1. Introduction
X-rays are the most common imaging technique used in medical diagnostics due to their widespread availability, speed, and cost-effectiveness [
1]. Specifically, chest X-rays (CXRs) are useful diagnostic imaging modalities in clinical practice, which aid medical professionals in detecting and monitoring of a wide range of thoracic diseases. Interpreting these images, however, requires radiological expertise and time, and remains susceptible to inter-reader variability [
2].
In addition to this, the high volume of CXR studies contributes to raising a radiologist workload [
3], increasing the risk of reporting delays and potential oversights, particularly for subtle findings. In resource-limited locations, the shortage of specialized radiologists further complicates timely and accurate diagnoses. These challenges indicate the need for improved solutions to enhance the efficiency, accuracy, and accessibility of CXR medical report generation [
4].
To address these challenges, deep learning systems learn to recognize patterns in medical images, helping detect pathologies like lung and heart diseases [
5]. Instead of relying solely on human experts, the technology can quickly analyze a large volume of X-rays, flagging potential issues for physicians to review.
It is, however, important that the Artificial Intelligence (AI) can also highlight the concerning areas on the image, making it easier for medical professionals to understand its findings, enhancing the clearer communication between the AI system and medical professionals and strengthening the trust in the model [
6].
Despite the progress in AI-driven CXR interpretation, existing solutions often lack a full integration of visual explanation, anatomical relevance, and structured report generation, limiting their clinical adoption.
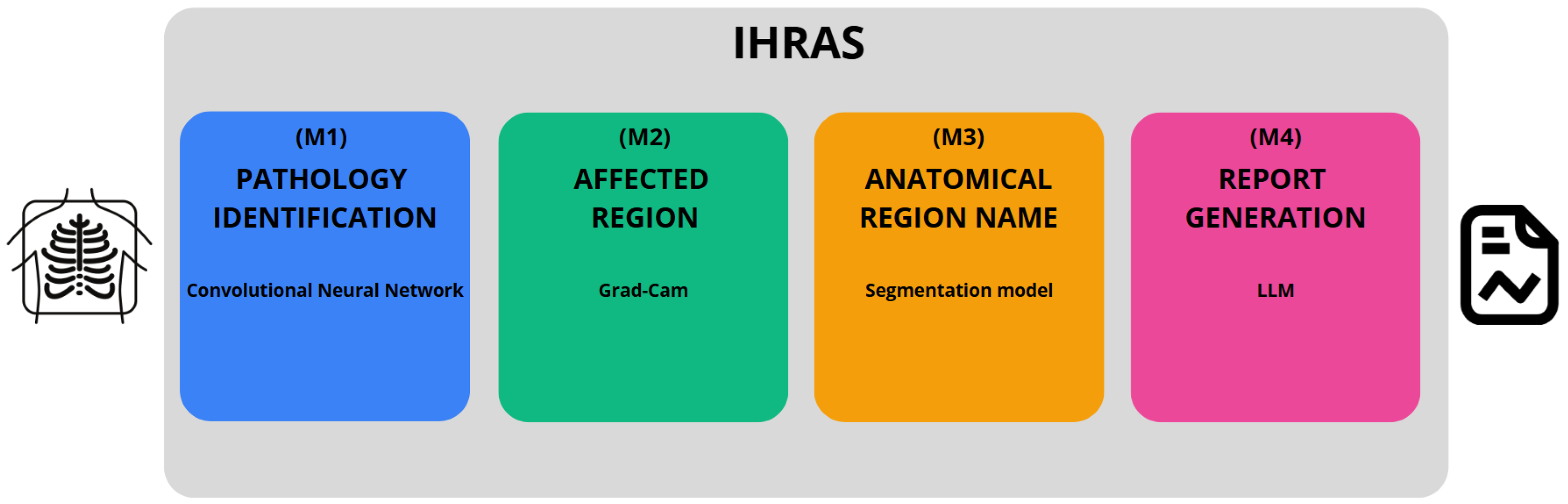
In this study, we introduce the Intelligent Humanized Radiology Analysis System (IHRAS), a modular architecture designed to overcome limitations in chest X-ray interpretation. IHRAS integrates disease classification, visual explanation, anatomical segmentation, and automated medical report generation. Upon receiving a chest X-ray image, the system classifies it into 14 common thoracic diseases using a deep Convolutional Neural Network (CNN). To enhance interpretability, it uses Gradient-weighted Class Activation Mapping (Grad-CAM) to highlight the spatial regions most relevant to the identified conditions. To enhance interpretability, We further segment these regions using a dedicated anatomical segmentation model, assigning them to clinically relevant anatomical structures. Ultimately, these findings are used to condition a Large Language Model (LLM) that generates a human-readable medical report using the Systematized Nomenclature of Medicine—Clinical Terms (SNOMED CT). IHRAS is tested on the NIH CXR dataset, due to its clinical relevance, diversity and standardization.
