A Real-Time Cell Image Segmentation Method Based on Multi-Scale Feature Fusion
Abstract
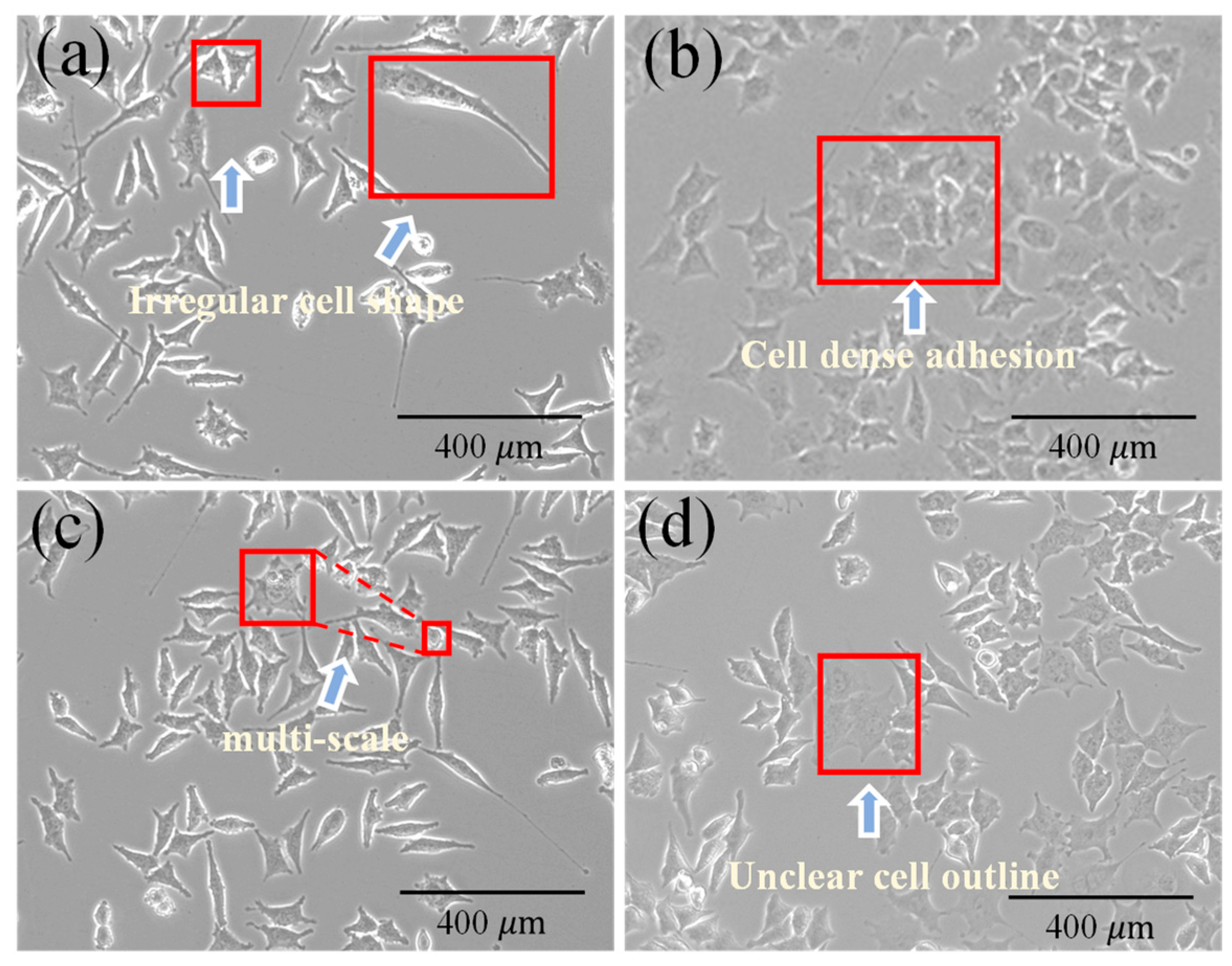
1. Introduction
- (1)
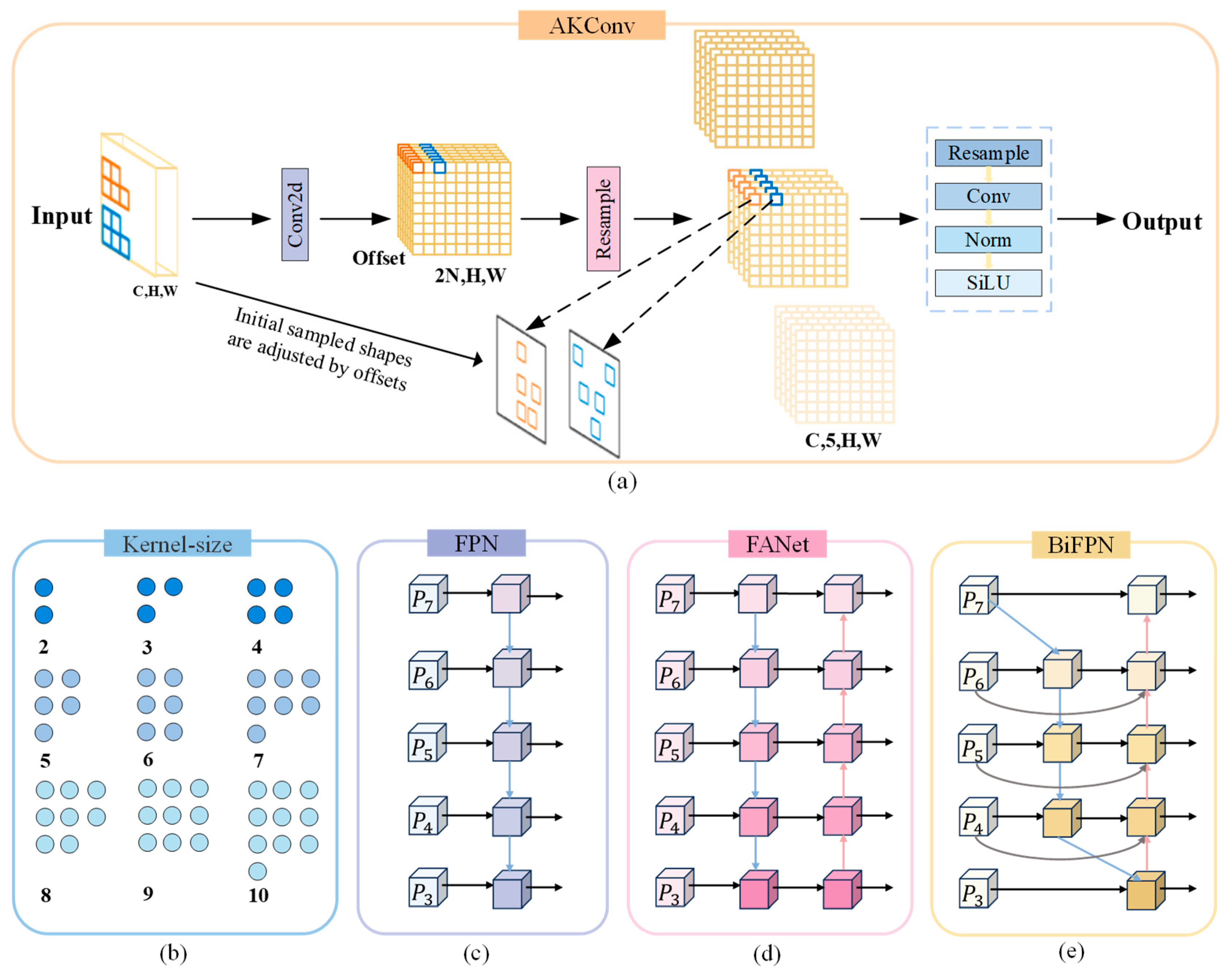
- An architecture is proposed to enable accurate and efficient cell segmentation. To overcome the inherent limitations of traditional FPNs in extracting multi-scale cellular features, this paper introduces a bidirectional feature pyramid network (BiFPN) with weighted fusion, which enhances the ability to extract multi-scale features through a bidirectional feature fusion strategy. Additionally, to mitigate the increased computational cost from the model improvements, adaptive kernel convolution (AKConv) is integrated into the convolutional layers. By incorporating learnable kernel offsets, this approach reduces parameter complexity while enhancing cell feature extraction ability. Through accurate and efficient cell segmentation, real-time and synchronized calculation of cell confluence and count is achieved, providing a powerful new tool for analyzing GSC growth dynamics.
- (2)
- The model was evaluated on both our GSCs dataset and DSB2018. Experimental results show superior segmentation accuracy and inference speed on GSCs, achieving optimal precision-efficiency balance. The public dataset tests confirm excellent generalization.
- (3)
- Glioma tissues were excised from murine brains and enzymatically digested to yield viable single-cell suspensions. From these suspensions, GSC-enriched adherent cultures were subsequently established under optimized media and culture conditions. Time-series images of the GSC cultures were then captured using an inverted fluorescence microscope to monitor morphological changes. Prior to model training, these images were preprocessed with a pipeline of Contrast-Limited Adaptive Histogram Equalization (CLAHE) and Gaussian blurring to enhance image quality.
