scOTM: A Deep Learning Framework for Predicting Single-Cell Perturbation Responses with Large Language Models
Abstract
1. Introduction
Related Works
2. Materials and Methods
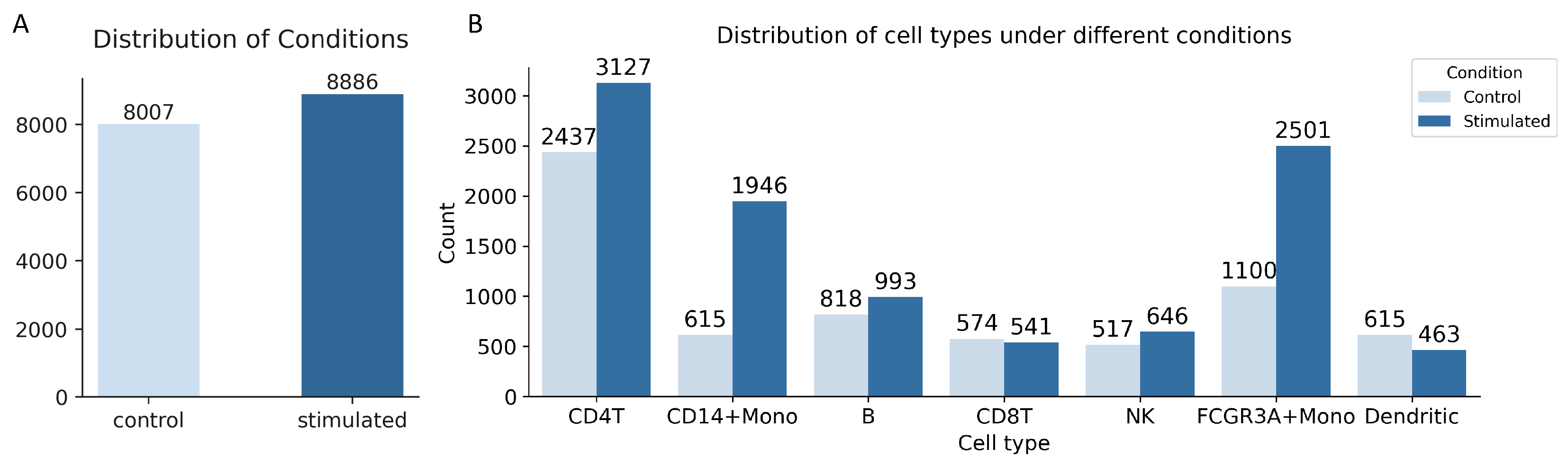
2.1. Datasets and Preprocessing
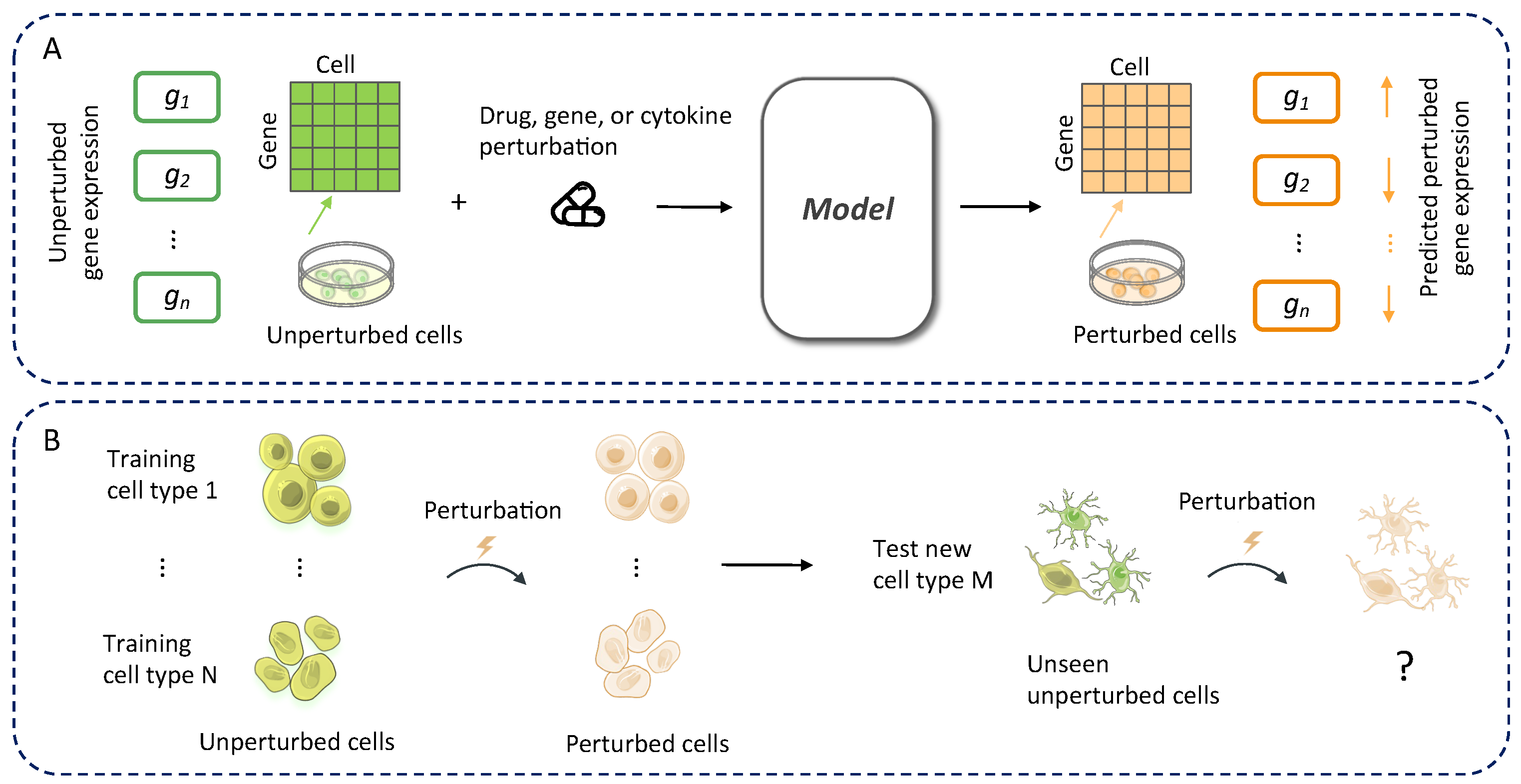
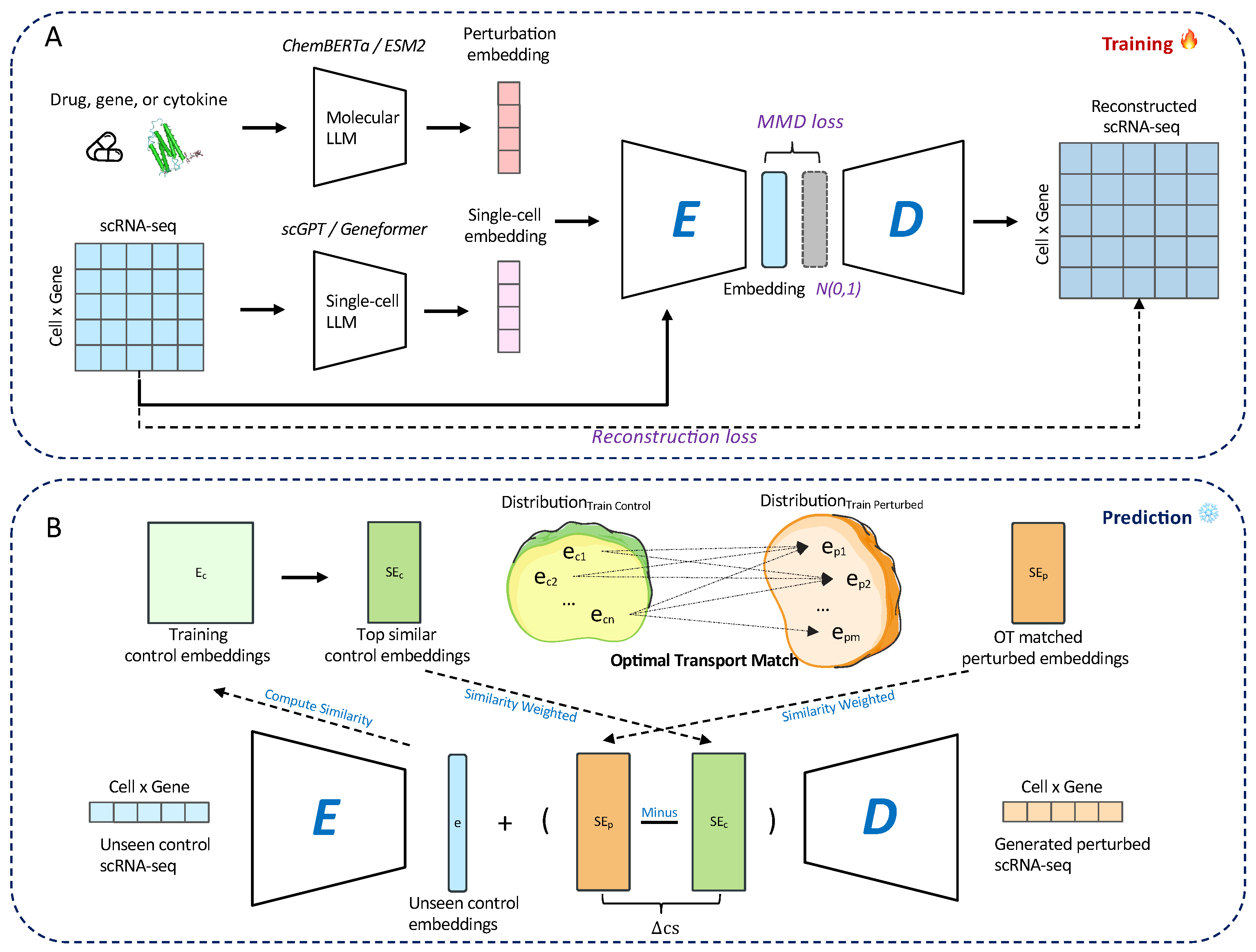
2.2. Model Framework
2.3. Optimal Transport-Based Alignment Between Cell States
2.4. Predicting the Perturbed State
3. Results
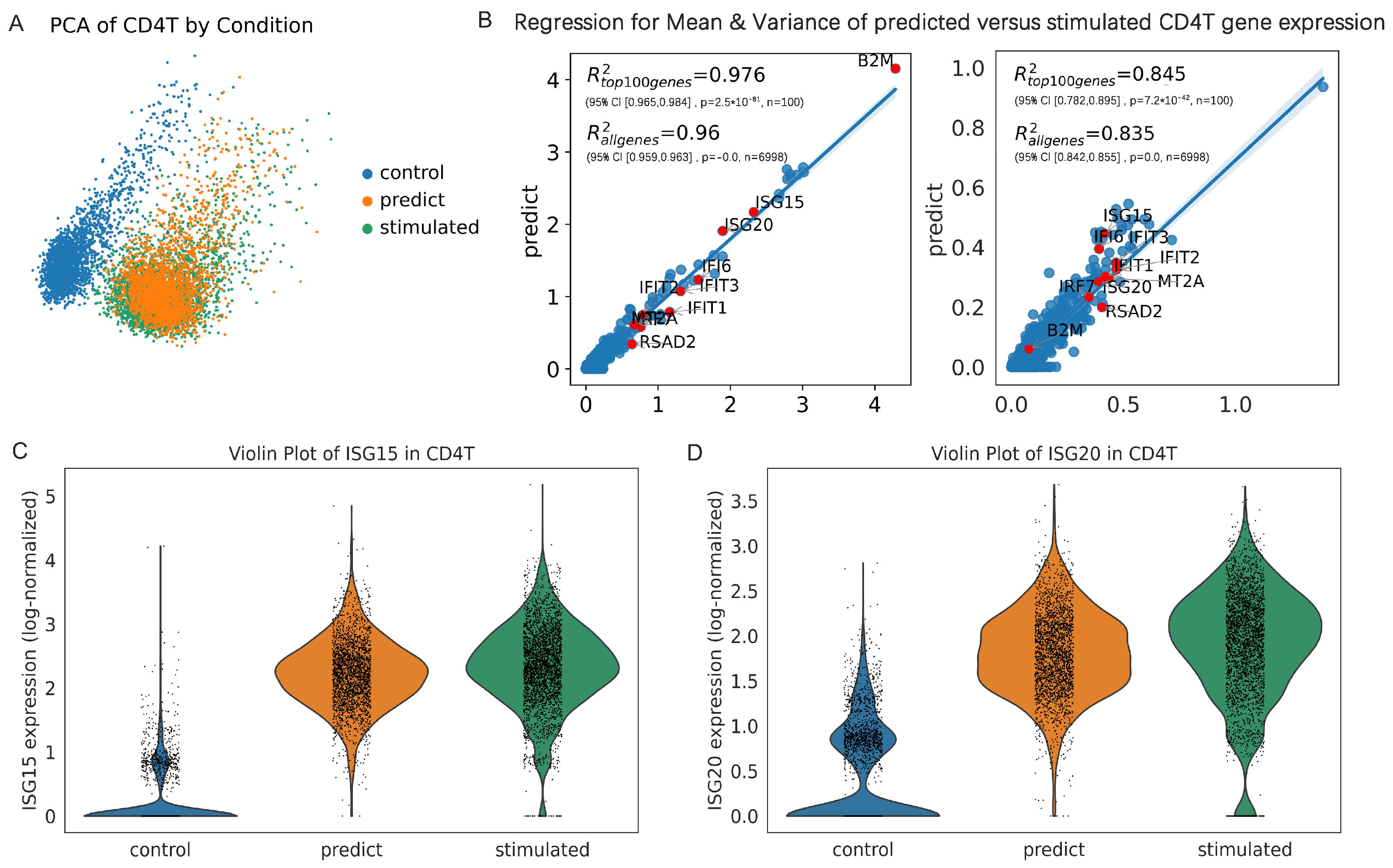
3.1. scOTM Accurately Predicts Perturbation Response Across Unseen Cell Types
3.2. scOTM Outperforms Alternative Approaches Across Unseen Cell Types
3.3. scOTM Enhances the Accuracy of Differentially Expressed Gene Identification
3.4. Interpretability of scOTM
3.5. Sensitivity Analysis Under Varying Data Scales
3.6. Ablation Analysis
3.6.1. Ablation Results and Effectiveness of MMD Loss
3.6.2. Ablation for Effectiveness of Combining LLM Embeddings
3.7. Computational Resources and Runtime
4. Conclusions and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marques, L.; Costa, B.; Pereira, M.; Silva, A.; Santos, J.; Saldanha, L.; Silva, I.; Magalhães, P.; Schmidt, S.; Vale, N. Advancing precision medicine: A review of innovative in silico approaches for drug development, clinical pharmacology and personalized healthcare. Pharmaceutics 2024, 16, 332. [Google Scholar] [CrossRef] [PubMed]
- Adduri, A.; Gautam, D.; Bevilacqua, B.; Imran, A.; Shah, R.; Naghipourfar, M.; Teyssier, N.; Ilango, R.; Nagaraj, S.; Ricci-Tam, C.; et al. Predicting cellular responses to perturbation across diverse contexts with STATE. bioRxiv 2025. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Brozovic, A.; Izraely, S.; Casas-Pais, A.; Witz, I.P.; Figueroa, A. Cancer drug resistance induced by EMT: Novel therapeutic strategies. Arch. Toxicol. 2021, 95, 2279–2297. [Google Scholar] [CrossRef] [PubMed]