While IHRAS demonstrates the possibility of automating radiological workflows, auditability is necessary to maintain accountability. The system’s decisions must be interpretable and traceable to allow clinicians to validate and understand AI outputs, mitigating the risk of over-reliance on automated systems.
Another consideration is the generalizability of the proposed system across diverse populations and imaging conditions. CXR datasets often reflect demographic and geographic biases that may lead to suboptimal performance in underrepresented groups [
7]. Therefore, an evaluation across multi-institutional datasets with diverse patient profiles is necessary to ensure consistent performance.
1.1. Contributions and Limitations of the Work
The primary contributions of this work lie in the development of a modular architecture for automated chest X-ray report generation that unifies classification, localization, segmentation, and language modeling. The system utilizes Grad-CAM to produce visual explanations of disease predictions, which are further enhanced by anatomical context through segmentation, thereby improving both interpretability and clinical relevance. This integrated framework represents a significant step forward in radiological image analysis and report generation.
Unlike prior architectures, IHRAS integrates classification, spatial reasoning, anatomical segmentation, and structured language modeling into a unified pipeline, thus providing a more complete approach.
Due to its modular nature, IHRAS may be adapted to generate medical reports beyond CXR, including X-rays of other body parts, such as abdomen and head, as well as other imaging modalities, such as blood cell microscopy and ocular imaging.
As limitation, the pretrained model achieved a modest F1-score for the multilabel classification problem, which, while comparable to other generalist models in the literature, reflects the challenge of diagnosing diverse pathologies simultaneously.
Furthermore, although IHRAS employs Grad-CAM visualizations to provide interpretable predictions, the lack of pixel-level annotations in the dataset prevents quantitative validation of the highlighted regions. Consequently, the accuracy of the attention maps cannot be formally assessed.
1.2. Organization of the Work
The remainder of this paper is structured as follows.
Section 2 reviews the related literature.
Section 3 presents the proposed modular architecture of IHRAS, whilst
Section 4 discusses the results obtained with the approach.
Section 5 concludes the paper.
3. Methodology
Our proposed architecture, developed in Python version 3.11.12, is depicted in
Figure 1, encompassing from the input of a CXR image to the generation of the medical report.
The IHRAS framework consists of four sequential modules. The first module, M1 (see
Figure 1), detects abnormalities in the CXR and is detailed in
Section 3.2. Modules M2 and M3, described in
Section 3.3, localize the regions influencing the classification model’s decision and map them to their corresponding anatomical terms, respectively. Finally, M4 integrates the outputs from M1 and M3, using an LLM to generate the medical report, as discussed in
Section 3.4.
3.1. The Dataset
The NIH CXR dataset, proposed by [
29], is a publicly available dataset containing 112,120 chest X-ray images from 30,805 unique patients, annotated with up to 14 thoracic pathology labels derived from associated radiology reports. It is chosen due to its large scale, multi-label annotations, and open accessibility, which enables reproducible evaluation.
To reduce computational costs and processing time while maintaining a representative sample of the dataset for the models comparison, we employed a stratified random sampling strategy. The population was partitioned into homogeneous strata based on four clinically relevant variables: (i) patient age (grouped into 25-year intervals); (ii) radiological findings; (iii) patient gender; and (iv) radiographic view position.
The sample size per stratum is determined based on Equation (
1), in which
Z represents the Z-score, corresponding to the desired confidence level;
p denotes the estimated proportion; and
e stands for the margin of error. The variable
n represents the sample size, whereas
N is the population size.
We selected 370 samples per stratum to achieve a 95% confidence level with a 5% margin of error for the largest strata (N = 9677). For smaller strata with fewer than 370 samples, all available samples were included. This stratified sampling approach preserves the original dataset’s clinical and demographic diversity. This results in a total of 50,133 selected X-ray images.