2. Materials and Methods
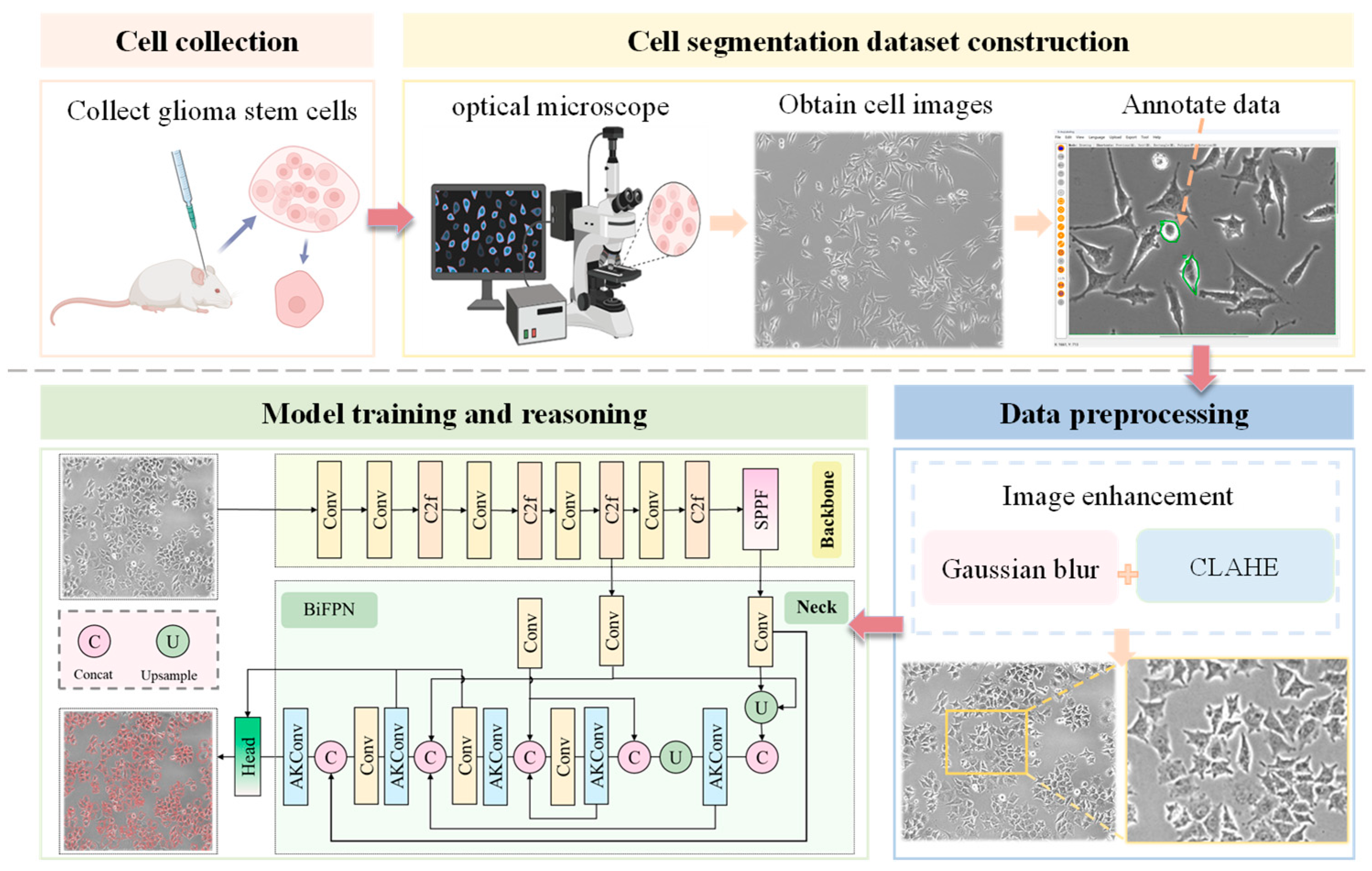
2.1. Data Collection and Dataset Construction
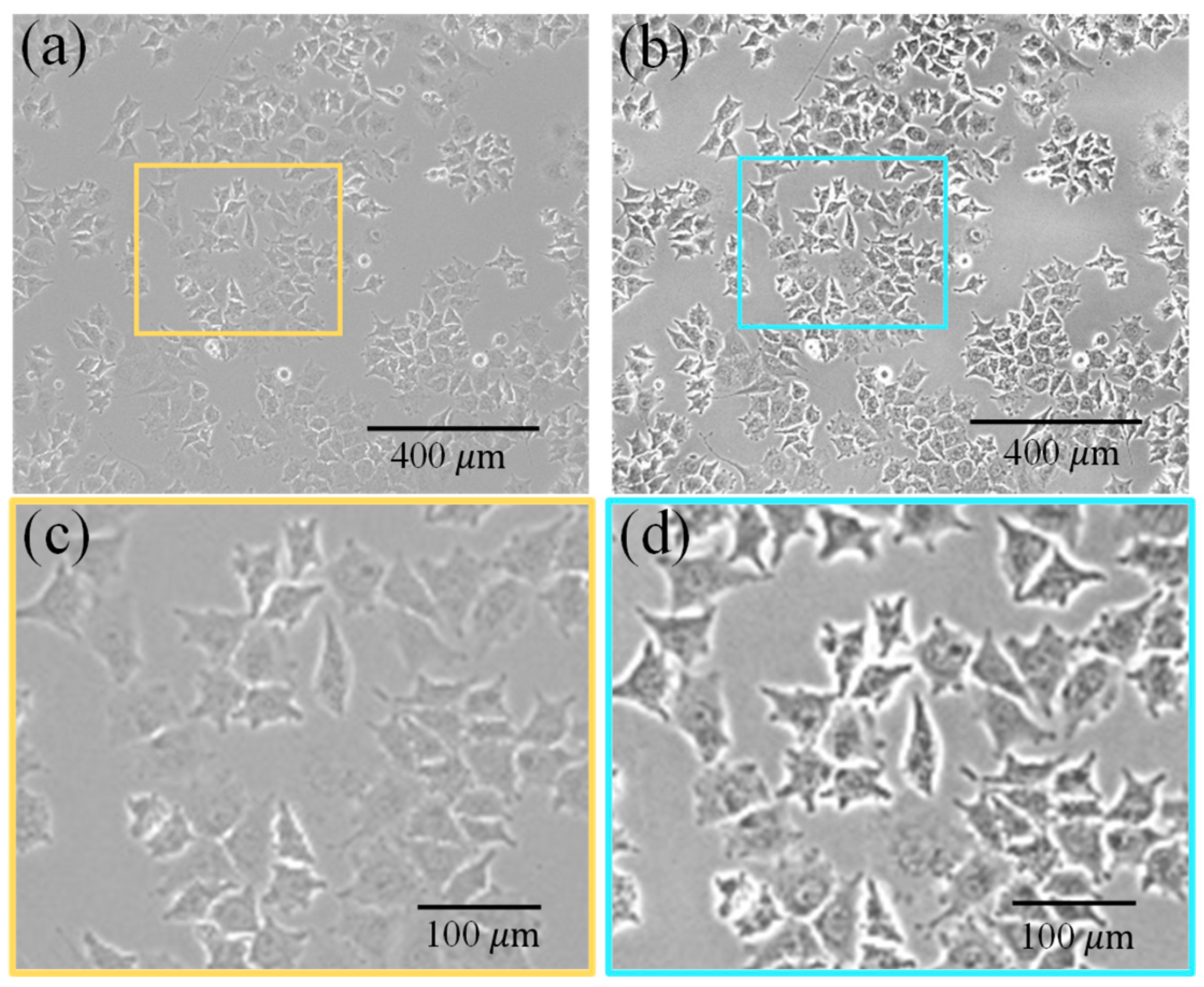
2.2. GSCs Image Preprocessing
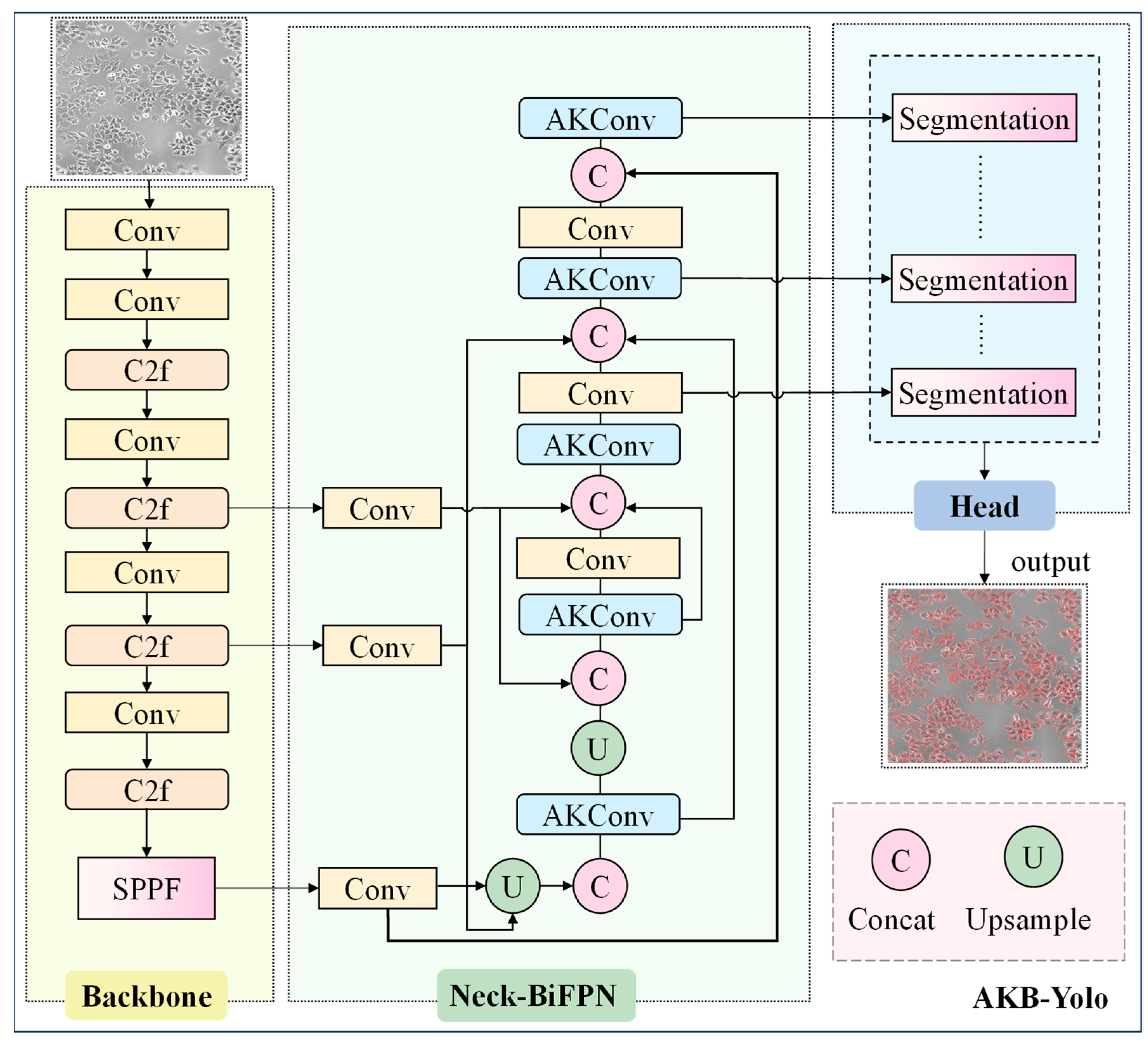
2.3. The Overview of Methods
2.4. Lightweight Improvement Based on AKConv Module
2.5. Using BiFPN Module in Neck Network
2.6. Anchor Box Optimization
2.7. Loss Functions
3. Experiment and Result
3.1. Experimental Settings
3.2. Evaluation Metrics
3.3. Ablation Study Results
3.4. Comparison Experimental Results
3.4.1. Quantitative Analysis Results
3.4.2. Qualitative Analysis Result
3.5. Analysis of Results on Public Dataset
- (1)
- The imaging parameters cover multi-level objective magnification and dual-mode imaging of fluorescence and bright field.
- (2)
- The biological samples include heterogeneous biological tissue samples such as human hepatocellular carcinoma cells (HepG2), mouse fibroblasts (3T3), and Drosophila embryo tissues.
- (3)
- The culture conditions involve complex environment simulations such as normoxia/hypoxia, different pH values, and metabolic states.
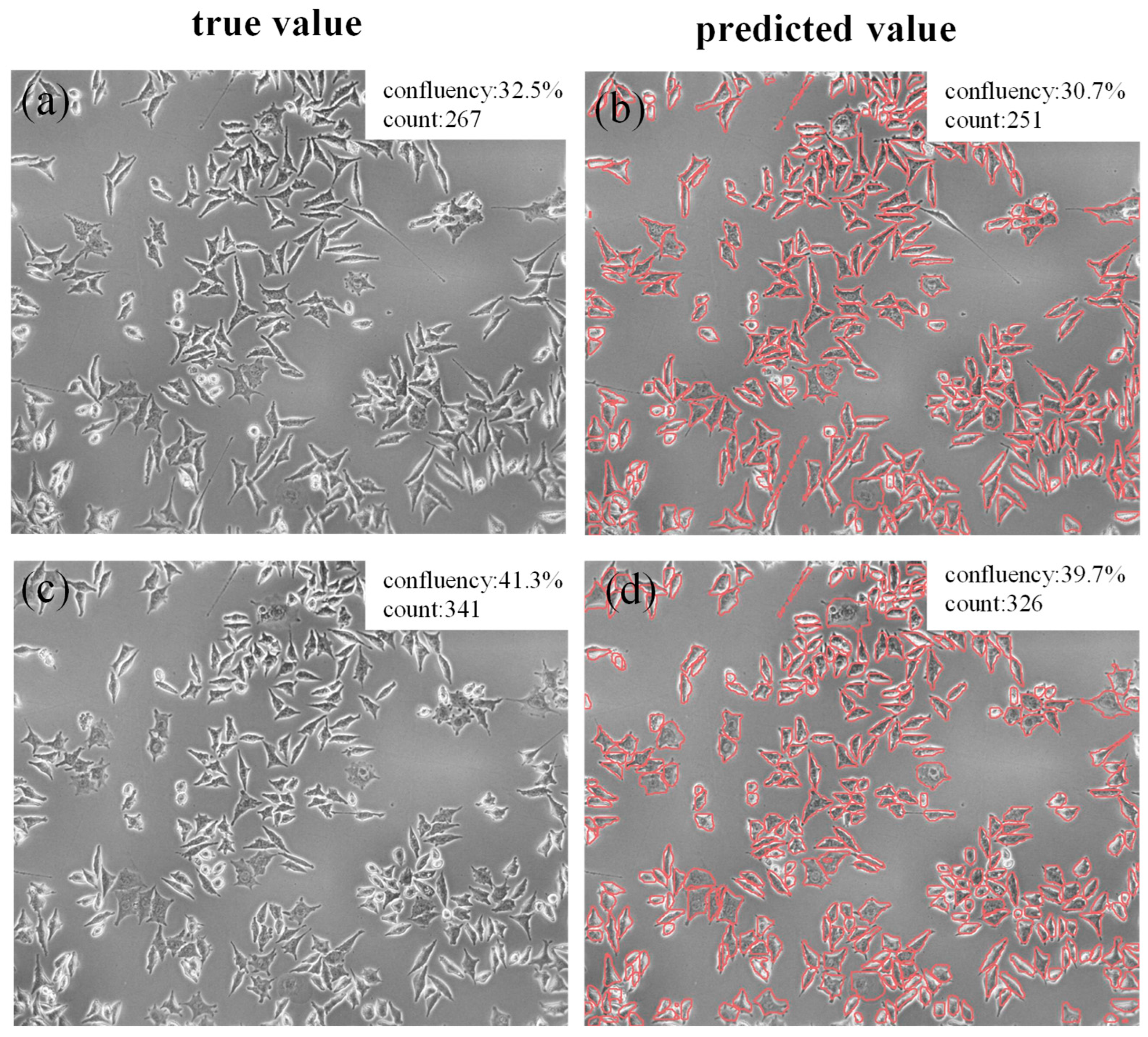
3.6. Confluence Calculation and Cell Counting Effect Analysis of Cellular Images
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, X.Q.; Liu, X.F.; Zhong, H.; Zhang, W.; Yu, S.Z.; Guan, R.F. Progress on Three-Dimensional Cell Culture Technology and Their Application. J. Biomed. Eng. 2023, 40, 602–608. [Google Scholar]
- Li, T.; Qin, X.; Ao, Q. Research Progress on Neural Cell Culture Systems. Curr. Neuropharmacol. 2025, 23, e1570159X360193. [Google Scholar] [CrossRef] [PubMed]