- Kasper, L.H.; Reder, A.T. Immunomodulatory activity of interferon-beta. Ann. Clin. Transl. Neurol. 2014, 1, 622–631. [Google Scholar] [CrossRef]
- Raftery, N.; Stevenson, N.J. Advances in anti-viral immune defence: Revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 2017, 74, 2525–2535. [Google Scholar] [CrossRef]
- Wang, W.; Xu, L.; Su, J.; Peppelenbosch, M.P.; Pan, Q. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 2017, 25, 573–584. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Boisson-Dupuis, S.; Chapgier, A.; Yang, K.; Bustamante, J.; Puel, A.; Picard, C.; Abel, L.; Jouanguy, E.; Casanova, J.L. Inborn errors of interferon (IFN)-mediated immunity in humans: Insights into the respective roles of IFN-α/β, IFN-γ, and IFN-λ in host defense. Immunol. Rev. 2008, 226, 29–40. [Google Scholar] [CrossRef]
- Gohil, S.H.; Iorgulescu, J.B.; Braun, D.A.; Keskin, D.B.; Livak, K.J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 244–256. [Google Scholar] [CrossRef]
- Lee, H.Z.; Kwitkowski, V.E.; Del Valle, P.L.; Ricci, M.S.; Saber, H.; Habtemariam, B.A.; Bullock, J.; Bloomquist, E.; Li Shen, Y.; Chen, X.H.; et al. FDA approval: Belinostat for the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Clin. Cancer Res. 2015, 21, 2666–2670. [Google Scholar] [CrossRef]
- Lavin, A.; Krakauer, D.; Zenil, H.; Gottschlich, J.; Mattson, T.; Brehmer, J.; Anandkumar, A.; Choudry, S.; Rocki, K.; Baydin, A.G.; et al. Simulation intelligence: Towards a new generation of scientific methods. arXiv 2021, arXiv:2112.03235. [Google Scholar]