3.2. Pathology Identification (M1)
Each inputted X-ray image may be classified as according to different pathologies, namely atelectasis, cardiomegaly, consolidation, edema, effusion, emphysema, fibrosis, hernia, infiltration, mass, nodule, pleural thickening, pneumonia, and pneumothorax. It is a multi label classification problem, that is, each image can be associated with multiple labels simultaneously.
Three pretrained deep learning models are evaluated, and the best performing model is selected for integration into the IHRAS framework, to ensure optimal diagnostic accuracy. These models are obtained from TorchXRayVision [
30], with their default configurations, and refer to a Densely Connected Convolutional Networks (DenseNet) [
31] trained on several CXR datasets; a DenseNet trained specifically on the NIH dataset; and a Residual Neural Network (ResNet) [
32] trained on several chest X-ray datasets. These models are selected due to their high accuracy in pathology classification [
33].
These models are evaluated based on some metrics. The first one is precision, which measures the proportion of correctly predicted positive instances among all predicted positives, as according to Equation (
2), where
,
,
, and
denote True Positives, False Positives, True Negatives, and False Negatives respectively. This metrics should be considered when the cost of false positives is high.
Recall, also known as sensitivity and defined by Equation (
3), quantifies the ability to identify true positives over all positive instances. It is a relevant metric when missing positive cases is undesirable.
The F1-score, determined by Equation (
4), represents the harmonic mean of precision and recall.
Ultimately, specificity, calculated as in Equation (
5), measures the ability to identify true negatives over all negative instances.
3.3. Affected Region and Its Anatomical Name (M2 and M3)
Since medical reports need to have clarity, Grad-CAM is used to enhance explainability, as it has been demonstrated to improve the interpretability of deep learning models in medical imaging [
34]. This technique generates heatmaps that indicate the regions of the image that most strongly influenced the model’s diagnostic conclusions. This spatial alignment with radiological markers allows clinicians to audit whether the model’s decisions are anatomically plausible, complying with requirements for transparency in medical AI.
To further enhance the interpretability of the model’s decisions, a segmentation model is employed to identify the anatomical name of the regions highlighted by Grad-CAM. This ensures adherence to standardized medical reporting, as the medical report references meaningful structures rather than relying on generic image coordinates or non-clinical descriptors.
To achieve this segmentation, we use the Structure-Aware Relation Network (SAR-Net) model proposed by [
35]. The SAR-Net model is evaluated on ChestX-Det, which is a subset of the NIH dataset, used in this work, achieving a Mean Intersection-Over-Union of 86.85%, outperforming comparable models [
36,
37,
38]. The anatomical structures it is trained to identify are the aorta, facies diaphragmatica, heart, left clavicle, left hilus pulmonis, left lung, left scapula, mediastinum, right clavicle, right hilus pulmonis, right lung, right scapula, spine, and weasand.
3.4. Report Generation (M4)
The extracted diagnostic data, comprising identified pathologies with associated probabilities and affected anatomical regions, alongside supplementary inputs such as patient demographics (age, gender) and radiographic projection, is processed by an LLM to generate a medical report. This approach provides a patient-aware report generation.
To select the LLM model for this task, we considered the BRIDGE benchmark, that is composed of 87 tasks based on clinical data from real-world sources, covering nine languages [
39]. The work compared a total of 52 different LLMs using multiple inference approaches, and DeepSeek-R1, an open-source model, achieved the highest score in radiology and ranks among the top 3 in the overall score, along with Gemini-1.5-Pro and GPT-4o. These general-purposed models achieved even better results than the medically fine-tuned ones. Because of these results, we use DeepSeek-R1 to generate the radiological report [
40].
The report generation employs the CRISPE framework (Capacity, Role, Insight, Statement, Personality, Experiment) for structured prompt engineering, which enhances the LLM’s output quality [
41]. This methodology ensures precise instruction by (C) defining the model’s clinical expertise boundaries; (R) establishing a radiologist role; (I) incorporating patient-specific contextual data and diagnosis; (S) specifying reporting requirements with SNOMED CT compliance; (P) maintaining professional tone consistency; and (E) limiting to a single response per prompt. The structure prompt following this framework is shown in
Table 2.
