Two-Dimensional Latent Space Manifold of Brain Connectomes Across the Spectrum of Clinical Cognitive Decline
Abstract
1. Introduction
1.1. Motivation and Objectives
- We propose a GNN-based deep learning framework that reveals a two-dimensional manifold of brain connectomes, capturing the continuum of clinical cognitive decline.
- We show that the learned manifold aligns with established clinical and anatomical patterns of dementia, offering an interpretable and neurologically grounded representation of disease progression.
- We find that the low-dimensional structure of cognitive decline reflects complex yet consistent alterations across the clinical cognitive decline spectrum.
1.2. Paper Structure
2. Materials and Methods
2.1. Brain Networks
2.2. Dataset
2.3. Brain Connectome Construction
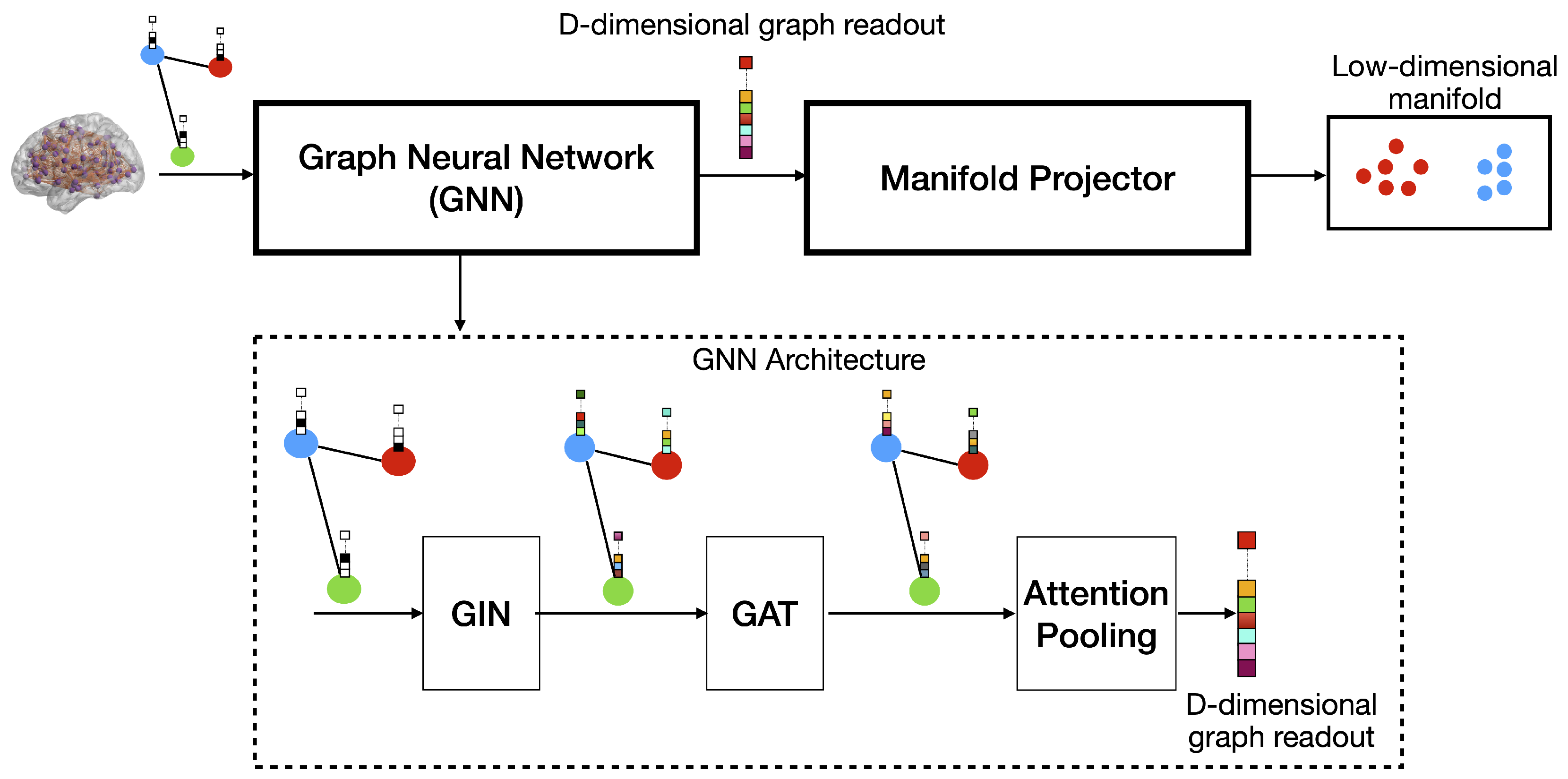
2.4. Graph Neural Networks
2.5. AI-Assisted Editing
3. Proposed Framework: Attention-Guided Graph Embedding and Manifold Projection
4. Results
4.1. Low-Dimensional Manifold of Brain Connectomes
4.2. Classifying Different Stages of Dementia
4.3. Ablation Study
- Substituting the attention-based graph readout mechanism with a simpler aggregation function, such as sum-pooling;
- Removing the graph attention (GAT) layer entirely;
- Replacing the Graph Isomorphism Network (GIN) layer with a Graph Convolutional Network (GCN);
- Eliminating the GIN layer altogether.