- Lotfollahi, M.; Wolf, F.A.; Theis, F.J. scGen predicts single-cell perturbation responses. Nat. Methods 2019, 16, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.M.; Mohammadi, F.; Sanchez-Aguila, C.; Zhan, P.J.; Liby, T.A.; Dane, M.A.; Meyer, A.S.; Heiser, L.M. Analysis and modeling of cancer drug responses using cell cycle phase-specific rate effects. Nat. Commun. 2023, 14, 3450. [Google Scholar] [CrossRef]
- Bunne, C.; Schiebinger, G.; Krause, A.; Regev, A.; Cuturi, M. Optimal transport for single-cell and spatial omics. Nat. Rev. Methods Prim. 2024, 4, 58. [Google Scholar] [CrossRef]
- Peidli, S.; Green, T.D.; Shen, C.; Gross, T.; Min, J.; Garda, S.; Yuan, B.; Schumacher, L.J.; Taylor-King, J.P.; Marks, D.S.; et al. scPerturb: Harmonized single-cell perturbation data. Nat. Methods 2024, 21, 531–540. [Google Scholar] [CrossRef]
- Lotfollahi, M.; Naghipourfar, M.; Theis, F.J.; Wolf, F.A. Conditional out-of-distribution generation for unpaired data using transfer VAE. Bioinformatics 2020, 36, i610–i617. [Google Scholar] [CrossRef]
- Kana, O.; Nault, R.; Filipovic, D.; Marri, D.; Zacharewski, T.; Bhattacharya, S. Generative modeling of single-cell gene expression for dose-dependent chemical perturbations. Patterns 2023, 4, 100817. [Google Scholar] [CrossRef]
- Sohn, K.; Lee, H.; Yan, X. Learning structured output representation using deep conditional generative models. Adv. Neural Inf. Process. Syst. 2015, 28, 3483–3491. [Google Scholar]
- Borgwardt, K.M.; Gretton, A.; Rasch, M.J.; Kriegel, H.P.; Schölkopf, B.; Smola, A.J. Integrating structured biological data by kernel maximum mean discrepancy. Bioinformatics 2006, 22, e49–e57. [Google Scholar] [CrossRef]