SNOMED CT enhances this architecture with the standardization of clinical terminology, improving data quality through consistent indexing, and with the advance of the continuity of care through interoperable patient records. It also facilitates clinical research with a structured data aggregation, and improves patient safety [
42]. Due to these benefits, the LLM is instructed to use SNOMED CT.
To evaluate the quality of the generated medical reports, we employed DeepEval [
43], an open-source framework for assessing LLM outputs using an LLM-as-a-judge approach. The metrics considered in the assessment are faithfulness, to evaluate whether the generated report is aligned with the findings from the previous IHRAS steps; answer relevancy, to measure how relevant the report is in relation to the prompt; hallucination to detect unsupported claims; toxicity and bias, to identify harmful or discriminatory language; and prompt alignment to assess adherence to the CRISPE-structured instructions.
4. Results and Discussion
This section presents the results obtained with the IHRAS pipeline, from the CXR image input up to the report generation.
4.1. Classification Models Evaluation
For the disease classification module, we evaluate three models, namely a DenseNet trained on multiple CXR datasets, a DenseNet fine-tuned specifically on the NIH dataset, and a ResNet trained across diverse CXR datasets. The best-performing model, based on evaluation metrics, is selected for integration into IHRAS.
4.1.1. Models Comparison
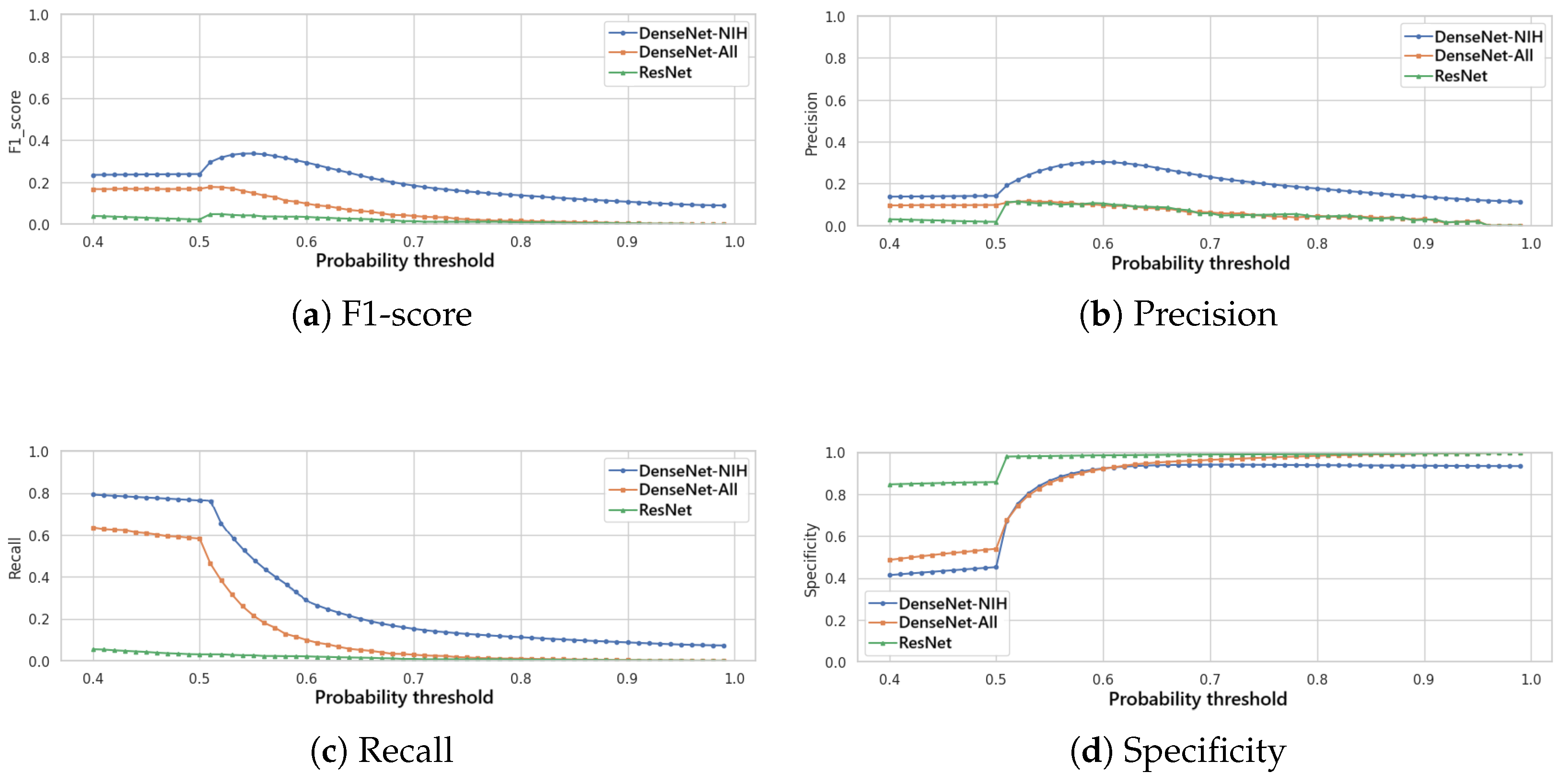
All evaluated models generate a probability score for each detectable pathology in the input X-ray image. To convert these continuous predictions into binary classifications (present/absent), an optimal decision threshold must be established.