5. Discussion
5.1. Cohort Composition and Methodological Considerations
5.2. Architectural Insights
5.3. Two-Dimensional Manifold Structure
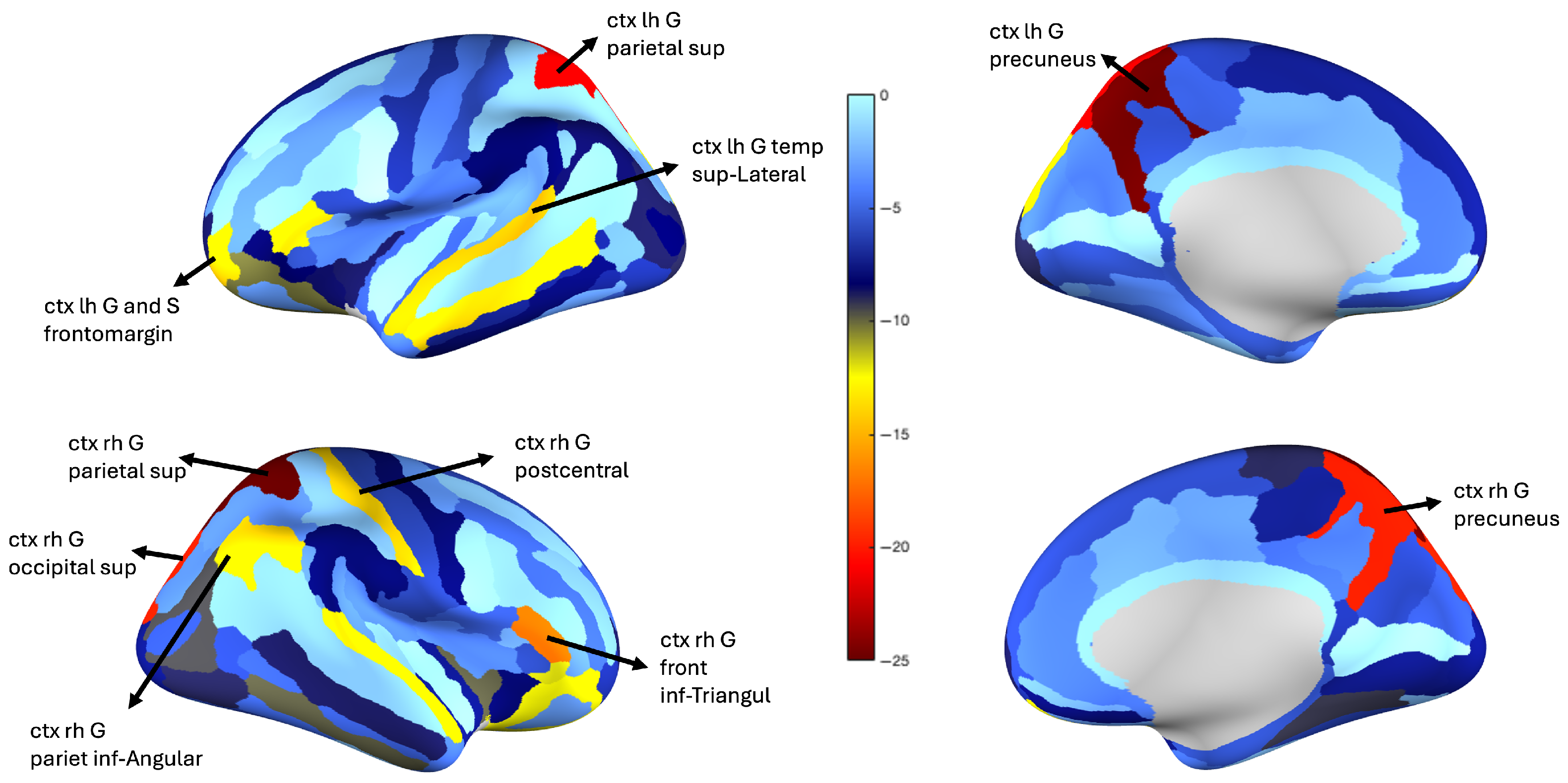
5.4. Neurological Perspective
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADD | Alzheimer’s Disease and Dementia |
| AUC | area under curve |
| BOLD | Blood-Oxygen-Level Dependent |
| CDR | Clinical Dementia Rating |
| CFI-S | Cognitive Functions Instrument—Subject form |
| CFI-SP | Cognitive Functions Instrument—Study Partner form |
| CI | cue index |
| CNN | Convolutional Neural Network |
| DAN | dorsal attention network |
| DMN | default mode network |
| DWI | diffusion-weighted imaging |
| EMCI | Early Mild Cognitive Impairment |
| EPI | Echo Planar Imaging |
| FA | fractional anisotropy |
| FCSRT | Free and Cued Selective Reminding Test |
| FFE | Fast Field Echo |
| fMRI | functional magnetic resonance imaging |
| fNets | functional networks |
| FOV | Field of View |
| GAT | Graph Attention Network |
| GCN | Graph Convolutional Network |
| GIN | Graph Isomorphism Network |
| GNN | Graph Neural Network |
| LLE | locally linear embedding |
| LOOCV | leave-one-out cross validation |
| LSTM | Long Short-Term Memory |
| MCI | Mild Cognitive Impairment |
| MLP | multi-layer perceptron |
| MRI | magnetic resonance imaging |
| NIA-AA | National Institute on Aging and Alzheimer’s Association |
| PC | Principal Component |
| PC1 | First Principal Component |
| PCA | Principal Component Analysis |
| RK4 | fourth-order Runge–Kutta |
| RNN | Recurrent Neural Network |
| ROC | receiver operating characteristic |
| rs-fMRI | resting-state functional magnetic resonance imaging |
| SCI | Subjective Cognitive Impairment |
| sNets | structural networks |
| SOB | sum of boxes |
| TE | Echo Time |
| TFE | Turbo Field Echo |
| TFR | total free recall |
| TR | Repetition Time |
| UMAP | Uniform Manifold Approximation and Projection |
| WL | Weisfeiler–Lehman |
References
- Morris, J.C. Early-Stage and Preclinical Alzheimer Disease. Alzheimer Dis. Assoc. Disord. 2005, 19, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O.; Tononi, G.; Kötter, R. The Human Connectome: A Structural Description of the Human Brain. PLoS Comput. Biol. 2005, 1, e42. [Google Scholar] [CrossRef]
- Yu, M.; Sporns, O.; Saykin, A.J. The human connectome in Alzheimer disease - relationship to biomarkers and genetics. Nat. Rev. Neurol. 2021, 17, 545–563. [Google Scholar] [CrossRef]
- Pievani, M.; Filippini, N.; van den Heuvel, M.P.; Cappa, S.F.; Frisoni, G.B. Brain connectivity in neurodegenerative diseases–from phenotype to proteinopathy. Nat. Rev. Neurol. 2014, 10, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Arigita, E.J.; Schoonheim, M.M.; Damoiseaux, J.S.; Rombouts, S.A.R.B.; Maris, E.; Barkhof, F.; Scheltens, P.; Stam, C.J. Loss of ‘Small-World’ Networks in Alzheimer’s Disease: Graph Analysis of fMRI Resting-State Functional Connectivity. PLoS ONE 2010, 5, e13788. [Google Scholar] [CrossRef]
