MRMS-CNNFormer: A Novel Framework for Predicting the Biochemical Recurrence of Prostate Cancer on Multi-Sequence MRI
Abstract
1. Introduction
- (1)
- We propose MRMS-CNNFormer, a novel analysis framework that integrates multi-region MRI (intratumoral, peritumoral, and periprostatic regions) with multi-sequence imaging (T2-weighted imaging with fat suppression (T2WI-FS) and diffusion-weighted imaging (DWI)) for the comprehensive characterization of the tumor microenvironment, addressing the limitations of conventional tumor-centric approaches.
- (2)
- We develop a hierarchical feature fusion architecture that combines CNN-based encoder for regional feature extraction with a transformer-based encoder for the cross-modal integration of imaging and clinical data, effectively capturing spatial relationships and complementary information across different anatomical regions, imaging sequences, and clinicopathological variables for enhanced prognostic assessment.
- (3)
- Experiments are conducted on a multi-center dataset, demonstrating that MRMS-CNNFormer outperforms both single-region models (AUC 0.835 vs. 0.658–0.803) and clinical-only models (AUC 0.835 vs. 0.612) in predicting BCR on the external testing dataset, providing a robust, non-invasive tool for personalized treatment planning in prostate cancer management.
2. Method
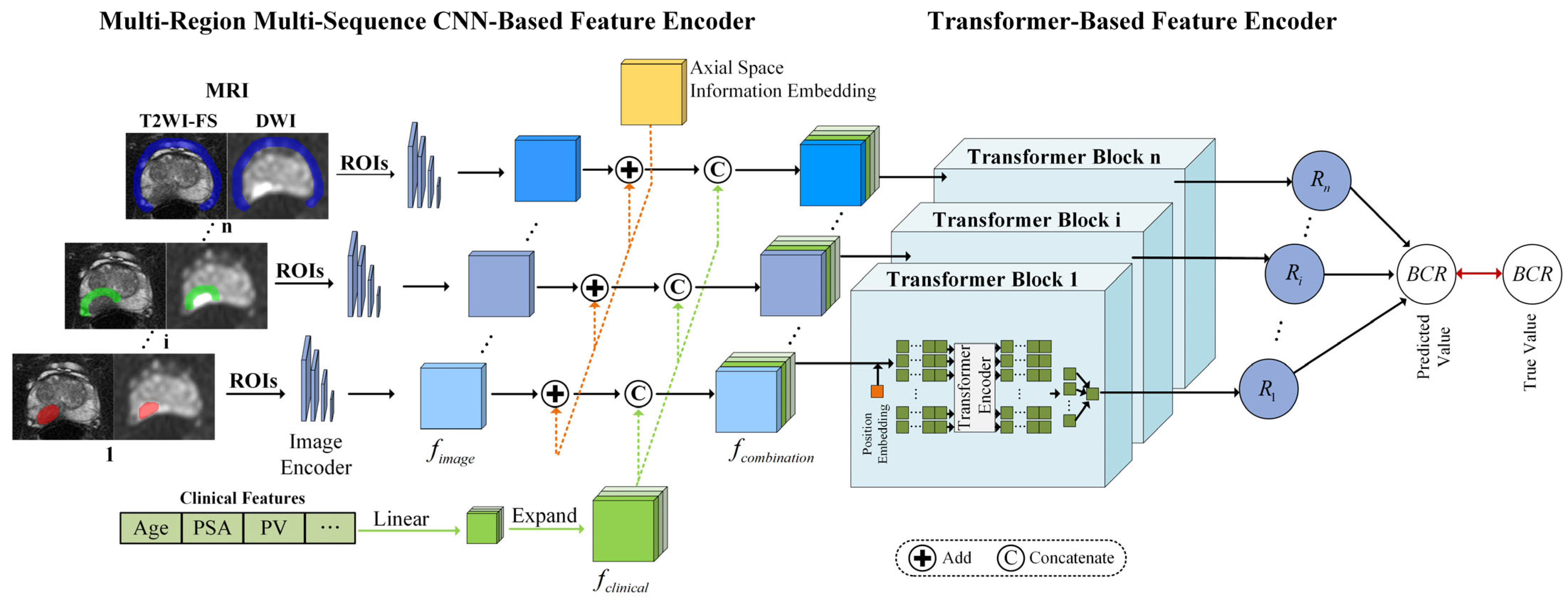
2.1. Multi-Region Multi-Sequence CNN-Based Feature Encoder
2.1.1. MRI Slice Feature Extraction
2.1.2. Axial Space Information Embedding
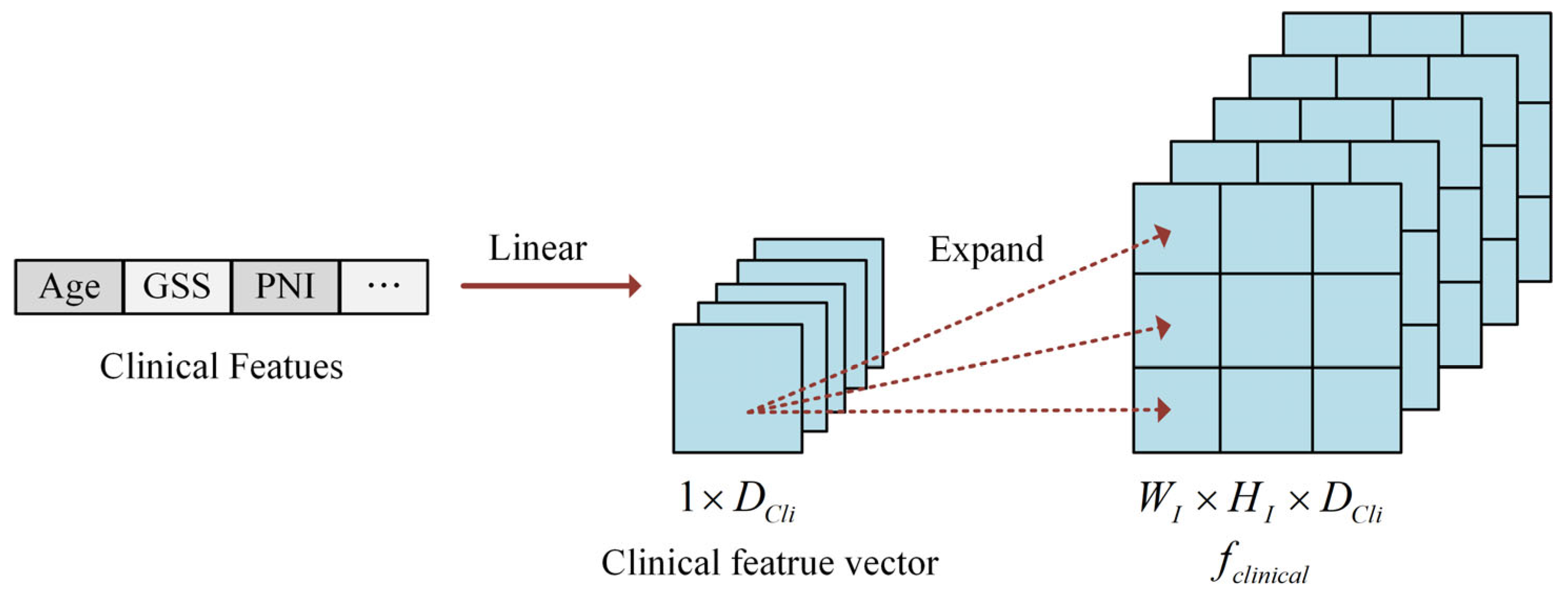
2.1.3. Clinical Feature Construction
2.2. Transformer-Based Feature Encoder
3. Materials and Experimental Configurations
3.1. Patient Cohorts
3.2. MRI Segmentation
3.3. Comparison of Results with Different Region Inputs and Clinical Data
3.4. Experimental Configuration
3.5. Statistical Analysis
4. Results
4.1. Patients
4.2. Performance of the Multi-Regional MRI-Based Models
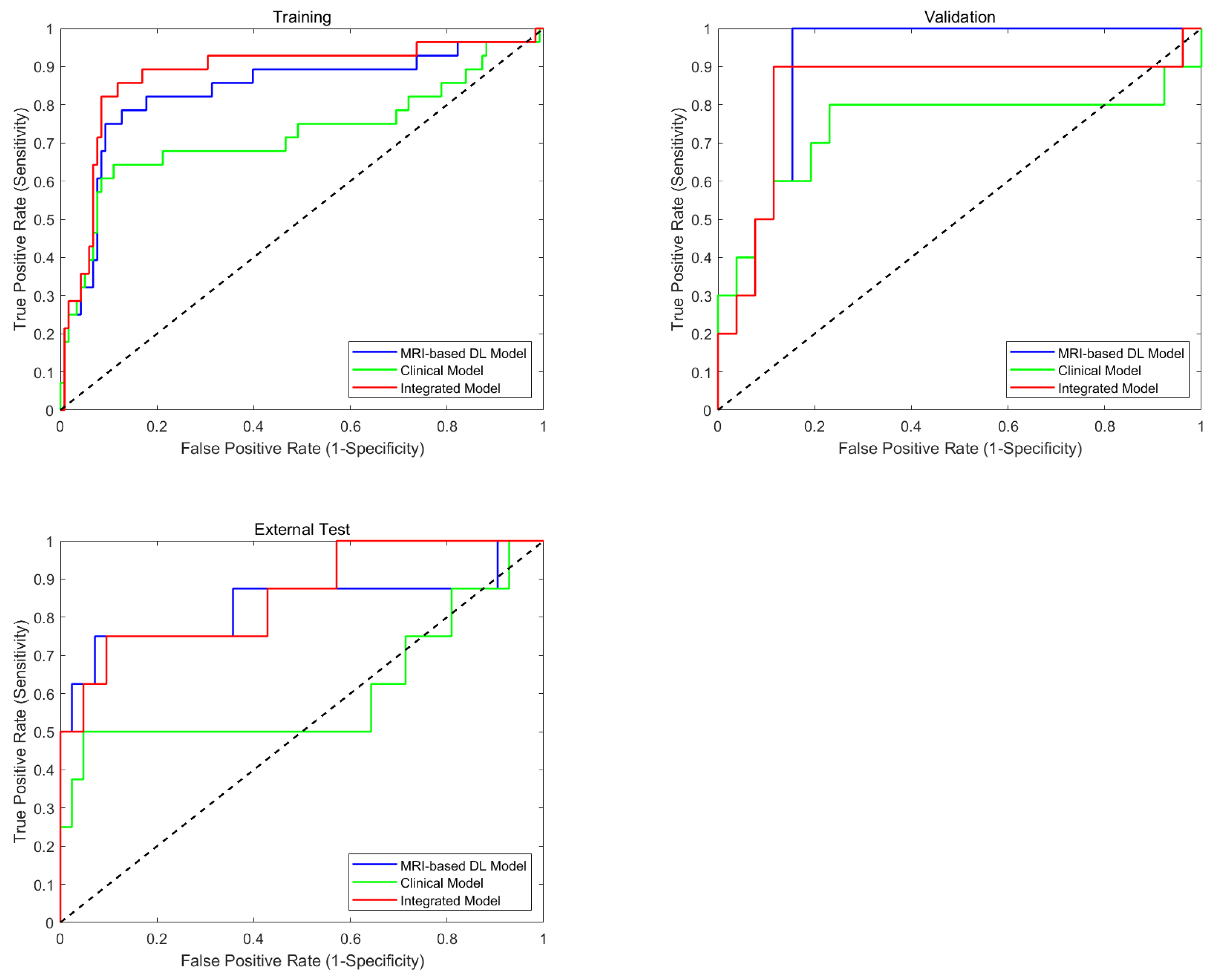
4.3. Performance of the MRI-Based Model, Clinical Model, and Integrated Model