- Cela, E.; Patterson, E.K.; Gill, S.E.; Cepinskas, G.; Fraser, D.D. Application of Human Plasma/Serum to Cell Culture In Vitro: A Translational Research Approach to Better Define Disease Mechanisms. Clin. Transl. Sci. 2025, 18, e70161. [Google Scholar] [CrossRef] [PubMed]
- Godbole, N.; Lai, A.; Carrion, F.; Scholz-Romero, K.; Ravichandran, A.; Croft, P.K.-D.; Reed, A.E.M.; Joshi, V.; Lakhani, S.R.; Masud, M.K.; et al. Extracellular Vesicle miRNAs from Three-Dimensional Ovarian Cancer In Vitro Models and Their Implication in Overall Cancer Survival. Heliyon 2025, 11, e42188. [Google Scholar] [CrossRef]
- Bunmi, K.A.; Adeyemo, K.S. A Review on Targeted Drug Development for Breast Cancer Using Innovative Active Pharmaceutical Ingredients (APIs). Curr. J. Appl. Sci. Technol. 2025, 44, 1–11. [Google Scholar] [CrossRef]
- Chen, R.; Qiu, K.Q.; Han, G.Q.; Kundu, B.K.; Ding, G.D.; Sun, Y.J.; Diao, J.J. Quantifying Cell Viability Through Organelle Ratiometric Probing. Chem. Sci. 2023, 14, 10236–10248. [Google Scholar] [CrossRef]
- Zhou, G.Z.; Wang, X.Z. Progress on On-Line Monitoring for Animal Cell Culture Based on PAT Technology. Chem. Ind. Eng. Prog. 2024, 43, 6356–6371. [Google Scholar]
- He, R.; Jie, P.; Hou, W.; Long, Y.; Zhou, G.; Wu, S.; Liu, W.; Lei, W.; Wen, W.; Wen, Y. Real-Time Artificial Intelligence-Assisted Detection and Segmentation of Nasopharyngeal Carcinoma Using Multimodal Endoscopic Data: A Multi-Center, Prospective Study. EClinicalMedicine 2025, 81, 103120. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wu, Y.; Hobson, C.M.; Su, Y.; Qian, S.; Krueger, E.; Christensen, R.; Kroeschell, G.; Bui, J.; Chaw, M.; et al. Deep Learning-Based Aberration Compensation Improves Contrast and Resolution in Fluorescence Microscopy. Nat. Commun. 2025, 16, 313. [Google Scholar] [CrossRef]