- Sun, Y.; Yin, Q.; Fang, R.; Yan, X.; Wang, Y.; Bezerianos, A.; Tang, H.; Miao, F.; Sun, J. Disrupted Functional Brain Connectivity and Its Association to Structural Connectivity in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease. PLoS ONE 2014, 9, e96505. [Google Scholar] [CrossRef]
- Dai, Z.; Yan, C.; Li, K.; Wang, Z.; Wang, J.; Cao, M.; Lin, Q.; Shu, N.; Xia, M.; Bi, Y.; et al. Identifying and Mapping Connectivity Patterns of Brain Network Hubs in Alzheimer’s Disease. Cereb. Cortex 2014, 25, 3723–3742. [Google Scholar] [CrossRef]
- Xiang, J.; Guo, H.; Rui, C.; Liang, H.; Chen, J. An abnormal resting-state functional brain network indicates progression towards Alzheimer’s disease. Neural Regen. Res. 2013, 8, 2789–2799. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Guo, X.; Ye, C.; Gao, N.; Fang, Y.; Ma, H.T. A Novel Multimodal MRI Analysis for Alzheimer’s Disease Based on Convolutional Neural Network. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 754–757. [Google Scholar] [CrossRef]
- Zamani, J.; Jafadideh, A.T. Predicting the conversion from mild cognitive impairment to Alzheimer’s disease using graph frequency bands and functional connectivity-based features. Res. Sq. 2024; preprints. [Google Scholar] [CrossRef]
- Durusoy, G.; Yıldırım, Z.; Dal, D.Y.; Ulasoglu-Yildiz, C.; Kurt, E.; Bayır, G.; Özacar, E.; Özarslan, E.; Demirtaş-Tatlıdede, A.; Bilgiç, B.; et al. B-Tensor: Brain Connectome Tensor Factorization for Alzheimer’s Disease. IEEE J. Biomed. Health Inform. 2021, 25, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Durusoy, G.; Karaaslanlı, A.; Dal, D.Y.; Yıldırım, Z.; Acar, B. Multi-modal brain tensor factorization: Preliminary results with AD patients. In Proceedings of the International Workshop on Connectomics in Neuroimaging, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 29–37. [Google Scholar]
- Long, Y.; Wu, M.; Liu, Y.; Fang, Y.; Kwoh, C.K.; Chen, J.; Luo, J.; Li, X. Pre-training graph neural networks for link prediction in biomedical networks. Bioinformatics 2022, 38, 2254–2262. [Google Scholar] [CrossRef]
- Lu, D.; Zhang, Q.; Zheng, C.; Li, J.; Yin, Z. DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction. Bioengineering 2024, 11, 1132. [Google Scholar] [CrossRef]
- Panaccione, F.P.; Mongardi, S.; Masseroli, M.; Pinoli, P. BioGAN: Enhancing Transcriptomic Data Generation with Biological Knowledge. Bioengineering 2025, 12, 658. [Google Scholar] [CrossRef]
- Kim, B.H.; Ye, J.C. Understanding Graph Isomorphism Network for rs-fMRI Functional Connectivity Analysis. Front. Neurosci. 2020, 14, 630. [Google Scholar] [CrossRef]
- Ktena, S.I.; Parisot, S.; Ferrante, E.; Rajchl, M.; Lee, M.; Glocker, B.; Rueckert, D. Metric learning with spectral graph convolutions on brain connectivity networks. Neuroimage 2018, 169, 431–442. [Google Scholar] [CrossRef]