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | Apparent Diffusion Coefficient |
| AUC | Area Under the Curve |
| BCR | Biochemical Recurrence |
| CAPRA | Cancer of the Prostate Risk Assessment |
| CI | Confidence Interval |
| CNN | Convolutional Neural Network |
| DCE | Dynamic Contrast-Enhanced |
| DSC | Dice Similarity Coefficient |
| DWI | Diffusion-Weighted Imaging |
| ECE | Extracapsular Extension |
| ERC | Endorectal Coil |
| FC | Fully Connected |
| FOV | Field of View |
| ITR | Intratumoral Region |
| MRI | Magnetic Resonance Imaging |
| mpMRI | multiparametric MRI |
| PACS | Picture Archiving and Communication System |
| PCa | Prostate Cancer |
| PPAC | Pelvic surface Phased Array Coil |
| PPR | Periprostatic Region |
| PSA | Prostate Specific Antigen |
| PTR | Peritumoral Region |
| ROC | Receiver Operating Characteristic |
| ROI | Region of Interest |
| RP | Radical Prostatectomy |
| SVI | Seminal Vesicle Invasion |
| T2WI-FS | T2-Weighted Imaging with Fat Suppression |
| TME | Tumor Microenvironment |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Tourinho-Barbosa, R.; Srougi, V.; Nunes-Silva, I.; Baghdadi, M.; Rembeyo, G.; Eiffel, S.S.; Barret, E.; Rozet, F.; Galiano, M.; Cathelineau, X.; et al. Biochemical Recurrence after Radical Prostatectomy: What Does It Mean? Int. Braz. J. Urol. 2018, 44, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.C.; Li, J.; Klink, J.C.; Kattan, M.W.; Klein, E.A.; Stephenson, A.J. Optimal Definition of Biochemical Recurrence After Radical Prostatectomy Depends on Pathologic Risk Factors: Identifying Candidates for Early Salvage Therapy. Eur. Urol. 2014, 66, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.I.; Parker, C.; Hope, T.A.; Paller, C.J. Best Approaches and Updates for Prostate Cancer Biochemical Recurrence. In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2022; pp. 352–359. [Google Scholar] [CrossRef]
- Matsukawa, A.; Yanagisawa, T.; Fazekas, T.; Miszczyk, M.; Tsuboi, I.; Kardoust Parizi, M.; Laukhtina, E.; Klemm, J.; Mancon, S.; Mori, K.; et al. Salvage Therapies for Biochemical Recurrence after Definitive Local Treatment: A Systematic Review, Meta-Analysis, and Network Meta-Analysis. Prostate Cancer Prostatic Dis. 2024. online ahead of print. [Google Scholar] [CrossRef]
