The Casual Associations Between Brain Functional Networks and Fibromyalgia: A Large-Scale Genetic Correlation and Mendelian Randomization Study
Abstract
1. Introduction
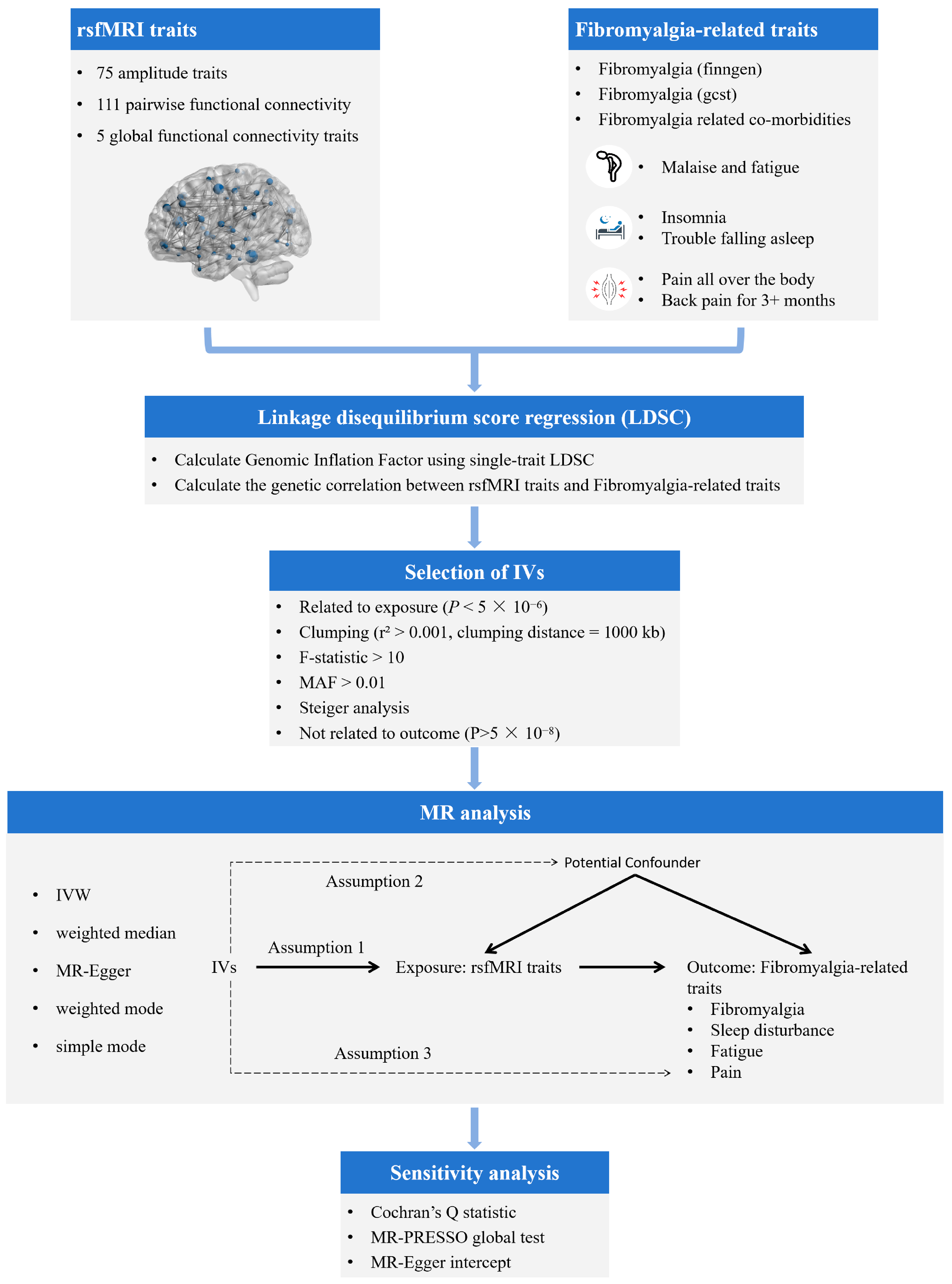
2. Materials and Methods
2.1. Data Source
2.1.1. GWASs of Brain rsfMRI Traits
2.1.2. GWASs of Fibromyalgia-Related Traits
2.2. Selection of Genetic Instruments
2.3. Genetic Correlation Analysis
2.4. Statistical Analysis of MR Study Design
3. Results
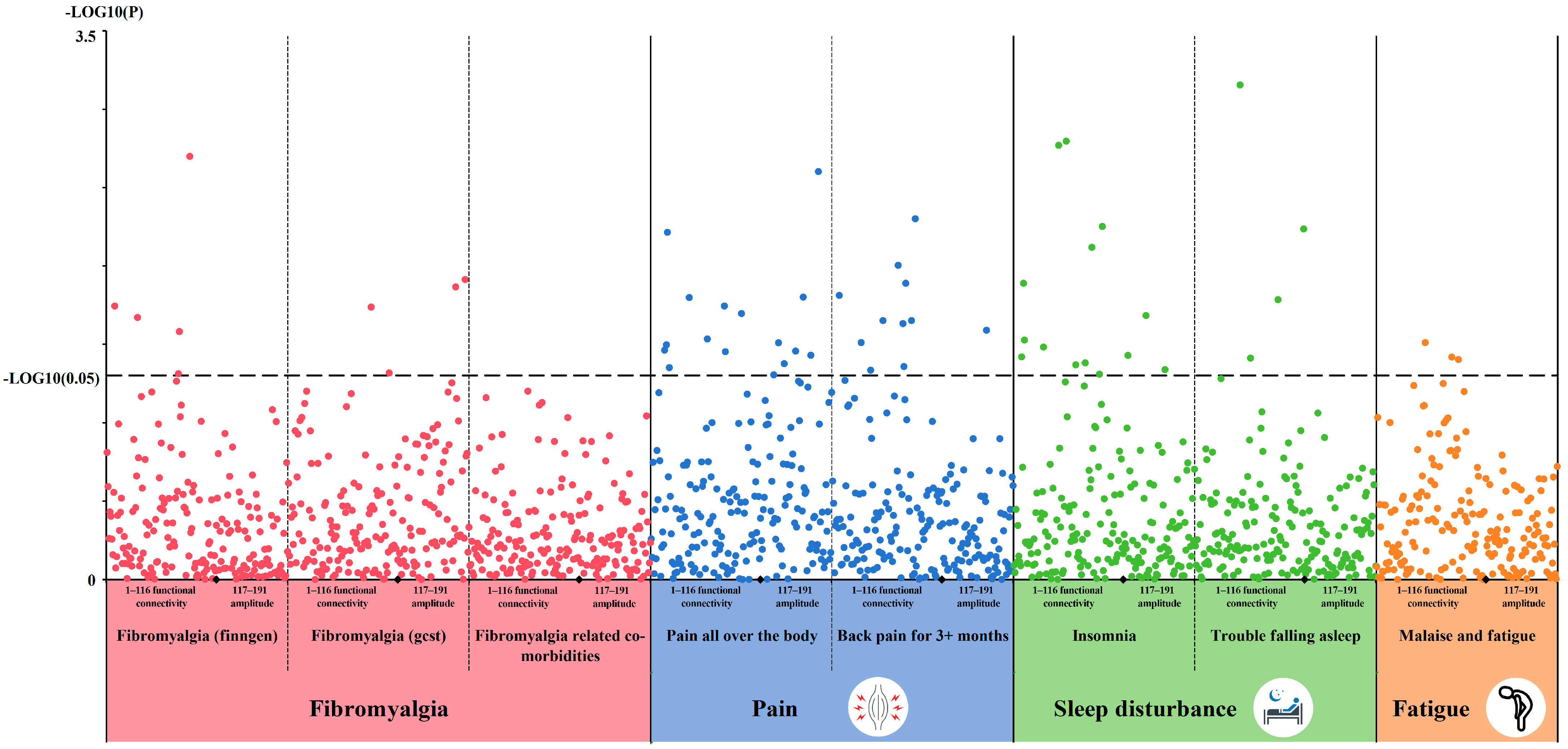
3.1. LDSC Results
3.2. Causal MR Associations Between rsfMRI Traits and Fibromyalgia-Related Traits
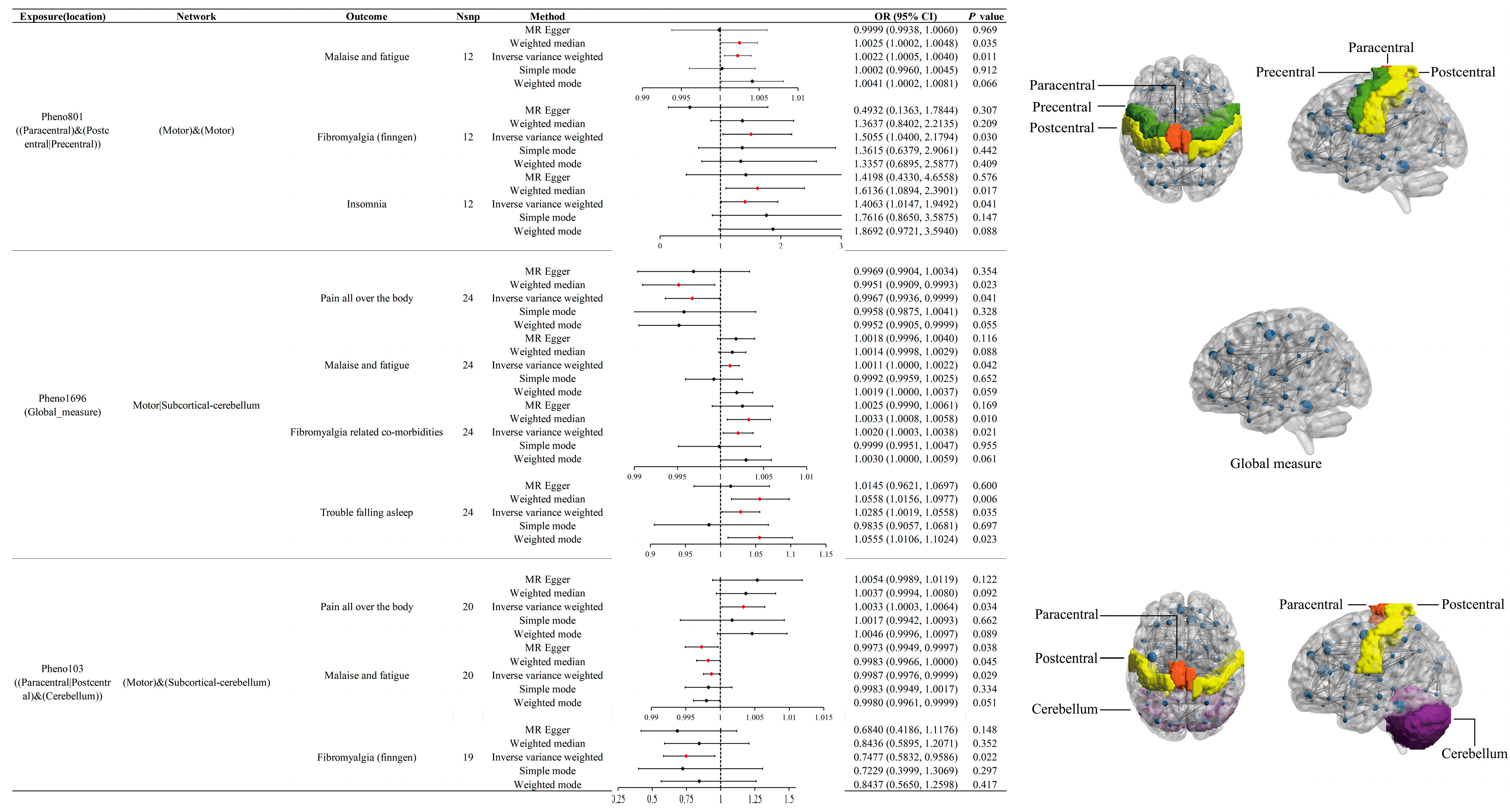
3.3. Effects of Pheno801 on Fibromyalgia-Related Traits
3.4. Effects of Pheno1696 on Fibromyalgia-Related Traits
3.5. Effects of Pheno103 on Fibromyalgia-Related Traits
3.6. Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarzi-Puttini, P.; Giorgi, V.; Marotto, D.; Atzeni, F. Fibromyalgia: An update on clinical characteristics, aetiopathogenesis and treatment. Nat. Rev. Rheumatol. 2020, 16, 645–660. [Google Scholar] [CrossRef]