Figure 2 presents the variation of the F1-score (
Figure 2a), of the precision (
Figure 2b), of the recall (
Figure 2c) and of the specificity (
Figure 2d) in function to the variation of this threshold for the three classification models. It is noted that the DenseNet trained specifically for the NIH dataset achieves the best F1-score, precision and recall for a given threshold, and, thus, this model is selected for the classification module of IHRAS in this work.
In this study, we employ the F1-score as our primary optimization metric, to balance false positives and false negatives. As seen in
Figure 2a, the optimal decision threshold of 0.55 maximizes the F1-score at 0.34. This threshold reflects a balance of clinical priorities, equitably weighting sensitivity (to avoid missed diagnoses) and precision (to reduce false alarms). The obtained F1-score is comparable to those of other multilabel classification models evaluated on the same and other datasets, as shown in
Table 3, especially considering the long-tailed characteristics of the used dataset [
44].
Although the F1 score achieved by the classification model selected for IHRAS is lower than the best reported in [
46], by Code-Free Deep Learning (CFDL) platforms, the chosen DenseNet offers significant advantages in terms of control and transparency, as users cannot audit the CFDL architecture and the explainability is hindered. Furthermore, the adopted DenseNet architecture outperformed all comparative studies in
Table 3 in at least one performance metric.
In scenarios where false negatives incur higher costs, the threshold should be lowered to prioritize recall, ensuring fewer missed cases. Conversely, when false positives are more detrimental, the threshold should be raised to maximize precision.
Figure 3 presents the normalized confusion matrix of the classification model, in which rows represent the true labels, while columns show predicted classifications. It indicates that the model tends to predict a CXR as atelectasis, infiltration and/or effusion, whilst labels such as pneumonia and hernia are more rarely predicted. This behavior possibly explains the modest F1-score obtained.
4.1.2. Binary Classification
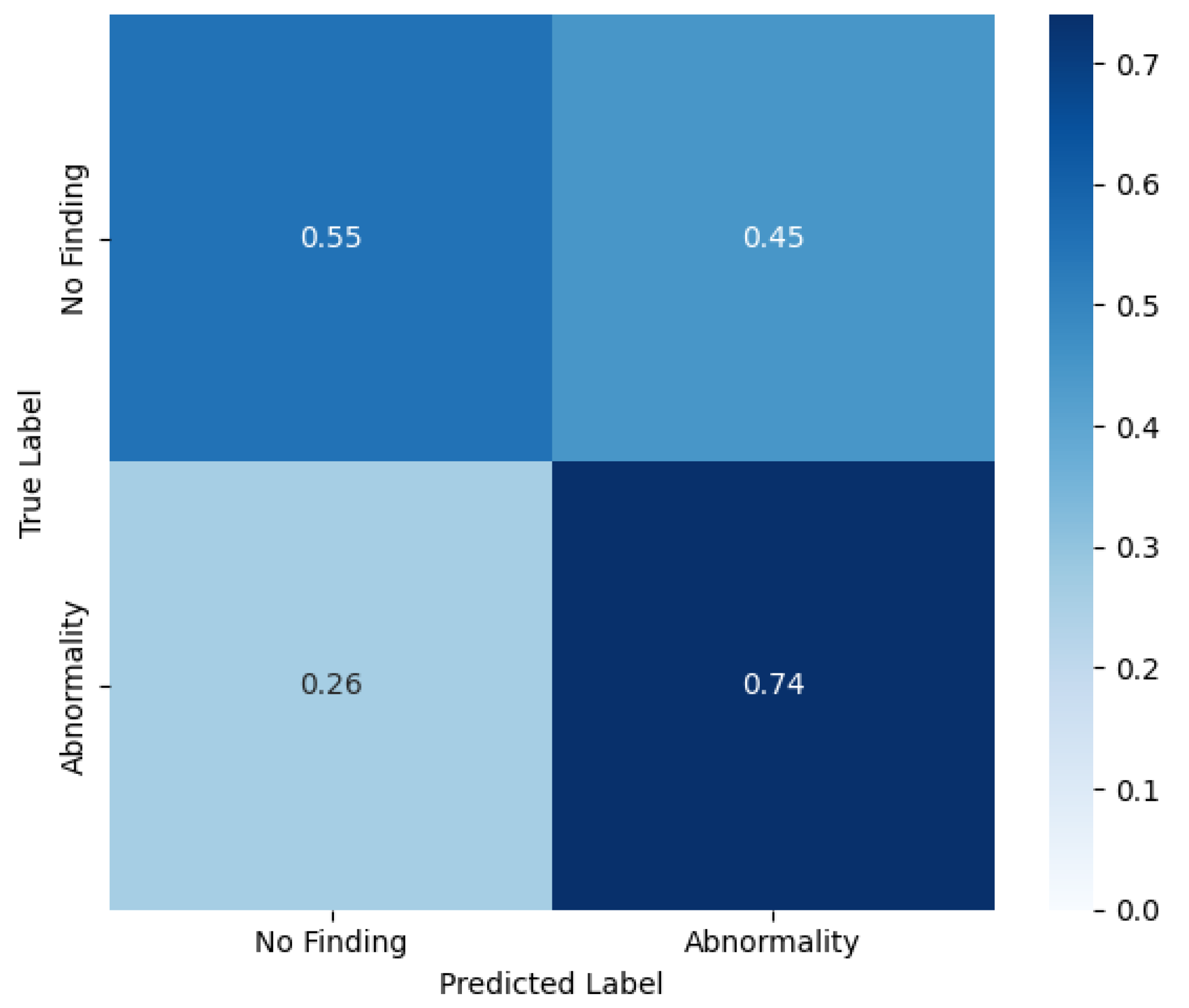