- Luu, N.; Zhang, S.; Lam, R.H.W.; Chen, W. Mechanical Constraints in Tumor Guide Emergent Spatial Patterns of Glioblastoma Cancer Stem Cells. Mechobiol. Med. 2024, 2, 100027. [Google Scholar] [CrossRef]
- Greenwald, A.C.; Darnell, N.G.; Hoefflin, R.; Simkin, D.; Mount, C.W.; Castro, L.N.G.; Harnik, Y.; Dumont, S.; Hirsch, D.; Nomura, M.; et al. Integrative Spatial Analysis Reveals a Multi-Layered Organization of Glioblastoma. Cell 2024, 187, 2485–2501.e26. [Google Scholar] [CrossRef]
- Lu, C.; Kang, T.; Zhang, J.W.; Yang, K. Combined Targeting of Glioblastoma Stem Cells of Different Cellular States Disrupts Malignant Progression. Nat. Commun. 2025, 16, 2974. [Google Scholar] [CrossRef]
- Petukhov, V.; Xu, R.J.; Soldatov, R.A.; Cadinu, P. Cell Segmentation in Imaging-Based Spatial Transcriptomics. Nat. Biotechnol. 2022, 40, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Navya, K.T.; Prakash, N.; Prasad, K.; Singh, B.M.K. Red Blood Cell Segmentation and Classification Using Hybrid Image Processing-Squeezenet Model. Multimed. Tools Appl. 2025. [Google Scholar] [CrossRef]
- Li, B.; Liu, F.; Lv, B.; Zhang, Y.; Gou, F.; Wu, J. Cytopathology Image Analysis Method Based on High-Resolution Medical Representation Learning in Medical Decision-Making System. Complex Intell. Syst. 2024, 10, 4253–4274. [Google Scholar] [CrossRef]
- Shi, L.; Feng, X.; Yue, M.; Song, D. Fluorescence In Situ Hybridization Cell Image Segmentation Method. Stud. Health Technol. Inform. 2023, 308, 216–224. [Google Scholar] [PubMed]
- Du, J.L.; Zhang, Y.Q.; Jin, X.Y.; Zhang, X. A Cell Image Segmentation Method Based on Edge Feature Residual Fusion. Methods 2023, 219, 111–118. [Google Scholar] [CrossRef]