- Chen, W.; Liao, Y.; Dai, R.; Dong, Y.; Huang, L. EEG-based emotion recognition using graph convolutional neural network with dual attention mechanism. Front. Comput. Neurosci. 2024, 18, 1416494. [Google Scholar] [CrossRef]
- Lecha, M.; Cavallo, A.; Dominici, F.; Levi, R.; Bue, A.D.; Isufi, E.; Morerio, P.; Battiloro, C. Directed Semi-Simplicial Learning with Applications to Brain Activity Decoding. arXiv 2025, arXiv:2505.17939. [Google Scholar] [CrossRef]
- Geravanchizadeh, M.; Shaygan Asl, A.; Danishvar, S. Selective Auditory Attention Detection Using Combined Transformer and Convolutional Graph Neural Networks. Bioengineering 2024, 11, 1216. [Google Scholar] [CrossRef]
- Qin, Z.; Li, Y.; Song, X.; Chai, L. Classification of Neuropsychiatric Disorders via Brain-Region-Selected Graph Convolutional Network. IEEE Trans. Neural Syst. Rehabil. Eng. 2025, 33, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zaman, A.; Wang, L.; Yan, J.; Zhu, D. A Cascaded Multi-modality Analysis in Mild Cognitive Impairment. In Proceedings of the Machine Learning in Medical Imaging, Shenzhen, China, 13 October 2019; Suk, H.I., Liu, M., Yan, P., Lian, C., Eds.; Springer: Cham, Switzerland, 2019; pp. 557–565. [Google Scholar]
- Raajasree, K.; Jaichandran, R. Relational Bi-level aggregation graph convolutional network with dynamic graph learning and puzzle optimization for Alzheimer’s classification. Comput. Biol. Med. 2025, 193, 110292. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Li, Q.; Wei, H.; Zhang, M.; Zhan, Y.; Zhou, X.S.; Xue, Z.; Shi, F. Dynamic Spectral Graph Convolution Networks with Assistant Task Training for Early MCI Diagnosis. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 639–646. [Google Scholar] [CrossRef]
- Sahu, B.; Panigrahi, A.; Pati, A.; Das, M.N.; Jain, P.; Sahoo, G.; Liu, H. Novel Hybrid Feature Selection Using Binary Portia Spider Optimization Algorithm and Fast mRMR. Bioengineering 2025, 12, 291. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, C.; Liu, H.; Zhang, Q.; Li, H. An Attention-Based Deep Learning Method for Schizophrenia Patients Classification Using DNA Methylation Data. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 172–175. [Google Scholar] [CrossRef]
- Dai, Y.; Niu, L.; Wei, L.; Tang, J. Feature Selection in High Dimensional Biomedical Data Based on BF-SFLA. Front. Neurosci. 2022, 16, 854685. [Google Scholar] [CrossRef]
- Akgüller, O.; Balcı, M.A.; Cioca, G. Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data. Diagnostics 2025, 15, 153. [Google Scholar] [CrossRef]
- Jiang, H.; Cao, P.; Xu, M.; Yang, J.; Zaiane, O. Hi-GCN: A hierarchical graph convolution network for graph embedding learning of brain network and brain disorders prediction. Comput. Biol. Med. 2020, 127, 104096. [Google Scholar] [CrossRef]
- Li, Y.; Mateos, G.; Zhang, Z. Learning to Model the Relationship Between Brain Structural and Functional Connectomes. IEEE Trans. Signal Inf. Process. Over Netw. 2022, 8, 830–843. [Google Scholar] [CrossRef]