- Pukl, M.; Keyes, S.; Keyes, M.; Guillaud, M.; Volavšek, M. Multi-Scale Tissue Architecture Analysis of Favorable-Risk Prostate Cancer: Correlation with Biochemical Recurrence. Investig. Clin. Urol. 2020, 61, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xie, K.; Logothetis, C.J.; Thompson, T.C.; Kim, J.; Huang, M.; Chang, D.W.; Gu, J.; Wu, X.; Ye, Y. Genetic Variants in Epithelial–Mesenchymal Transition Genes as Predictors of Clinical Outcomes in Localized Prostate Cancer. Carcinogenesis 2020, 41, 1057–1064. [Google Scholar] [CrossRef]
- Jeyapala, R.; Kamdar, S.; Olkhov-Mitsel, E.; Zlotta, A.; Fleshner, N.; Visakorpi, T.; Van Der Kwast, T.; Bapat, B. Combining CAPRA-S with Tumor IDC/C Features Improves the Prognostication of Biochemical Recurrence in Prostate Cancer Patients. Clin. Genitourin. Cancer 2022, 20, e217–e226. [Google Scholar] [CrossRef]
- Livingston, A.J.; Dvergsten, T.; Morgan, T.N. Initial Postoperative Prostate Specific Antigen and PSA Velocity Are Important Indicators of Underlying Malignancy After Simple Prostatectomy. J. Endourol. 2023, 37, 1057–1062. [Google Scholar] [CrossRef]
- Zhai, T.-S.; Hu, L.-T.; Ma, W.-G.; Chen, X.; Luo, M.; Jin, L.; Zhou, Z.; Liu, X.; Kang, Y.; Kang, Y.-X.; et al. Peri-Prostatic Adipose Tissue Measurements Using MRI Predict Prostate Cancer Aggressiveness in Men Undergoing Radical Prostatectomy. J. Endocrinol. Investig. 2021, 44, 287–296. [Google Scholar] [CrossRef]
- Manceau, C.; Beauval, J.-B.; Lesourd, M.; Almeras, C.; Aziza, R.; Gautier, J.-R.; Loison, G.; Salin, A.; Tollon, C.; Soulié, M.; et al. MRI Characteristics Accurately Predict Biochemical Recurrence after Radical Prostatectomy. J. Clin. Med. 2020, 9, 3841. [Google Scholar] [CrossRef]