- Spaeth, M. Epidemiology, costs, and the economic burden of fibromyalgia. Arthritis Res. Ther. 2009, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- White, K.P.; Speechley, M.; Harth, M.; Ostbye, T. The London Fibromyalgia Epidemiology Study: The prevalence of fibromyalgia syndrome in London, Ontario. J. Rheumatol. 1999, 26, 1570–1576. [Google Scholar] [PubMed]
- Queiroz, L.P. Worldwide epidemiology of fibromyalgia. Curr. Pain Headache Rep. 2013, 17, 356. [Google Scholar] [CrossRef]
- Lachaine, J.; Beauchemin, C.; Landry, P.-A. Clinical and economic characteristics of patients with fibromyalgia syndrome. Clin. J. Pain 2010, 26, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Dukes, E.; Martin, S.; Edelsberg, J.; Oster, G. Characteristics and healthcare costs of patients with fibromyalgia syndrome. Int. J. Clin. Pract. 2007, 61, 1498–1508. [Google Scholar] [CrossRef]
- Guymer, E.K.; Littlejohn, G.O.; Brand, C.K.; Kwiatek, R.A. Fibromyalgia onset has a high impact on work ability in Australians. Intern. Med. J. 2016, 46, 1069–1074. [Google Scholar] [CrossRef]
- Mu, C.; Dang, X.; Luo, X.-J. Mendelian randomization analyses reveal causal relationships between brain functional networks and risk of psychiatric disorders. Nat. Hum. Behav. 2024, 8, 1417–1428. [Google Scholar] [CrossRef]