Although IHRAS is designed to identify and differentiate between several thoracic pathologies, we also evaluate its performance in a binary classification task distinguishing between abnormal and normal chest X-rays. In this binary setting, all images exhibiting at least one pathological finding, regardless of the specific diagnosis, are grouped under the abnormal class, whereas those explicitly labeled as no findings constitute the normal class. This evaluation enables the assessment of the model’s capability to serve as a general screening tool, flagging potentially abnormal cases for further review. It also provides insight into the system’s general sensitivity to pathological signals irrespective of specific diagnoses.
In this binary classification, the same probability threshold of 0.55 is used, achieving a precision of 0.93, a recall of 0.74, a specificity of 0.55 and a F1-score of 0.83. The confusion matrix of this scenario is shown in
Figure 4.
These results indicate the adaptability of IHRAS, demonstrating its capability to identify specific thoracic diseases and to function as a screening tool. This dual functionality broadens its potential applications in clinical workflows, from rapid triage to decision support.
4.1.3. Sample Demographics
It has been observed by [
7] that AI models underdiagnose pathologies in marginalized groups, concluding that there is a significant disparity when comparing “black female” patients with “white male”. To assess this disparity within the 0.55 threshold DenseNet model adopted by IHRAS, the demographic and clinical characteristics of the sampled dataset are presented in
Figure 5.
Figure 5a reveals apparent anomalies in the age data, including patients recorded as over 400 years old. These outliers do not compromise the diagnosis, as age is not considered in the analysis, being only included as metadata in the report for informational purposes. The median age for both genders, however, is 50 years, suggesting these extreme outliers are likely artifacts of data entry errors rather than representative of the true distribution. The sampled dataset also contains slightly more males than females (
Figure 5b) and Postero-Anterior (PA) views than Antero-Posterior (AP) (
Figure 5c).