- Dinis Fernandes, C.; Dinh, C.V.; Walraven, I.; Heijmink, S.W.; Smolic, M.; Van Griethuysen, J.J.M.; Simões, R.; Losnegård, A.; Van Der Poel, H.G.; Pos, F.J.; et al. Biochemical Recurrence Prediction after Radiotherapy for Prostate Cancer with T2w Magnetic Resonance Imaging Radiomic Features. Phys. Imaging Radiat. Oncol. 2018, 7, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Rajwa, P.; Mori, K.; Huebner, N.A.; Martin, D.T.; Sprenkle, P.C.; Weinreb, J.C.; Ploussard, G.; Pradere, B.; Shariat, S.F.; Leapman, M.S. The Prognostic Association of Prostate MRI PI-RADSTM v2 Assessment Category and Risk of Biochemical Recurrence after Definitive Local Therapy for Prostate Cancer: A Systematic Review and Meta-Analysis. J. Urol. 2021, 206, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, E.; Na, I.; Kim, C.K.; Seo, S.I.; Park, H. Novel Multiparametric Magnetic Resonance Imaging-Based Deep Learning and Clinical Parameter Integration for the Prediction of Long-Term Biochemical Recurrence-Free Survival in Prostate Cancer after Radical Prostatectomy. Cancers 2023, 15, 3416. [Google Scholar] [CrossRef]
- Aguirre, D.A.; Cardona Ortegón, J.D. Unlocking the Benefits of Multiparametric MRI for Predicting Prostate Cancer Recurrence. Radiology 2023, 309, e232819. [Google Scholar] [CrossRef]
- Bourbonne, V.; Fournier, G.; Vallières, M.; Lucia, F.; Doucet, L.; Tissot, V.; Cuvelier, G.; Hue, S.; Le Penn Du, H.; Perdriel, L.; et al. External Validation of an MRI-Derived Radiomics Model to Predict Biochemical Recurrence after Surgery for High-Risk Prostate Cancer. Cancers 2020, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shiradkar, R.; Leo, P.; Algohary, A.; Fu, P.; Tirumani, S.H.; Mahran, A.; Buzzy, C.; Obmann, V.C.; Mansoori, B.; et al. A Novel Imaging Based Nomogram for Predicting Post-Surgical Biochemical Recurrence and Adverse Pathology of Prostate Cancer from Pre-Operative Bi-Parametric MRI. EBioMedicine 2021, 63, 103163. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Kim, C.K.; Park, S.Y.; Park, B.K.; Lee, H.M.; Cho, S.W. Prostate Cancer: Role of Pretreatment Multiparametric 3-T MRI in Predicting Biochemical Recurrence After Radical Prostatectomy. Am. J. Roentgenol. 2014, 202, W459–W465. [Google Scholar] [CrossRef]