- Gamgam, G.; Kabakcioglu, A.; Yüksel Dal, D.; Acar, B. Disentangled Attention Graph Neural Network for Alzheimer’s Disease Diagnosis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 219–228. [Google Scholar]
- Kim, S.; Kim, M.; Lee, J.E.; Park, B.Y.; Park, H. Prognostic model for predicting Alzheimer’s disease conversion using functional connectome manifolds. Alzheimers. Res. Ther. 2024, 16, 217. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, Q.; Fu, Z.; Zeng, D.; Han, Y.; Li, S. Functional gradients reveal altered functional segregation in patients with amnestic mild cognitive impairment and Alzheimer’s disease. Cereb. Cortex 2023, 33, 10836–10847. [Google Scholar] [CrossRef]
- Sheng, J.; Xin, Y.; Zhang, Q.; Yang, Z.; Wang, L.; Zhang, Q.; Wang, B. Novel Alzheimer’s disease subtypes based on functional brain connectivity in human connectome project. Sci. Rep. 2024, 14, 14821. [Google Scholar] [CrossRef] [PubMed]
- Lamontagne-Caron, R.; Desrosiers, P.; Potvin, O.; Doyon, N.; Duchesne, S. Predicting cognitive decline in a low-dimensional representation of brain morphology. Sci. Rep. 2023, 13, 16793. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Lowe, V.; Graff-Radford, J.; Botha, H.; Barnard, L.; Wiepert, D.; Murphy, M.C.; Murray, M.; Senjem, M.; Gunter, J.; et al. A computational model of neurodegeneration in Alzheimer’s disease. Nat. Commun. 2022, 13, 1643. [Google Scholar] [CrossRef]
- van der Haar, D.; Moustafa, A.; Warren, S.L.; Alashwal, H.; van Zyl, T. An Alzheimer’s disease category progression sub-grouping analysis using manifold learning on ADNI. Sci. Rep. 2023, 13, 10483. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.; Pan, R.; Lee, D.; Thiyyagura, P.; Chen, K.; Alzheimer’s Disease Neuroimaging Initiative. Visualizing Alzheimer’s disease progression in low dimensional manifolds. Heliyon 2019, 5, e02216. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Lemos, R.; Simões, M.R.; Santiago, B.; Santana, I. The free and cued selective reminding test: Validation for mild cognitive impairment and Alzheimer’s disease. J. Neuropsychol. 2015, 9, 242–257. [Google Scholar] [CrossRef]
- Jessen, F.; Wolfsgruber, S.; Wiese, B.; Bickel, H.; Mösch, E.; Kaduszkiewicz, H.; Pentzek, M.; Riedel-Heller, S.G.; Luck, T.; Fuchs, A.; et al. AD dementia risk in late MCI, in early MCI, and in subjective memory impairment. Alzheimer’s Dement. 2014, 10, 76–83. [Google Scholar] [CrossRef]
- Tench, C.; Morgan, P.; Wilson, M.; Blumhardt, L. White matter mapping using diffusion tensor MRI. Magn. Reson. Med. 2002, 47, 967–972. [Google Scholar] [CrossRef]
- Destrieux, C.; Fischl, B.; Dale, A.; Halgren, E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage 2010, 53, 1–15. [Google Scholar] [CrossRef]