- Nair, R.P.H.; Menon, R.; Kemkemer, R. Generalised Image Processing Method for Quantitative Analysis of Nucleus, Cell and Focal Adhesion Clusters. Curr. Dir. Biomed. Eng. 2021, 7, 558–561. [Google Scholar] [CrossRef]
- He, G.S.; Shi, L.L.; Zou, S.S.; Hong, H.H.; Wen, L.H.; Shi, Z.Z. Research on Mesenchymal Stem Cells Segmentation Based on Adaptive Threshold. J. Electron. Meas. Instrum. 2019, 33, 18–23. [Google Scholar]
- Shen, D.H.; E, X.; Zhang, L.C.; Yan, Q.; Hou, J. An Edge Detection Algorithm Based on Gradient for Cell Image. Inf. Technol. 2018, 6–9. [Google Scholar] [CrossRef]
- Hu, Y.B.; Liu, J.P. Cell Image Segmentation of Gastric Cancer Based on Region-Growing and Watersheds. Comput. Digit. Eng. 2006, 34, 151–153. [Google Scholar] [CrossRef]
- Greenwald, N.F.; Miller, G.; Moen, E.; Kong, A.; Kagel, A.; Dougherty, T.; Fullaway, C.C.; McIntosh, B.J.; Leow, K.X.; Schwartz, M.S.; et al. Whole-Cell Segmentation of Tissue Images with Human-Level Performance Using Large-Scale Data Annotation and Deep Learning. Nat. Biotechnol. 2022, 40, 555–565. [Google Scholar] [CrossRef]
- Tiryaki, V.M.; Ayres, V.M.; Ahmed, I.; Shreiber, D.I. Sub-Micro Scale Cell Segmentation Using Deep Learning. Cytometry A 2022, 101, 507–520. [Google Scholar] [CrossRef]
- Pirayesh, Z.; Mohammad-Rahimi, H.; Ghasemi, N.; Motamedian, S.R.; Sadeghi, T.S.; Koohi, H.; Rokhshad, R.; Lotfi, S.M.; Najafi, A.; Alajaji, S.A.; et al. Deep Learning-Based Image Classification and Segmentation on Digital Histopathology for Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. J. Oral Pathol. Med. 2024, 53, 551–566. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. arXiv 2017, arXiv:1703.06870. [Google Scholar]
- Johnson, J. Adapting Mask-RCNN for Automatic Nucleus Segmentation. arXiv 2018, arXiv:1805.00500. [Google Scholar]