Figure 5d show that the imbalance for the pathologies count is significantly greater, thus suggesting the long-tailed characteristics of the dataset. However, since this section focuses on assessing a pretrained model’s diagnostic performance across diverse subgroups, rather than training a new model, such imbalances reflect clinical variability rather than methodological limitations.
Upon comparing
Figure 3 and
Figure 5d, it is evident that the classification model exhibits a bias toward predicting the most frequently occurring pathologies in the dataset, such as infiltration, effusion, and atelectasis, while underrepresenting rarer conditions like pneumonia and hernia. This pattern suggests that the model’s predictions are influenced by class imbalance, which is a common challenge in classification problems in healthcare and in other domains [
44,
47,
49].
The comparison of the evaluation metrics for these different demographics and clinical characteristics is presented in
Table 4, which indicates no significant disparities between different demographic and clinical groups.
We quantify pairwise differences in model performance using Cohen’s
h effect size measure, computed for all pairs of values within the same category. As shown in
Table 5, the maximum observed
h values across all categories is of 0.1003 (Findings/Recall), which is below Cohen’s threshold of 0.2 for small effects [
50]. This demonstrates statistically negligible variation in model performance across the demographic subgroups, image acquisition parameters and clinical findings.
4.2. Report Generation Evaluation
The metrics associated to the generated medical reports are presented in
Table 6. This evaluation assesses the LLM’s ability to generate accurate reports based on the classification results and on the identified affected anatomical region provided to it, rather than comparing against the actual annotated pathology. The performance of the disease classification model has been validated in
Section 4.1.
Instead, this analysis tests the LLM’s capacity to faithfully translate input clinical data into coherent reports, to maintain contextual relevance, and to adhere to clinical reporting standards.
The results shown in
Table 6 demonstrate capabilities for safe clinical deployment. They indicate that the IHRAS LLM module is capable of operating within the healthcare workflow, where accuracy, safety, and consistency are fundamental.
The faithfulness scores validate that reports accurately reflect diagnostic inputs, ensuring reliability in communicating critical findings, which is a fundamental requirement for medical decision-making. The strong performance in answer relevancy indicates that the reports have successfully fulfilled the prompt, while perfect hallucination, toxicity, and bias scores confirm the absence of fabricated claims and harmful content, addressing patient safety and ethical concerns. The prompt alignment results, though slightly lower than other metrics, still reflect robust adherence to structured clinical reporting standards.