- Moyer, D.; Gutman, B.A.; Faskowitz, J.; Jahanshad, N.; Thompson, P.M. Continuous representations of brain connectivity using spatial point processes. Med. Image Anal. 2017, 41, 32–39. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural message passing for Quantum chemistry. In Proceedings of the 34th International Conference on Machine Learning, ICML’17, Sydney, NSW, Australia, 6–11 August 2017; Volume 70, pp. 1263–1272. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How Powerful are Graph Neural Networks? In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019.
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Yun, S.; Jeong, M.; Kim, R.; Kang, J.; Kim, H.J. Graph Transformer Networks. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Wallach, H., Larochelle, H., Beygelzimer, A., d’Alché-Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; Volume 32. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Hu, W.; Liu, B.; Gomes, J.; Zitnik, M.; Liang, P.; Pande, V.; Leskovec, J. Strategies for Pre-training Graph Neural Networks. In Proceedings of the International Conference on Learning Representations, Virtual, 26 April–1 May 2020. [Google Scholar]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Zhang, M.; Cui, Z.; Neumann, M.; Chen, Y. An end-to-end deep learning architecture for graph classification. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence and Thirtieth Innovative Applications of Artificial Intelligence Conference and Eighth AAAI Symposium on Educational Advances in Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; AAAI Press: Washington, DC, USA, 2018. [Google Scholar]
- Vinyals, O.; Bengio, S.; Kudlur, M. Order Matters: Sequence to sequence for sets. In Proceedings of the 4th International Conference on Learning Representations, ICLR 2016, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Li, Y.; Tarlow, D.; Brockschmidt, M.; Zemel, R.S. Gated Graph Sequence Neural Networks. In Proceedings of the 4th International Conference on Learning Representations, ICLR 2016, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Oono, K.; Suzuki, T. Graph Neural Networks Exponentially Lose Expressive Power for Node Classification. In Proceedings of the International Conference on Learning Representations, Virtual, 26 April–1 May 2020. [Google Scholar]
- Cai, C.; Wang, Y. A Note on Over-Smoothing for Graph Neural Networks. arXiv 2020, arXiv:2006.13318. [Google Scholar] [CrossRef]
- Hari, E.; Kizilates-Evin, G.; Kurt, E.; Bayram, A.; Ulasoglu-Yildiz, C.; Gurvit, H.; Demiralp, T. Functional and structural connectivity in the Papez circuit in different stages of Alzheimer’s disease. Clin. Neurophysiol. 2023, 153, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Villemagne, V.L.; Pike, K.E.; Chételat, G.; Ellis, K.A.; Mulligan, R.S.; Bourgeat, P.; Ackermann, U.; Jones, G.; Szoeke, C.; Salvado, O.; et al. Longitudinal assessment of Aβ and cognition in aging and Alzheimer disease. Ann. Neurol. 2011, 69, 181–192. [Google Scholar] [CrossRef]