- Santambrogio, F. Optimal Transport for Applied Mathematicians; Birkhäuser: Cham, Switzerland, 2015; Volume 87. [Google Scholar]
- Bunne, C.; Stark, S.G.; Gut, G.; Del Castillo, J.S.; Levesque, M.; Lehmann, K.V.; Pelkmans, L.; Krause, A.; Rätsch, G. Learning single-cell perturbation responses using neural optimal transport. Nat. Methods 2023, 20, 1759–1768. [Google Scholar] [CrossRef]
- Makkuva, A.; Taghvaei, A.; Oh, S.; Lee, J. Optimal transport mapping via input convex neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2020; pp. 6672–6681. [Google Scholar]
- Jiang, Q.; Chen, S.; Chen, X.; Jiang, R. scPRAM accurately predicts single-cell gene expression perturbation response based on attention mechanism. Bioinformatics 2024, 40, btae265. [Google Scholar] [CrossRef]
- Li, Q.; Hu, Z.; Wang, Y.; Li, L.; Fan, Y.; King, I.; Jia, G.; Wang, S.; Song, L.; Li, Y. Progress and opportunities of foundation models in bioinformatics. Briefings Bioinform. 2024, 25, bbae548. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, C.; Maan, H.; Pang, K.; Luo, F.; Duan, N.; Wang, B. scGPT: Toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 2024, 21, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Subramaniam, M.; Targ, S.; Nguyen, M.; Maliskova, L.; McCarthy, E.; Wan, E.; Wong, S.; Byrnes, L.; Lanata, C.M.; et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018, 36, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, D.; Benz, A.; Lieberman, R.; Gigante, S.; Chow, A.; Holbrook, R.; Cannoodt, R.; Luecken, M.; Open Problems—Single-Cell Perturbations. Kaggle. 2023. Available online: https://kaggle.com/competitions/open-problems-single-cell-perturbations (accessed on 17 August 2025).