- Zhong, Q.-Z.; Long, L.-H.; Liu, A.; Li, C.-M.; Xiu, X.; Hou, X.-Y.; Wu, Q.-H.; Gao, H.; Xu, Y.-G.; Zhao, T.; et al. Radiomics of Multiparametric MRI to Predict Biochemical Recurrence of Localized Prostate Cancer After Radiation Therapy. Front. Oncol. 2020, 10, 731. [Google Scholar] [CrossRef]
- Algohary, A.; Shiradkar, R.; Pahwa, S.; Purysko, A.; Verma, S.; Moses, D.; Shnier, R.; Haynes, A.-M.; Delprado, W.; Thompson, J.; et al. Combination of Peri-Tumoral and Intra-Tumoral Radiomic Features on Bi-Parametric MRI Accurately Stratifies Prostate Cancer Risk: A Multi-Site Study. Cancers 2020, 12, 2200. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Q.; Yang, Z.; Li, Y.; Lee, C.H.; Shen, L. Recent Progress in Transformer-Based Medical Image Analysis. Comput. Biol. Med. 2023, 164, 107268. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, D. Transformers and Their Application to Medical Image Processing: A Review. J. Radiat. Res. Appl. Sci. 2023, 16, 100680. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Tang, Y.; Wang, C.; Landman, B.A.; Zhou, S.K. Transforming Medical Imaging with Transformers? A Comparative Review of Key Properties, Current Progresses, and Future Perspectives. Med. Image Anal. 2023, 85, 102762. [Google Scholar] [CrossRef]
- Kumar, S.S. Advancements in Medical Image Segmentation: A Review of Transformer Models. Comput. Electr. Eng. 2025, 123, 110099. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, Y.; Liu, F. TransMed: Transformers Advance Multi-Modal Medical Image Classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar]
- Zhang, H.; Li, X.; Zhang, Y.; Huang, C.; Wang, Y.; Yang, P.; Duan, S.; Mao, N.; Xie, H. Diagnostic Nomogram Based on Intralesional and Perilesional Radiomics Features and Clinical Factors of Clinically Significant Prostate Cancer. Magn. Reson. Imaging 2021, 53, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Crum, W.R.; Camara, O.; Hill, D.L.G. Generalized Overlap Measures for Evaluation and Validation in Medical Image Analysis. IEEE Trans. Med. Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef]