- Jung, H.; Lodhi, B.; Kang, J. An Automatic Nuclei Segmentation Method Based on Deep Convolutional Neural Networks for Histopathology Images. BMC Biomed. Eng. 2019, 1, 24. [Google Scholar] [CrossRef]
- Fujita, S.; Han, X.H. Cell Detection and Segmentation in Microscopy Images with Improved Mask R-CNN. In Computer Vision—ACCV 2020 Workshops; Sato, I., Han, B., Eds.; Lecture Notes in Computer Science series 12628; Springer Nature: Berlin, Germany, 2020; Volume 12627, pp. 58–70. [Google Scholar]
- Bancher, B.; Mahbod, A.; Ellinger, I. Improving Mask R-CNN for Nuclei Instance Segmentation in Hematoxylin & Eosin-Stained Histological Images. In Proceedings of the MICCAI Workshop on Computational Pathology, Strasbourg, France, 27 September–1 October 2021; Atzori, M., Burlutskiy, N., Ciompi, F., Zhang, L., Minhas, F., Müller, H., Peng, T., Rajpoot, N., Torben-Nielsen, B., van der Laak, J., et al., Eds.; PMLR: New York, NY, USA, 2021; Volume 156, pp. 20–35. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Chen, W.; Song, H.; Dai, C.; Huang, Z.; Wu, A.; Shan, G.; Liu, H.; Jiang, A.; Liu, X.; Ru, C.; et al. CP-Net: Instance-Aware Part Segmentation Network for Biological Cell Parsing. Med. Image Anal. 2024, 97, 103243. [Google Scholar] [CrossRef]
- Naji, H.; Sancéré, L.; Simon, A.G.; Büttner, R.; Eich, M.-L.; Lohneis, P.; Bożek, K. HoLy-Net: Segmentation of Histological Images of Diffuse Large B-Cell Lymphoma. Comput. Biol. Med. 2024, 170, 107978. [Google Scholar] [CrossRef] [PubMed]
- Salem, D.; Li, Y.; Xi, P.; Phenix, H.; Cuperlovic-Culf, M.; Kærn, M. YeastNet: Deep-Learning-Enabled Accurate Segmentation of Budding Yeast Cells in Bright-Field Microscopy. Appl. Sci. 2021, 11, 2692. [Google Scholar] [CrossRef]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLO, Version 8.0.0; Ultralytics: Frederick, MD, USA, 2023.