- Owen, J.A.; Simon, B. How well do we understand the neural origins of the fMRI BOLD signal? Trends Neurosci. 2002, 25, 27–31. [Google Scholar] [CrossRef]
- Cifre, I.; Sitges, C.; Fraiman, D.; Muñoz, M.Á.; Balenzuela, P.; González-Roldán, A.; Martínez-Jauand, M.; Birbaumer, N.; Chialvo, D.R.; Montoya, P. Disrupted functional connectivity of the pain network in fibromyalgia. Psychosom. Med. 2012, 74, 55–62. [Google Scholar] [CrossRef]
- Napadow, V.; LaCount, L.; Park, K.; As-Sanie, S.; Clauw, D.J.; Harris, R.E. Intrinsic Brain Connectivity in Fibromyalgia is Associated with Chronic Pain Intensity. Arthritis Rheum. 2010, 62, 2545–2555. [Google Scholar] [CrossRef]
- Aoe, T.; Kawanaka, R.; Ohsone, F.; Hara, A.; Yokokawa, T. Functional connectivity associated with attention networks differs among subgroups of fibromyalgia patients: An observational case-control study. Sci. Rep. 2024, 14, 10197. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, B.J. From the garden to the clinic: How Mendelian randomization is shaping up atherosclerotic cardiovascular disease prevention strategies. Eur. Heart J. 2022, 43, 4447–4449. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.K.; Loh, P.-R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Yang, G.; Xie, W.; Li, B.; Zhao, G.; Li, J.; Xiao, W.; Li, Y. Casual associations between brain structure and sarcopenia: A large-scale genetic correlation and mendelian randomization study. Aging Cell 2024, 23, e14252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, T.; Smith, S.M.; Xiong, D.; Wang, X.; Yang, Y.; Luo, T.; Zhu, Z.; Shan, Y.; Matoba, N.; et al. Common variants contribute to intrinsic human brain functional networks. Nat. Genet. 2022, 54, 508–517. [Google Scholar] [CrossRef]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Sipilä, T.P.; Kristiansson, K.; Donner, K.M.; Reeve, M.P.; Laivuori, H.; Aavikko, M.; Kaunisto, M.A.; et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023, 613, 508–518. [Google Scholar] [CrossRef]
- Neale Lab [Internet]. UK Biobank. Available online: http://www.nealelab.is/uk-biobank (accessed on 18 September 2024).
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Güneş, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef]
- Access Results|FinnGen [Internet]. Available online: https://www.finngen.fi/en/access_results (accessed on 18 September 2024).