- Chithrananda, S.; Grand, G.; Ramsundar, B. ChemBERTa: Large-scale self-supervised pretraining for molecular property prediction. arXiv 2020, arXiv:2010.09885. [Google Scholar]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Peyré, G.; Cuturi, M. Computational optimal transport: With applications to data science. Found. Trends® Mach. Learn. 2019, 11, 355–607. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2021. [Google Scholar]
- Fisher, R.A. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika 1915, 10, 507–521. [Google Scholar] [CrossRef]
- Crow, M.K. Type I interferon in the pathogenesis of lupus. J. Immunol. 2014, 192, 5459–5468. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The reactome pathway knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Pizarroso, J.; Portela, J.; Muñoz, A. NeuralSens: Sensitivity analysis of neural networks. J. Stat. Softw. 2022, 102, 1–36. [Google Scholar] [CrossRef]
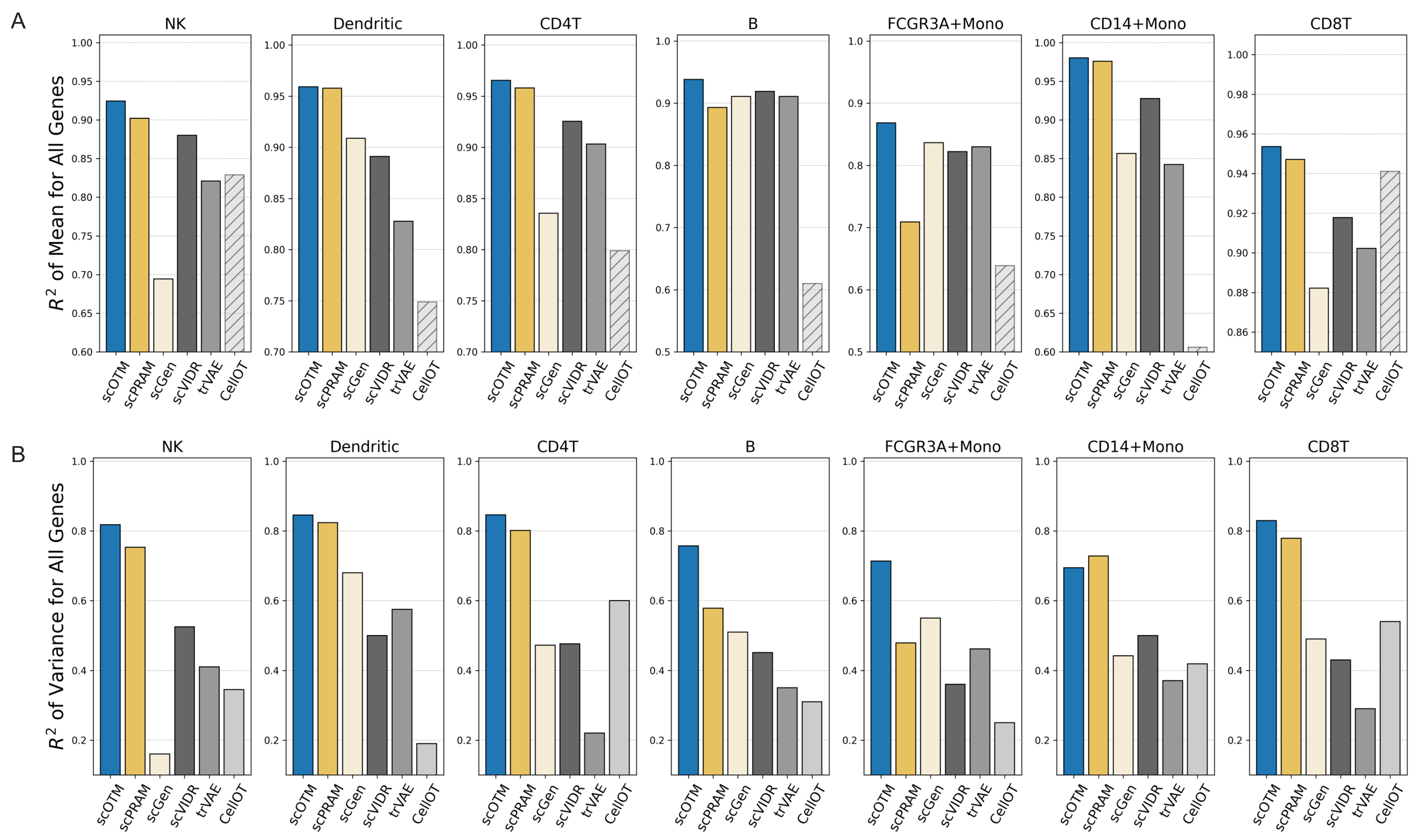
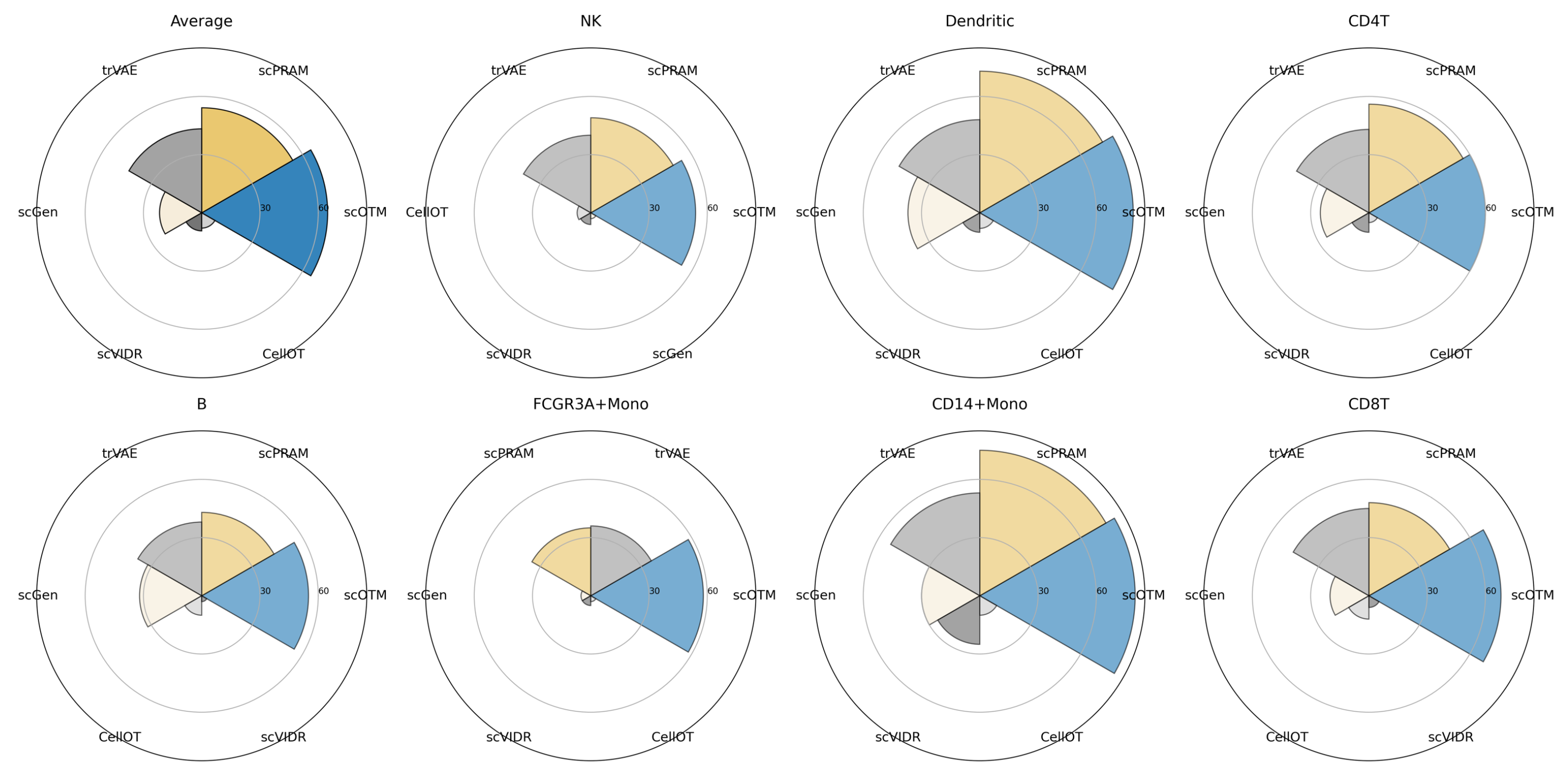

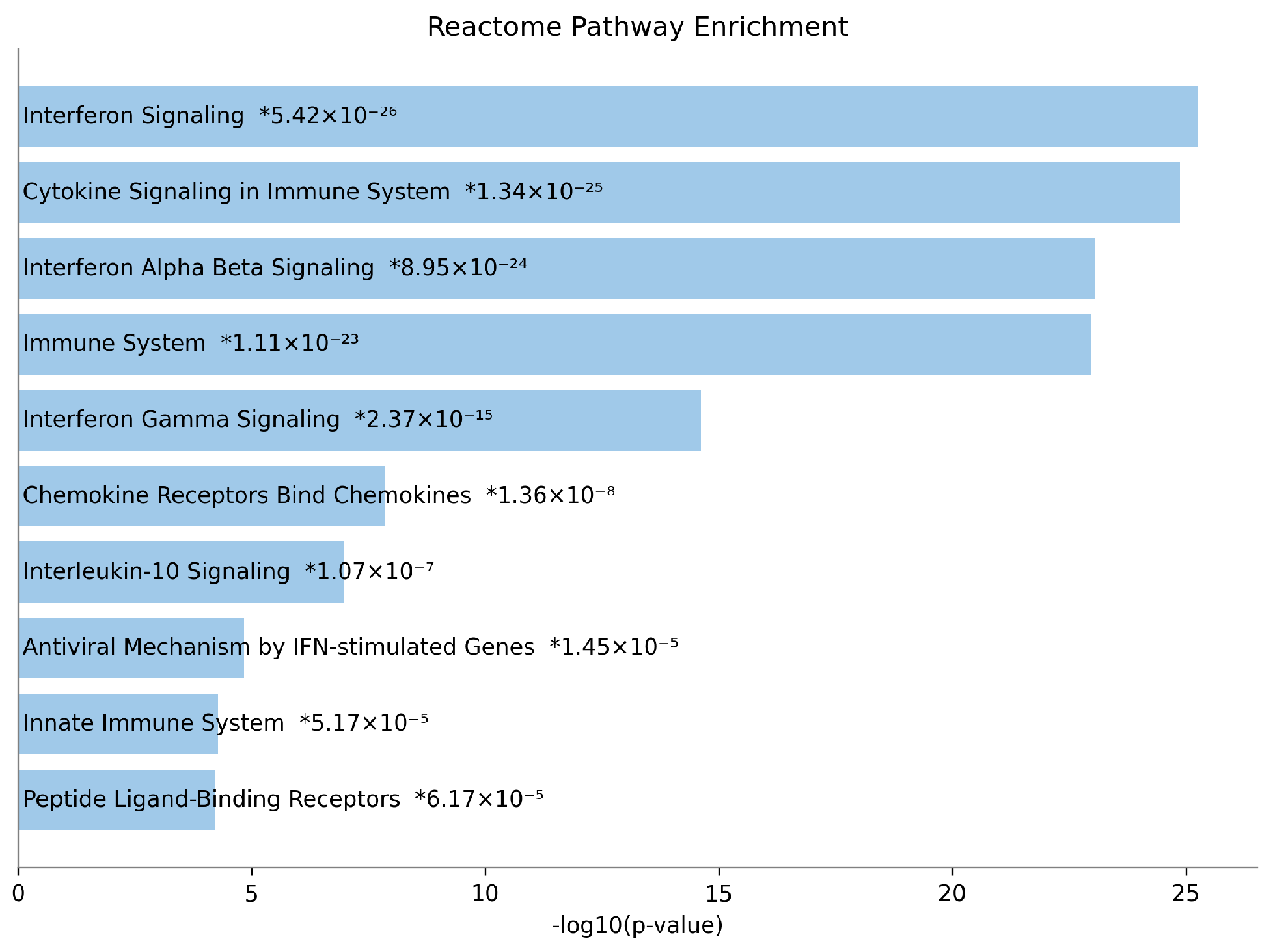