- Lin, T.-Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Ghaisi, G.; Lin, T.-Y.; Pang, R.; Le, Q.V. NAS-FPN: Learning Scalable Feature Pyramid Architecture for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 7034–7043. [Google Scholar]
- Kang, M.; Ting, C.-M.; Ting, F.F.; Phan, R.C.-W. ASF-YOLO: A Novel YOLO Model with Attentional Scale Sequence Fusion for Cell Instance Segmentation. Image Vision Comput. 2024, 147, 105057. [Google Scholar] [CrossRef]
- Wu, X.H. Glioma Grade Discrimination Based on Class Imbalance Learning with Terahertz Spectral Data. Infrared Phys. Technol. 2025, 147, 105809. [Google Scholar] [CrossRef]
- Li, G.W.; Jin, Y.P.; Sheng, M.F. The miR-192/EGR1-HOXB9 Loop Inhibits Immune Evasion in Glioma by Arresting Their NSC Phenotypes. Int. Immunopharmacol. 2025, 152, 114453. [Google Scholar] [CrossRef]
- Jin, P.; Bai, X. Exploring the Roles and Clinical Potential of Exosome-Derived Non-Coding RNAs in Glioma. IBRO Neurosci. Rep. 2025, 18, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhao, W.; Zhang, L.; Wu, S.; Xue, S.; Cao, H.; Xu, B.; Li, X.; Hu, N.; Jiang, T.; et al. Centromere Protein U Mediates the Ubiquitination and Degradation of RPS3 to Facilitate Temozolomide Resistance in Glioblastoma. Drug Resist. Updat. 2025, 80, 101214. [Google Scholar] [CrossRef] [PubMed]
- Maza, E.J.L.; Pavoni, J.F.; Leoni, R.F. Early and Late Structural Brain Changes After Radiation Therapy: An MRI Study in Glioma Patients. J. Neuro-Oncol. 2025, 173, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Meehan, B.; Adnani, L.; Zhu, X.; Tawil, N.; Garnier, D.; Nakano, I.; Huang, S.; Rak, J. Curative Timed NK Cell-Based Immunochemotherapy Aborts Brain Tumour Recurrence Driven by Mesenchymal Glioma Stem Cells. Acta Neuropathol. Commun. 2025, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Dimou, J. Akt Inhibition Is Effective Against PTEN-Deleted, Chemoirradiation-Resistant Glioblastoma Stem Cells. Growth Factors 2025, 43, 20–36. [Google Scholar] [CrossRef]
- Poorva, P.; Mast, J.; Cao, B.; Pollok, K.E.; Shen, J. Killing the Killers: Natural Killer Cell Therapy Targeting Glioma Stem Cells in High-Grade Glioma. Mol. Ther. 2025, 33, 2462–2478. [Google Scholar] [CrossRef]
- Lin, J. Glioma Stem Cell Membrane-Camouflaged Photothermal Nanozyme for Synergistic Antitumor via Dual-Targeted Drug Delivery Across Blood-Brain Barrier. Chem. Eng. J. 2025, 507, 160181. [Google Scholar] [CrossRef]
- Zhang, X. AKConv: Convolutional Kernel with Arbitrary Sampled Shapes and Arbitrary Number of Parameters. arXiv 2023, arXiv:2311.11587. [Google Scholar]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10781–10790. [Google Scholar]
- Woo, S.; Debnath, S.; Hu, R.; Chen, X.; Liu, Z.; Kweon, I.S.; Xie, S. ConvNeXt V2: Co-Designing and Scaling ConvNets with Masked Autoencoders. arXiv 2023, arXiv:2301.00808. [Google Scholar]
- Wang, X.; Zhang, R.; Kong, T.; Li, L.; Shen, C. SOLOv2: Dynamic, Faster and Stronger. arXiv 2020, arXiv:2003.10152. [Google Scholar]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. YOLACT: Real-Time Instance Segmentation. arXiv 2019, arXiv:1904.02689. [Google Scholar]


| Training Parameters | Numeric |
|---|---|
| Image size | 640 × 640 |
| Batch size | 16 |
| Epoch | 200 |
| SDG momentum | 0.9 |
| SDG initial learning rate | 0.001 |
| SDG weight decay | 0.0005 |
| Model | Box | Mask | Param (KB) | Weight (MB) | FPS | ||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | mAP50 | Precision | Recall | mAP50 | ||||
| yolov8s-seg | 0.934 | 0.9 | 0.923 | 0.933 | 0.895 | 0.917 | 11,780 | 23.9 | 13 |
| YOLOv8s-seg + Soft-NMS | 0.942 | 0.91 | 0.934 | 0.941 | 0.903 | 0.93 | 11,993 | 23.9 | 13.68 |
| YOLOv8s-seg + BiFPN + Soft-NMS | 0.953 | 0.923 | 0.947 | 0.95 | 0.905 | 0.944 | 12,035 | 22.1 | 19.84 |
| YOLOv8s-seg + BiFPN + Soft-NMS + Akconv | 0.962 | 0.913 | 0.957 | 0.956 | 0.906 | 0.95 | 10,364 | 21.1 | 38 |
| Number of Layers | Box | Mask | Param (KB) | Weight (MB) | FLOPs (G) | FPS | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | mAP50 | Precision | Recall | mAP50 | |||||