These metrics, however, were obtained through an LLM-as-judge evaluation process, which, while efficient for automated assessment, introduces limitations regarding clinical validity.
4.3. IHRAS Case Studies
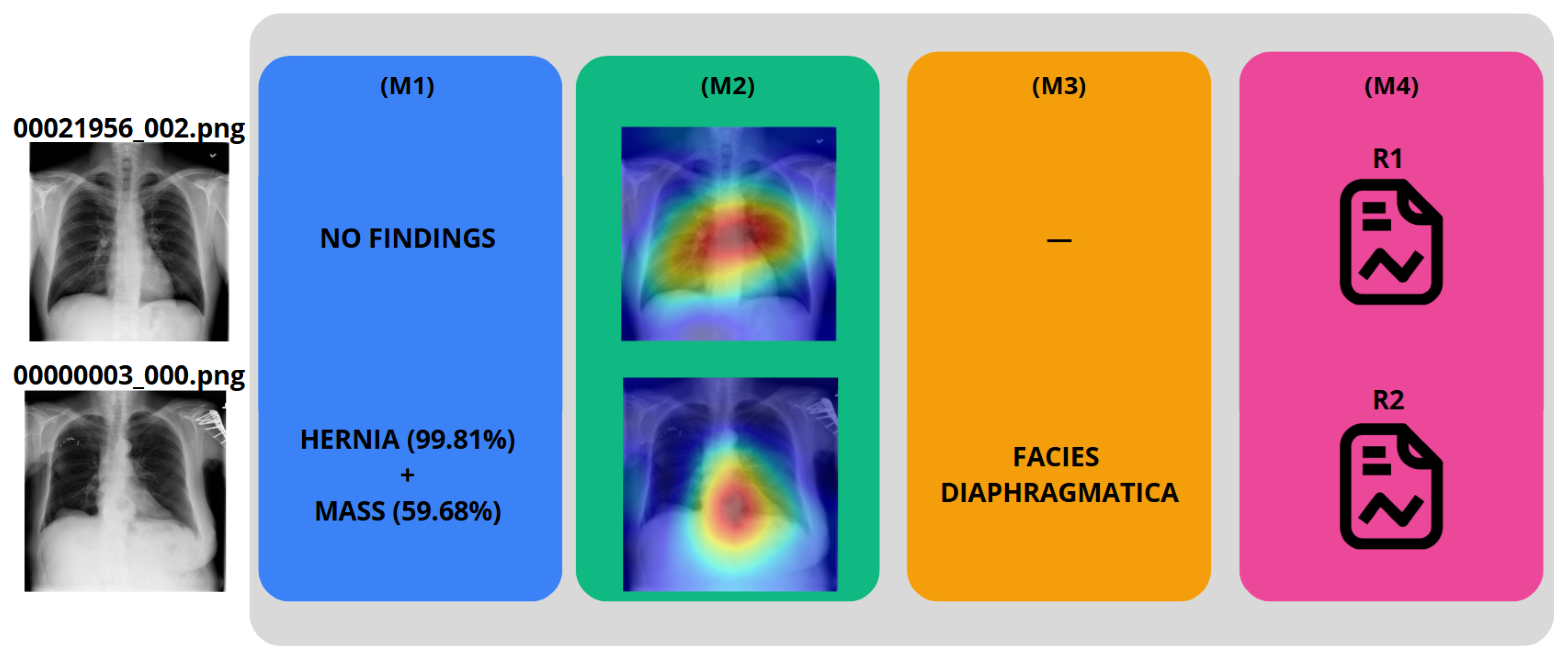
To demonstrate the IHRAS workflow,
Figure 6 depicts two example CXR images from the NIH dataset inputted into the system, with the generated reports R1 and R2 presented in
Table 7.
The CXR 00021956_002.png is correctly classified as a healthy image, whereas it wrongly attributes a mass diagnosis for 00000003_000.png, although the hernia is correctly identified. Such misdiagnosis is taken into account in
Section 4.1.
Additionally, in medical imaging, timely diagnosis is fundamental for effective patient care, particularly in urgent scenarios. Rapid report generation ensures that clinicians receive relevant information without delay. In that regard, IHRAS completes the CXR initial classification in 570 ± 85 ms, generates the GradCAM heatmap generation in 1878 ± 221 ms, performs segmentation in 12,746 ± 713 ms, and generates the medical report generation in 4216 ± 402 ms. These processing times suggest the system’s ability to deliver rapid results.
Since Grad-CAM (M2) visualizations and LLM-generated reports (M4) are conditioned on the model’s predictions (M1), their utility is linked to the quality of those outputs. However, the system’s interpretability components offer importante information regarding model behavior, indicating regions of interest and generating plausible hypotheses that can assist clinicians in decision-making. Rather than presenting these outputs as definitive diagnoses, IHRAS may be used as an assistive tool that complements clinical judgment.
5. Conclusions
This work introduced IHRAS, a modular and integrated system designed to enhance the automation and interpretability of chest X-ray analysis by combining disease classification, visual explainability, anatomical segmentation, and structured medical report generation via Large Language Models. IHRAS identifies thoracic pathologies with associated anatomical relevance and produces clinically coherent reports adhering to SNOMED CT standards.
The system demonstrated consistent performance across demographic and clinical subgroups in the NIH ChestX-ray dataset, indicating its potential for equitable clinical deployment. The integration of Grad-CAM for visual explanations and SAR-Net for anatomical localization contributes significantly to transparency and trust in AI-driven radiology.
However, the system’s performance is currently constrained by limitations, including the modest F1-score in the multi-label classification task, which likely arises from the imbalanced data problem, and the lack of evaluation of the Grad-CAM. Hence, as future work, these modules of the IHRAS architecture should be further studied, with the aim of improving its performance, including clinician-based evaluations of the complete system. Future works should also investigate the use of the proposed architecture in other medical imaging applications, such as Computed Tomography scans, Magnetic Resonance Imaging, and ultrasound, or to other anatomical regions beyond the thorax.