.23 ± 3.81 | 84.48 | |
| MLP combine | 92.77 ± 2.88 | 90.45 ±2.35 | 81.88 ± 3.91 | 78.49 ± 3.52 | 85.90 | |
| Ours | 93.09 ± 2.15 | 90.88 ± 2.63 | 82.73 ± 4.74 | 79.42 ± 3.19 | 86.53 | |
| (b) HD95 (unit: mm) | ||||||
| Iteration-based | Demons | 4.35 ± 1.42 | 5.47 ± 1.21 | 8.96 ± 3.41 | 9.25 ± 3.01 | 7.01 |
| Elastix | 4.15 ± 1.17 | 5.17 ± 1.36 | 8.59 ± 4.01 | 8.93 ± 3.26 | 6.71 | |
| INR-based | MLP only | 3.40 ± 1.43 | 4.26 ± 0.97 | 7.01 ± 3.92 | 7.33 ± 3.98 | 5.50 |
| KAN only | 3.32 ± 1.56 | 4.24 ± 1.14 | 6.94 ± 4.01 | 7.28 ± 3.83 | 5.45 | |
| MLP combine | 3.13 ± 1.16 | 4.16 ± 0.93 | 6.27 ± 3.03 | 7.21 ± 3.15 | 5.19 | |
| Ours | 2.97 ± 1.03 | 4.06 ± 1.01 | 6.11 ± 3.14 | 7.23 ± 2.79 | 5.09 | |
| Method | Mean Jacobian | Std. Dev. | Min Value | Max Value | Moderate (%) |
|---|---|---|---|---|---|
| (a) Demons | |||||
| Bladder | 0.946 | 0.119 | 0.240 | 2.460 | 85.612 |
| Femur | 0.993 | 0.017 | 0.560 | 1.590 | 99.619 |
| Global | 0.986 | 0.019 | 0.110 | 3.370 | 98.713 |
| Hip | 0.991 | 0.017 | 0.500 | 1.700 | 99.401 |
| Rectum | 1.122 | 0.131 | 0.290 | 2.510 | 80.334 |
| (b) Elastix | |||||
| Bladder | 0.951 | 0.120 | 0.270 | 2.410 | 88.308 |
| Femur | 0.995 | 0.018 | 0.580 | 1.590 | 99.770 |
| Global | 0.989 | 0.019 | 0.130 | 3.130 | 98.776 |
| Hip | 0.994 | 0.016 | 0.500 | 1.700 | 99.524 |
| Rectum | 1.113 | 0.121 | 0.290 | 2.450 | 81.051 |
| (c) MLP only | |||||
| Bladder | 0.976 | 0.115 | 0.310 | 2.200 | 88.540 |
| Femur | 0.998 | 0.012 | 0.580 | 1.550 | 99.839 |
| Global | 0.993 | 0.015 | 0.160 | 2.950 | 99.046 |
| Hip | 0.997 | 0.013 | 0.510 | 1.500 | 99.629 |
| Rectum | 1.080 | 0.118 | 0.350 | 2.280 | 82.397 |
| (d) KAN only | |||||
| Bladder | 0.976 | 0.115 | 0.320 | 2.170 | 89.214 |
| Femur | 0.998 | 0.012 | 0.580 | 1.550 | 99.844 |
| Global | 0.994 | 0.015 | 0.160 | 2.920 | 99.117 |
| Hip | 0.998 | 0.013 | 0.510 | 1.490 | 99.635 |
| Rectum | 1.080 | 0.116 | 0.350 | 2.260 | 83.481 |
| (e) MLP combine | |||||
| Bladder | 0.977 | 0.115 | 0.330 | 2.100 | 89.975 |
| Femur | 0.998 | 0.011 | 0.580 | 1.530 | 99.844 |
| Global | 0.994 | 0.014 | 0.160 | 2.890 | 99.227 |
| Hip | 0.998 | 0.012 | 0.50 | 1.480 | 99.644 |
| Rectum | 1.079 | 0.113 | 0.360 | 2.260 | 85.792 |
| (f) Ours | |||||
| Bladder | 0.978 | 0.113 | 0.330 | 2.060 | 90.007 |
| Femur | 0.998 | 0.012 | 0.590 | 1.530 | 99.859 |
| Global | 0.995 | 0.013 | 0.160 | 2.870 | 99.343 |
| Hip | 0.998 | 0.011 | 0.520 | 1.480 | 99.656 |
| Rectum | 1.078 | 0.112 | 0.370 | 2.250 | 86.133 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, P.; Zhang, C.; Yang, Z.; Yin, F.-F.; Liu, M. An Implicit Registration Framework Integrating Kolmogorov–Arnold Networks with Velocity Regularization for Image-Guided Radiation Therapy. Bioengineering 2025, 12, 1005. https://doi.org/10.3390/bioengineering12091005
Sun P, Zhang C, Yang Z, Yin F-F, Liu M. An Implicit Registration Framework Integrating Kolmogorov–Arnold Networks with Velocity Regularization for Image-Guided Radiation Therapy. Bioengineering. 2025; 12(9):1005. https://doi.org/10.3390/bioengineering12091005
Chicago/Turabian StyleSun, Pulin, Chulong Zhang, Zhenyu Yang, Fang-Fang Yin, and Manju Liu. 2025. "An Implicit Registration Framework Integrating Kolmogorov–Arnold Networks with Velocity Regularization for Image-Guided Radiation Therapy" Bioengineering 12, no. 9: 1005. https://doi.org/10.3390/bioengineering12091005
APA StyleSun, P., Zhang, C., Yang, Z., Yin, F.-F., & Liu, M. (2025). An Implicit Registration Framework Integrating Kolmogorov–Arnold Networks with Velocity Regularization for Image-Guided Radiation Therapy. Bioengineering, 12(9), 1005. https://doi.org/10.3390/bioengineering12091005