| 3 | 0.944 | 0.899 | 0.93 | 0.94 | 0.891 | 0.927 | 10,953 | 22.2 | 43.2 | 22.63 |
| 4 | 0.946 | 0.904 | 0.939 | 0.942 | 0.893 | 0.933 | 10,920 | 22.2 | 42.8 | 22.94 |
| 5 | 0.953 | 0.923 | 0.947 | 0.95 | 0.905 | 0.944 | 10,887 | 22.1 | 42.6 | 19.84 |
| Location | Box | Mask | Param (KB) | Weight (MB) | FLOPs (G) | FPS | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | mAP50 | Precision | Recall | mAP50 | |||||
| Backbone | 0.941 | 0.888 | 0.93 | 0.941 | 0.881 | 0.925 | 10,480 | 21.3 | 41.9 | 17 |
| neck | 0.943 | 0.899 | 0.937 | 0.942 | 0.895 | 0.933 | 10,561 | 21.4 | 42.4 | 35.34 |
| Backbone + neck | 0.883 | 0.824 | 0.888 | 0.877 | 0.812 | 0.871 | 10,487 | 20.2 | 40.8 | 24.04 |
| Model | Box | Mask | Param (KB) | FLOPs (G) | Weight (MB) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | mAP50 (%) | Precision (%) | Recall (%) | mAP50 (%) | ||||
| YOLOv3tiny-seg | 0.642 | 0.566 | 0.615 | 0.403 | 0.404 | 0.345 | 14,099 | 32.7 | 28.4 |
| YOLOv5s-seg | 0.903 | 0.867 | 0.907 | 0.894 | 0.846 | 0.888 | 9765 | 37.8 | 19.9 |
| YOLOv5n-seg | 0.81 | 0.657 | 0.754 | 0.784 | 0.6 | 0.702 | 2755 | 11.0 | 5.8 |
| YOLOv8s-seg | 0.934 | 0.9 | 0.923 | 0.933 | 0.895 | 0.917 | 11,780 | 42.4 | 23.9 |
| YOLOv8n-seg | 0.936 | 0.821 | 0.895 | 0.932 | 0.806 | 0.883 | 3258 | 12.0 | 6.8 |
| YOLOv10n-seg | 0.954 | 0.742 | 0.852 | - | - | - | 2695 | 6.7 | 5.8 |
| YOLOv10s-seg | 0.954 | 0.75 | 0.86 | - | - | - | 8036 | 21.6 | 16.5 |
| YOLOv11s-seg | 0.934 | 0.889 | 0.919 | 0.925 | 0.882 | 0.911 | 10,067 | 35.5 | 20.5 |
| (Ours) | 0.962 | 0.913 | 0.957 | 0.956 | 0.906 | 0.95 | 10,364 | 41.4 | 21.1 |
| Model | Param (KB) | Box | Mask | ||
|---|---|---|---|---|---|
| Precision | mAP50 | Precision | mAP50 | ||
| Mask R-CNN | 43,997 | 0.776 | 0.715 | 0.737 | 0.691 |
| ConvNeXt-V2 | 28,676 | 0.732 | 0.699 | 0.697 | 0.631 |
| SOLO | 45,925 | - | - | 0.698 | 0.457 |
| SOLOv2 | 46,299 | - | - | 0.683 | 0.564 |
| YOLACT | 47,365 | 0.687 | 0.592 | 0.651 | 0.464 |
| YOLOv8s-seg | 11,780 | 0.934 | 0.923 | 0.933 | 0.917 |
| ASF-YOLO | 11,957 | 0.941 | 0.923 | 0.93 | 0.914 |
| (Ours) | 10,364 | 0.962 | 0.957 | 0.956 | 0.95 |
| Model | Box | Mask | Param (KB) | FLOPs (G) | ||||
|---|---|---|---|---|---|---|---|---|
| Precision | Recall | mAP50 | Precision | Recall | mAP50 | |||
| YOLOv3tiny-seg | 0.924 | 0.839 | 0.894 | 0.792 | 0.71 | 0.716 | 14,099 | 32.7 |
| YOLOv5s-seg | 0.919 | 0.852 | 0.922 | 0.913 | 0.864 | 0.906 | 9765 | 37.8 |
| YOLOv5n-seg | 0.914 | 0.841 | 0.898 | 0.911 | 0.856 | 0.899 | 2755 | 11.0 |
| YOLOv8s-seg | 0.936 | 0.863 | 0.919 | 0.926 | 0.85 | 0.91 | 11,780 | 42.4 |
| YOLOv8n-seg | 0.925 | 0.843 | 0.905 | 0.917 | 0.831 | 0.895 | 3258 | 12.0 |
| YOLOv10n-seg | 0.911 | 0.853 | 0.915 | - | - | - | 2694 | 6.7 |
| YOLOv10s-seg | 0.922 | 0.869 | 0.925 | - | - | - | 8036 | 24.4 |
| YOLOv11s-seg | 0.937 | 0.876 | 0.928 | 0.932 | 0.864 | 0.915 | 10,067 | 35.5 |
| YOLOv8s-ASF | 0.941 | 0.891 | 0.923 | 0.93 | 0.863 | 0.914 | 11,957 | 44.3 |
| (Ours) | 0.949 | 0.875 | 0.943 | 0.929 | 0.865 | 0.935 | 10,365 | 41.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, Y.; Li, Z.; Song, Y.; Chen, S.; Mao, Z.; Liu, Z.; Liao, G.; Nie, L. A Real-Time Cell Image Segmentation Method Based on Multi-Scale Feature Fusion. Bioengineering 2025, 12, 843. https://doi.org/10.3390/bioengineering12080843
Zhang X, Zhang Y, Li Z, Song Y, Chen S, Mao Z, Liu Z, Liao G, Nie L. A Real-Time Cell Image Segmentation Method Based on Multi-Scale Feature Fusion. Bioengineering. 2025; 12(8):843. https://doi.org/10.3390/bioengineering12080843
Chicago/Turabian StyleZhang, Xinyuan, Yang Zhang, Zihan Li, Yujiao Song, Shuhan Chen, Zhe Mao, Zhiyong Liu, Guanglan Liao, and Lei Nie. 2025. "A Real-Time Cell Image Segmentation Method Based on Multi-Scale Feature Fusion" Bioengineering 12, no. 8: 843. https://doi.org/10.3390/bioengineering12080843
APA StyleZhang, X., Zhang, Y., Li, Z., Song, Y., Chen, S., Mao, Z., Liu, Z., Liao, G., & Nie, L. (2025). A Real-Time Cell Image Segmentation Method Based on Multi-Scale Feature Fusion. Bioengineering, 12(8), 843. https://doi.org/10.3390/bioengineering12080843