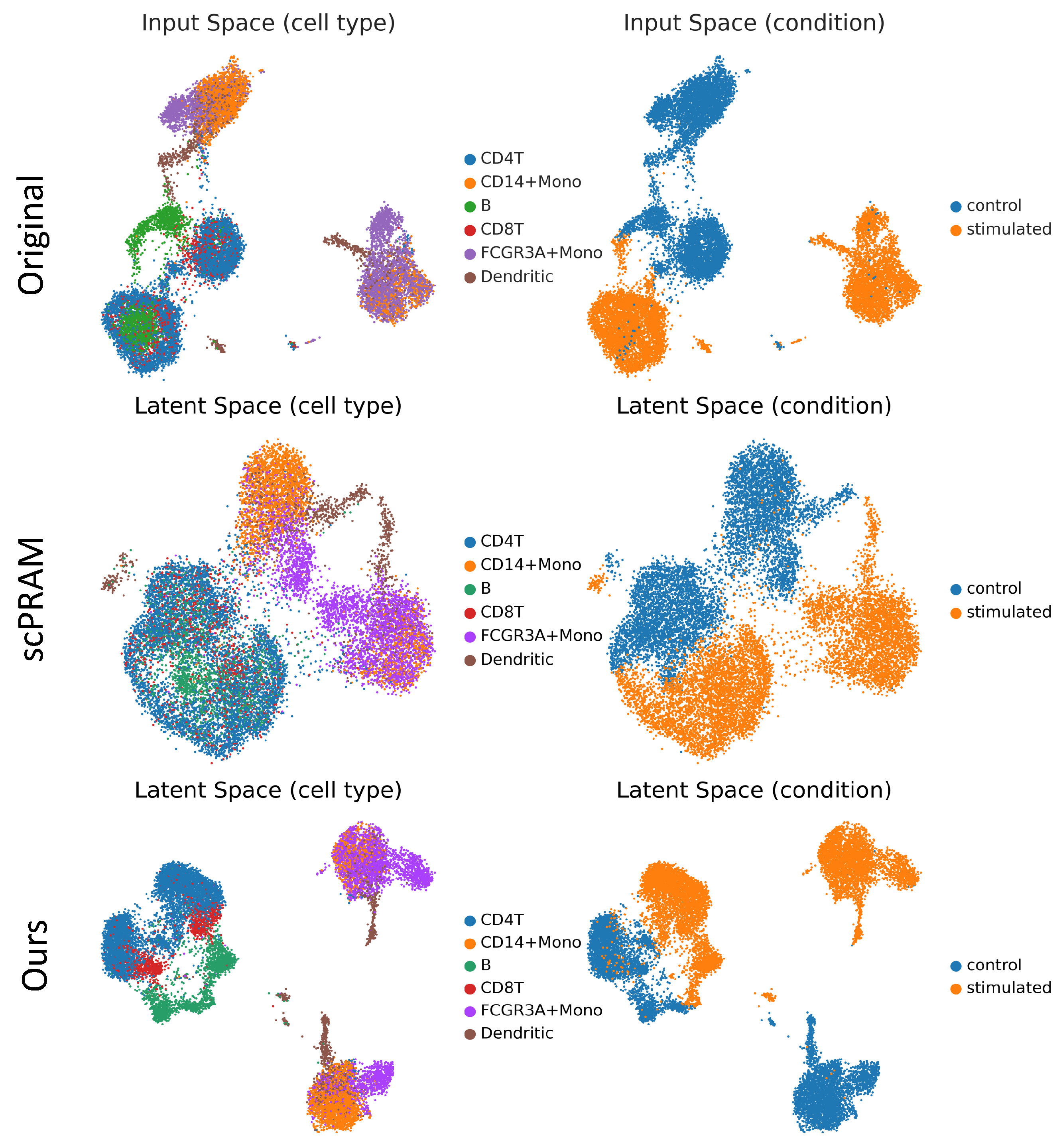
| Cell Type | 0.1 | 0.3 | 0.5 | 0.7 | 1.0 |
|---|---|---|---|---|---|
| (A) Mean of All Genes | |||||
| NK | 0.8471 | 0.8947 | 0.9242 | 0.9111 | 0.9205 |
| Dendritic | 0.9025 | 0.9384 | 0.9493 | 0.9467 | 0.9638 |
| CD4T | 0.8782 | 0.9232 | 0.9421 | 0.9438 | 0.9608 |
| B | 0.8552 | 0.8925 | 0.9342 | 0.9372 | 0.9349 |
| FCGR3A+ Mono | 0.6311 | 0.7805 | 0.7641 | 0.6633 | 0.9292 |
| CD14+ Mono | 0.9451 | 0.9612 | 0.9676 | 0.9674 | 0.9678 |
| CD8T | 0.8913 | 0.9313 | 0.9443 | 0.9464 | 0.9546 |
| (B) Common DEGs among Top 100 DEGs | |||||
| NK | 47 | 50 | 54 | 56 | 56 |
| Dendritic | 65 | 74 | 73 | 75 | 78 |
| CD4T | 48 | 50 | 55 | 55 | 55 |
| B | 43 | 51 | 55 | 57 | 58 |
| FCGR3A+ Mono | 33 | 40 | 40 | 36 | 59 |
| CD14+ Mono | 76 | 80 | 82 | 81 | 81 |
| CD8T | 50 | 60 | 61 | 62 | 62 |
| Cell Type | Common DEGs (Top 100) | of Regression Analyses | ||
|---|---|---|---|---|
| w/o MMD | with MMD | Expr. Mean | Expr. Variance | |
| NK | 52 | 54 | 0.8998 → 0.9225 | 0.7941 → 0.8172 |