- Zabihollahy, F.; Naim, S.; Wibulpolprasert, P.; Reiter, R.E.; Raman, S.S.; Sung, K. Understanding Spatial Correlation Between Multiparametric MRI Performance and Prostate Cancer. Magn. Reson. Imaging 2024, 60, 2184–2195. [Google Scholar] [CrossRef]
- Lin, M.; Yu, X.; Ouyang, H.; Luo, D.; Zhou, C. Consistency of T2WI-FS/ASL Fusion Images in Delineating the Volume of Nasopharyngeal Carcinoma. Sci. Rep. 2015, 5, 18431. [Google Scholar] [CrossRef]
- Koh, D.-M.; Collins, D.J. Diffusion-Weighted MRI in the Body: Applications and Challenges in Oncology. Am. J. Roentgenol. 2007, 188, 1622–1635. [Google Scholar] [CrossRef]
- Sun, C.; Chatterjee, A.; Yousuf, A.; Antic, T.; Eggener, S.; Karczmar, G.S.; Oto, A. Comparison of T2-Weighted Imaging, DWI, and Dynamic Contrast-Enhanced MRI for Calculation of Prostate Cancer Index Lesion Volume: Correlation with Whole-Mount Pathology. Am. J. Roentgenol. 2019, 212, 351–356. [Google Scholar] [CrossRef]
- De Kok, J.W.T.M.; De La Hoz, M.Á.A.; De Jong, Y.; Brokke, V.; Elbers, P.W.G.; Thoral, P.; Castillejo, A.; Trenor, T.; Castellano, J.M.; Bronchalo, A.E.; et al. A Guide to Sharing Open Healthcare Data under the General Data Protection Regulation. Sci. Data 2023, 10, 404. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef]
- Mukasheva, A.; Koishiyeva, D.; Sergazin, G.; Sydybayeva, M.; Mukhammejanova, D.; Seidazimov, S. Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet. In Proceedings of the EEPES 2024, Kavala, Greece, 19–21 June 2024; MDPI: Basel, Switzerland, 2024; p. 16. [Google Scholar]
- Aboussaleh, I.; Riffi, J.; Fazazy, K.E.; Mahraz, M.A.; Tairi, H. Efficient U-Net Architecture with Multiple Encoders and Attention Mechanism Decoders for Brain Tumor Segmentation. Diagnostics 2023, 13, 872. [Google Scholar] [CrossRef]