- Zorina-Lichtenwalter, K.; Bango, C.I.; Van Oudenhove, L.; Čeko, M.; Lindquist, M.A.; Grotzinger, A.D.; Keller, M.C.; Friedman, N.P.; Wager, T.D. Genetic risk shared across 24 chronic pain conditions: Identification and characterization with genomic structural equation modeling. PAIN 2023, 164, 2239–2252. [Google Scholar] [CrossRef]
- Aissani, B. Confounding by linkage disequilibrium. J. Hum. Genet. 2014, 59, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Martin, R.M.; Lewis, S.J.; Kazmi, N.; Robinson, T.M.; Albanes, D.; Aleksandrova, K.; et al. Physical activity and risks of breast and colorectal cancer: A Mendelian randomisation analysis. Nat. Commun. 2020, 11, 597. [Google Scholar] [CrossRef]
- Zhang, L.; Xiong, Y.; Zhang, J.; Feng, Y.; Xu, A. Systematic proteome-wide Mendelian randomization using the human plasma proteome to identify therapeutic targets for lung adenocarcinoma. J. Transl. Med. 2024, 22, 330. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.-R.; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3; Duncan, L.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Lee, C.H.; Cook, S.; Lee, J.S.; Han, B. Comparison of Two Meta-Analysis Methods: Inverse-Variance-Weighted Average and Weighted Sum of Z-Scores. Genom. Inf. 2016, 14, 173–180. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Patkar, A.A.; Bilal, L.; Masand, P.S. Management of fibromyalgia. Curr. Psychiatry Rep. 2003, 5, 218–224. [Google Scholar] [CrossRef]
- Koroschetz, J.; Rehm, S.E.; Gockel, U.; Brosz, M.; Freynhagen, R.; Tölle, T.R.; Baron, R. Fibromyalgia and neuropathic pain--differences and similarities. A comparison of 3057 patients with diabetic painful neuropathy and fibromyalgia. BMC Neurol. 2011, 11, 55. [Google Scholar] [CrossRef]
- Rahman, A.; Underwood, M.; Carnes, D. Fibromyalgia. BMJ 2014, 348, g1224. [Google Scholar] [CrossRef] [PubMed]
- Clauw, D.J. Fibromyalgia and related conditions. Mayo Clin. Proc. 2015, 90, 680–692. [Google Scholar] [CrossRef]
- Kleinman, L.; Mannix, S.; Arnold, L.M.; Burbridge, C.; Howard, K.; McQuarrie, K.; Pitman, V.; Resnick, M.; Roth, T.; Symonds, T. Assessment of sleep in patients with fibromyalgia: Qualitative development of the fibromyalgia sleep diary. Health Qual. Life Outcomes 2014, 12, 111. [Google Scholar] [CrossRef]
- Littlejohn, G. Neurogenic neuroinflammation in fibromyalgia and complex regional pain syndrome. Nat. Rev. Rheumatol. 2015, 11, 639–648. [Google Scholar] [CrossRef]
- Woolf, C.J. Central sensitization: Implications for the diagnosis and treatment of pain. PAIN 2011, 152, S2–S15. [Google Scholar] [CrossRef] [PubMed]
- Nijs, J.; Meeus, M.; Van Oosterwijck, J.; Ickmans, K.; Moorkens, G.; Hans, G.; De Clerck, L.S. In the mind or in the brain? Scientific evidence for central sensitisation in chronic fatigue syndrome. Eur. J. Clin. Investig. 2012, 42, 203–212. [Google Scholar] [CrossRef]
- Hsiao, F.-J.; Wang, S.-J.; Lin, Y.-Y.; Fuh, J.-L.; Ko, Y.-C.; Wang, P.-N.; Chen, W.-T. Altered insula-default mode network connectivity in fibromyalgia: A resting-state magnetoencephalographic study. J. Headache Pain 2017, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Cagnie, B.; Coppieters, I.; Denecker, S.; Six, J.; Danneels, L.; Meeus, M. Central sensitization in fibromyalgia? A systematic review on structural and functional brain MRI. Semin. Arthritis Rheum. 2014, 44, 68–75. [Google Scholar] [CrossRef]
- Jensen, K.B.; Kosek, E.; Petzke, F.; Carville, S.; Fransson, P.; Marcus, H.; Williams, S.C.R.; Choy, E.; Giesecke, T.; Mainguy, Y.; et al. Evidence of dysfunctional pain inhibition in Fibromyalgia reflected in rACC during provoked pain. PAIN 2009, 144, 95–100. [Google Scholar] [CrossRef]