- Ay, U.; Gürvit, I.H. Alterations in large-scale intrinsic connectivity networks in the Parkinson’s disease-associated cognitive impairment continuum: A systematic review. Noro Psikiyatr. Ars. 2022, 59, S57–S66. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Petersen, R.C.; Caracciolo, B.; Brayne, C.; Gauthier, S.; Jelic, V.; Fratiglioni, L. Mild cognitive impairment: A concept in evolution. J. Intern. Med. 2014, 275, 214–228. [Google Scholar] [CrossRef]
- Mesulam, M.M. Principles of Behavioral and Cognitive Neurology; Oxford University Press: Oxford, UK, 2000. [Google Scholar] [CrossRef]
- Yuksel Dal, D.; Yıldırım, Z.; Gurvit, H.; Kabakçıoğlu, A.; Acar, B. Reorganization of brain connectivity across the spectrum of clinical cognitive decline. Neurol. Sci. 2024, 45, 5719–5730. [Google Scholar] [CrossRef]
- Seeley, W.W.; Crawford, R.K.; Zhou, J.; Miller, B.L.; Greicius, M.D. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009, 62, 42–52. [Google Scholar] [CrossRef]
- Buckner, R.L.; Snyder, A.Z.; Shannon, B.J.; LaRossa, G.; Sachs, R.; Fotenos, A.F.; Sheline, Y.I.; Klunk, W.E.; Mathis, C.A.; Morris, J.C.; et al. Molecular, Structural, and Functional Characterization of Alzheimer’s Disease: Evidence for a Relationship between Default Activity, Amyloid, and Memory. J. Neurosci. 2005, 25, 7709–7717. [Google Scholar] [CrossRef] [PubMed]



| Task | ROC-AUC (Mean ± Std) |
|---|---|
| ADD/MCI | 0.81 ± 0.025 |
| ADD/SCI | 0.93 ± 0.005 |
| MCI/SCI | 0.55 ± 0.048 |
| Method | Task | ROC-AUC (Mean ± Std) |
|---|---|---|
| Baseline Model | ||
| GIN+GAT+Attention Pool | ADD/MCI | 0.81 ± 0.025 |
| ADD/SCI | 0.93 ± 0.005 | |
| Ablated Models | ||
| GIN+GAT+Sum Pool | ADD/MCI | 0.80 ± 0.018 |
| ADD/SCI | 0.90 ± 0.011 | |
| GIN+Attention Pool | ADD/MCI | 0.79 ± 0.014 |
| ADD/SCI | 0.90 ± 0.007 | |
| GCN+GAT+Attention Pool | ADD/MCI | 0.62 ± 0.032 |
| ADD/SCI | 0.78 ± 0.012 | |
| GAT+Attention Pool | ADD/MCI | 0.74 ± 0.010 |
| ADD/SCI | 0.84 ± 0.016 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayır, G.; Yüksel Dal, D.; Harı, E.; Ay, U.; Gurvit, H.; Kabakçıoğlu, A.; Acar, B. Two-Dimensional Latent Space Manifold of Brain Connectomes Across the Spectrum of Clinical Cognitive Decline. Bioengineering 2025, 12, 819. https://doi.org/10.3390/bioengineering12080819
Bayır G, Yüksel Dal D, Harı E, Ay U, Gurvit H, Kabakçıoğlu A, Acar B. Two-Dimensional Latent Space Manifold of Brain Connectomes Across the Spectrum of Clinical Cognitive Decline. Bioengineering. 2025; 12(8):819. https://doi.org/10.3390/bioengineering12080819
Chicago/Turabian StyleBayır, Güneş, Demet Yüksel Dal, Emre Harı, Ulaş Ay, Hakan Gurvit, Alkan Kabakçıoğlu, and Burak Acar. 2025. "Two-Dimensional Latent Space Manifold of Brain Connectomes Across the Spectrum of Clinical Cognitive Decline" Bioengineering 12, no. 8: 819. https://doi.org/10.3390/bioengineering12080819
APA StyleBayır, G., Yüksel Dal, D., Harı, E., Ay, U., Gurvit, H., Kabakçıoğlu, A., & Acar, B. (2025). Two-Dimensional Latent Space Manifold of Brain Connectomes Across the Spectrum of Clinical Cognitive Decline. Bioengineering, 12(8), 819. https://doi.org/10.3390/bioengineering12080819