- Abdelhalim, I.; Badawy, M.A.; Abou El-Ghar, M.; Ghazal, M.; Contractor, S.; Van Bogaert, E.; Gondim, D.; Silva, S.; Khalifa, F.; El-Baz, A. Multi-Branch CNNFormer: A Novel Framework for Predicting Prostate Cancer Response to Hormonal Therapy. BioMed. Eng. OnLine 2024, 23, 131. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Wang, Y.; Fan, P.; Xu, T.; Han, P.; Deng, Y.; Song, Y.; Wang, X.; Zhang, M. Bi-Parametric MRI-Based Quantification Radiomics Model for the Noninvasive Prediction of Histopathology and Biochemical Recurrence after Prostate Cancer Surgery: A Multicenter Study. Abdom. Radiol. 2025. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Kebaili, A.; Lapuyade-Lahorgue, J.; Ruan, S. Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. J. Imaging 2023, 9, 81. [Google Scholar] [CrossRef]
- Yang, X.; Chen, Y.; Yue, X.; Ma, C.; Yang, P. Local Linear Embedding Based Interpolation Neural Network in Pancreatic Tumor Segmentation. Appl. Intell. 2022, 52, 8746–8756. [Google Scholar] [CrossRef]
- Wang, R.; Guo, J.; Zhou, Z.; Wang, K.; Gou, S.; Xu, R.; Sher, D.; Wang, J. Locoregional Recurrence Prediction in Head and Neck Cancer Based on Multi-Modality and Multi-View Feature Expansion. Phys. Med. Biol. 2022, 67, 125004. [Google Scholar] [CrossRef]
- Jiang, K.; Peng, P.; Lian, Y.; Xu, W. The Encoding Method of Position Embeddings in Vision Transformer. J. Vis. Commun. Image Represent. 2022, 89, 103664. [Google Scholar] [CrossRef]
- Zheng, W.; Gong, G.; Tian, J.; Lu, S.; Wang, R.; Yin, Z.; Li, X.; Yin, L. Design of a Modified Transformer Architecture Based on Relative Position Coding. Int. J. Comput. Intell. Syst. 2023, 16, 168. [Google Scholar] [CrossRef]
- Su, J.; Ahmed, M.; Lu, Y.; Pan, S.; Bo, W.; Liu, Y. RoFormer: Enhanced Transformer with Rotary Position Embedding. Neurocomputing 2024, 568, 127063. [Google Scholar] [CrossRef]
- Lei, M.; Zhang, Y.; Deng, E.; Ni, Y.; Xiao, Y.; Zhang, Y.; Zhang, J. Intelligent Recognition of Joints and Fissures in Tunnel Faces Using an Improved Mask Region-based Convolutional Neural Network Algorithm. Comput. Aided Civ. Infrastruct. Eng. 2024, 39, 1123–1142. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, Z.; Wang, Z. META-Unet: Multi-Scale Efficient Transformer Attention Unet for Fast and High-Accuracy Polyp Segmentation. IEEE Trans. Autom. Sci. Eng. 2024, 21, 4117–4128. [Google Scholar] [CrossRef]