| Dendritic | 71 | 79 | 0.9604 → 0.9648 | 0.8300 → 0.8042 |
| CD4T | 60 | 60 | 0.9606 → 0.9610 | 0.8488 → 0.8352 |
| B | 53 | 55 | 0.9246 → 0.9409 | 0.7174 → 0.7460 |
| FCGR3A+ Mono | 39 | 58 | 0.7259 → 0.8754 | 0.5271 → 0.7019 |
| CD14+ Mono | 80 | 81 | 0.9720 → 0.9743 | 0.6967 → 0.7027 |
| CD8T | 68 | 68 | 0.9537 → 0.9536 | 0.8072 → 0.8160 |
| Mean | 60.43 | 65 | 0.9141 → 0.9418 | 0.7459 → 0.7748 |
| Cell Type | Common DEGs (Top 100) | of Regression Analyses | ||
|---|---|---|---|---|
| w/o LLM | with LLM | Expr. Mean | Expr. Variance | |
| NK | 54 | 55 | 0.9209 → 0.9244 | 0.8279 → 0.8377 |
| Dendritic | 75 | 77 | 0.9546 → 0.9608 | 0.7410 → 0.7485 |
| CD4T | 60 | 60 | 0.9554 → 0.9575 | 0.8223 → 0.8346 |
| B | 55 | 59 | 0.9230 → 0.9342 | 0.7097 → 0.7554 |
| FCGR3A+ Mono | 61 | 63 | 0.8940 → 0.9301 | 0.7080 → 0.7783 |
| CD14+ Mono | 83 | 81 | 0.9732 → 0.9661 | 0.6906 → 0.6916 |
| CD8T | 65 | 65 | 0.9533 → 0.9506 | 0.8259 → 0.8031 |