- Perrot, S. Fibromyalgia: A misconnection in a multiconnected world? Eur. J. Pain 2019, 23, 866–873. [Google Scholar] [CrossRef]
- Truini, A.; Tinelli, E.; Gerardi, M.C.; Calistri, V.; Iannuccelli, C.; La Cesa, S.; Tarsitani, L.; Mainero, C.; Sarzi-Puttini, P.; Cruccu, G.; et al. Abnormal resting state functional connectivity of the periaqueductal grey in patients with fibromyalgia. Clin. Exp. Rheumatol. 2016, 34 (Suppl. 96), S129–S133. [Google Scholar] [PubMed]
- Vanneste, S.; Ost, J.; Van Havenbergh, T.; De Ridder, D. Resting state electrical brain activity and connectivity in fibromyalgia. PLoS ONE 2017, 12, e0178516. [Google Scholar] [CrossRef] [PubMed]
- Giesecke, T.; Gracely, R.H.; Grant, M.A.B.; Nachemson, A.; Petzke, F.; Williams, D.A.; Clauw, D.J. Evidence of augmented central pain processing in idiopathic chronic low back pain. Arthritis Rheum. 2004, 50, 613–623. [Google Scholar] [CrossRef]
- Pujol, J.; López-Solà, M.; Ortiz, H.; Vilanova, J.C.; Harrison, B.J.; Yücel, M.; Soriano-Mas, C.; Cardoner, N.; Deus, J. Mapping brain response to pain in fibromyalgia patients using temporal analysis of FMRI. PLoS ONE 2009, 4, e5224. [Google Scholar] [CrossRef] [PubMed]
- Burgmer, M.; Pogatzki-Zahn, E.; Gaubitz, M.; Stüber, C.; Wessoleck, E.; Heuft, G.; Pfleiderer, B. Fibromyalgia unique temporal brain activation during experimental pain: A controlled fMRI Study. J. Neural Transm. 2010, 117, 123–131. [Google Scholar] [CrossRef]
- Markus, B.; Esther, P.-Z.; Markus, G.; Erik, W.; Gereon, H.; Bettina, P. Altered brain activity during pain processing in fibromyalgia. NeuroImage 2009, 44, 502–508. [Google Scholar] [CrossRef]
- Schmidt-Wilcke, T.; Luerding, R.; Weigand, T.; Jürgens, T.; Schuierer, G.; Leinisch, E.; Bogdahn, U. Striatal grey matter increase in patients suffering from fibromyalgia--a voxel-based morphometry study. PAIN 2007, 132 (Suppl. 1), S109–S116. [Google Scholar] [CrossRef]
- Lutz, J.; Jäger, L.; de Quervain, D.; Krauseneck, T.; Padberg, F.; Wichnalek, M.; Beyer, A.; Stahl, R.; Zirngibl, B.; Morhard, D.; et al. White and gray matter abnormalities in the brain of patients with fibromyalgia: A diffusion-tensor and volumetric imaging study. Arthritis Rheum. 2008, 58, 3960–3969. [Google Scholar] [CrossRef]
- Pacheco-Barrios, K.; Lima, D.; Pimenta, D.; Slawka, E.; Navarro-Flores, A.; Parente, J.; Rebello-Sanchez, I.; Cardenas-Rojas, A.; Gonzalez-Mego, P.; Castelo-Branco, L.; et al. Motor cortex inhibition as a fibromyalgia biomarker: A meta-analysis of transcranial magnetic stimulation studies. Brain Netw. Modul. 2022, 1, 88–101. [Google Scholar] [CrossRef]
- Gracely, R.H.; Petzke, F.; Wolf, J.M.; Clauw, D.J. Functional magnetic resonance imaging evidence of augmented pain processing in fibromyalgia. Arthritis Rheum. 2002, 46, 1333–1343. [Google Scholar] [CrossRef]
- Cook, D.B.; Lange, G.; Ciccone, D.S.; Liu, W.-C.; Steffener, J.; Natelson, B.H. Functional imaging of pain in patients with primary fibromyalgia. J. Rheumatol. 2004, 31, 364–378. [Google Scholar] [PubMed]
- Gentile, E.; Ricci, K.; Delussi, M.; Brighina, F.; de Tommaso, M. Motor Cortex Function in Fibromyalgia: A Study by Functional Near-Infrared Spectroscopy. Pain Res. Treat. 2019, 2019, 2623161. [Google Scholar] [CrossRef] [PubMed]
- Castillo Saavedra, L.; Mendonca, M.; Fregni, F. Role of the primary motor cortex in the maintenance and treatment of pain in fibromyalgia. Med. Hypotheses 2014, 83, 332–336. [Google Scholar] [CrossRef]
- Park, S.H.; Michael, A.M.; Baker, A.K.; Lei, C.; Martucci, K.T. Enhanced motor network engagement during reward gain anticipation in fibromyalgia. Cortex 2024, 173, 161–174. [Google Scholar] [CrossRef]
- Coombes, S.A.; Misra, G. Pain and motor processing in the human cerebellum. PAIN 2016, 157, 117–127. [Google Scholar] [CrossRef]
- Saab, C.Y.; Willis, W.D. The cerebellum: Organization, functions and its role in nociception. Brain Res. Brain Res. Rev. 2003, 42, 85–95. [Google Scholar] [CrossRef]
- Diano, M.; D’Agata, F.; Cauda, F.; Costa, T.; Geda, E.; Sacco, K.; Duca, S.; Torta, D.M.; Geminiani, G.C. Cerebellar Clustering and Functional Connectivity During Pain Processing. Cerebellum 2016, 15, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Agoalikum, E.; Wu, H.; Klugah-Brown, B.; Maes, M. Brain structural differences between fibromyalgia patients and healthy control subjects: A source-based morphometric study. Sci. Rep. 2025, 15, 17446. [Google Scholar] [CrossRef]
- Xin, M.; Qu, Y.; Peng, X.; Zhu, D.; Cheng, S. A systematic review and meta-analysis of voxel-based morphometric studies of fibromyalgia. Front. Neurosci. 2023, 17, 1164145. [Google Scholar] [CrossRef]
- Pineda-Pardo, J.A.; Obeso, I.; Guida, P.; Dileone, M.; Strange, B.A.; Obeso, J.A.; Oliviero, A.; Foffani, G. Static magnetic field stimulation of the supplementary motor area modulates resting-state activity and motor behavior. Commun. Biol. 2019, 2, 397. [Google Scholar] [CrossRef]
- Wei, W.; Zhu, T.; Wang, X.; Li, L.; Zou, Q.; Lv, Y. Altered Topological Organization in the Sensorimotor Network After Application of Different Frequency rTMS. Front. Neurosci. 2019, 13, 1377. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Tang, F.; Wang, Y.; Yan, Y.; Dong, L.; Wang, T.; Zhu, D.; Tian, M.; Lin, X.; Shi, J. Effect of cerebellum stimulation on cognitive recovery in patients with Alzheimer disease: A randomized clinical trial. Brain Stimul. 2022, 15, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, J.M.; Barch, D.M. Cognition and resting-state functional connectivity in schizophrenia. Neurosci. Biobehav. Rev. 2016, 61, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ebrahim, S. Mendelian randomization: Prospects, potentials, and limitations. Int. J. Epidemiol. 2004, 33, 30–42. [Google Scholar] [CrossRef]
- Bowden, J.; Holmes, M.V. Meta-analysis and Mendelian randomization: A review. Res. Synth. Methods 2019, 10, 486–496. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Yang, G.; Deng, Z.; Yang, S.; Li, Y.; Xiao, W.; Lu, B.; Zhu, X. The Casual Associations Between Brain Functional Networks and Fibromyalgia: A Large-Scale Genetic Correlation and Mendelian Randomization Study. Bioengineering 2025, 12, 692. https://doi.org/10.3390/bioengineering12070692
Hu Y, Yang G, Deng Z, Yang S, Li Y, Xiao W, Lu B, Zhu X. The Casual Associations Between Brain Functional Networks and Fibromyalgia: A Large-Scale Genetic Correlation and Mendelian Randomization Study. Bioengineering. 2025; 12(7):692. https://doi.org/10.3390/bioengineering12070692
Chicago/Turabian StyleHu, Yiqun, Guang Yang, Zhenhan Deng, Shengwu Yang, Yusheng Li, Wenfeng Xiao, Bangbao Lu, and Xiongbai Zhu. 2025. "The Casual Associations Between Brain Functional Networks and Fibromyalgia: A Large-Scale Genetic Correlation and Mendelian Randomization Study" Bioengineering 12, no. 7: 692. https://doi.org/10.3390/bioengineering12070692
APA StyleHu, Y., Yang, G., Deng, Z., Yang, S., Li, Y., Xiao, W., Lu, B., & Zhu, X. (2025). The Casual Associations Between Brain Functional Networks and Fibromyalgia: A Large-Scale Genetic Correlation and Mendelian Randomization Study. Bioengineering, 12(7), 692. https://doi.org/10.3390/bioengineering12070692