| Mean | 64.71 | 65.71 | 0.9392 → 0.9462 | 0.7608 → 0.7785 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Lu, T.; Chen, X.; Yao, Z.; Wong, K.-C. scOTM: A Deep Learning Framework for Predicting Single-Cell Perturbation Responses with Large Language Models. Bioengineering 2025, 12, 884. https://doi.org/10.3390/bioengineering12080884
Wang Y, Lu T, Chen X, Yao Z, Wong K-C. scOTM: A Deep Learning Framework for Predicting Single-Cell Perturbation Responses with Large Language Models. Bioengineering. 2025; 12(8):884. https://doi.org/10.3390/bioengineering12080884
Chicago/Turabian StyleWang, Yuchen, Tianchi Lu, Xingjian Chen, Zhongyu Yao, and Ka-Chun Wong. 2025. "scOTM: A Deep Learning Framework for Predicting Single-Cell Perturbation Responses with Large Language Models" Bioengineering 12, no. 8: 884. https://doi.org/10.3390/bioengineering12080884
APA StyleWang, Y., Lu, T., Chen, X., Yao, Z., & Wong, K.-C. (2025). scOTM: A Deep Learning Framework for Predicting Single-Cell Perturbation Responses with Large Language Models. Bioengineering, 12(8), 884. https://doi.org/10.3390/bioengineering12080884