- Du, D.; Shiri, I.; Yousefirizi, F.; Salmanpour, M.R.; Lv, J.; Wu, H.; Zhu, W.; Zaidi, H.; Lu, L.; Rahmim, A. Impact of Harmonization and Oversampling Methods on Radiomics Analysis of Multi-Center Imbalanced Datasets: Application to PET-Based Prediction of Lung Cancer Subtypes. EJNMMI Phys. 2025, 12, 34. [Google Scholar] [CrossRef]
- Seoni, S.; Shahini, A.; Meiburger, K.M.; Marzola, F.; Rotunno, G.; Acharya, U.R.; Molinari, F.; Salvi, M. All You Need Is Data Preparation: A Systematic Review of Image Harmonization Techniques in Multi-Center/Device Studies for Medical Support Systems. Comput. Methods Programs Biomed. 2024, 250, 108200. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Total (n = 232) | Internal Dataset (n = 182) | External Dataset (n = 50) | p Value |
|---|---|---|---|---|
| Age, years (IQR) | 70.0 (65.0, 76.0) | 71.0 (65.0, 76.0) | 68.0 (62.8, 73.0) | <0.05 † |
| Gleason score sum, (n, %) | <0.05 * | |||
| <7 | 43 (18.5) | 37 (20.3) | 3 (6.0) | |
| =7 | 87 (37.5) | 73 (40.1) | 16 (32.0) | |
| >7 | 102 (44.0) | 72 (39.6) | 31 (62.0) | |
| ISUP grade, (n, %) | <0.05 * | |||
| 1 | 40 (17.2) | 37 (20.3) | 3 (6.0) | |
| 2 | 56 (24.1) | 47 (25.8) | 9 (18.0) | |
| 3 | 33 (14.2) | 26 (14.3) | 7 (14.0) | |
| 4 | 46 (19.8) | 35 (19.2) | 11 (22.0) | |
| 5 | 57 (24.6) | 37 (20.3) | 20 (40.0) | |
| Clinical stage, (n, %) | <0.05 ‡ | |||
| cT2x | 190 (81.9) | 156 (85.7) | 34 (68.0) | |
| cT3x | 42 (18.1) | 26 (14.3) | 16 (32.0) | |
| Pathological stage, (n, %) | <0.05 ‡ | |||
| pT2 | 143 (61.6) | 103 (56.6) | 41 (82.0) | |
| pT3x | 89 (38.4) | 79 (43.4) | 9 (18.0) | |
| Perineural invasion (PNI), (n, %) | 0.193 ‡ | |||
| Positive | 125 (53.9) | 94 (51.6) | 88 (48.4) | |
| Negative | 107 (46.1) | 31 (62.0) | 19 (38.0) | |
| Biochemical recurrence (BCR), (n, %) | 0.443 ‡ | |||
| Yes | 46 (19.8) | 38 (20.9) | 8 (16.0) | |
| No | 186 (80.2) | 144 (79.1) | 42(84.0) | |
| CAPRA Score, (n, %) | <0.05 ‡ | |||
| ≥6 | 128 (55.2) | 93 (51.1) | 37 (74.0) | |
| <6 | 104 (44.8) | 89 (48.9) | 13 (26.0) |
| Training (95% CI) (n = 146) | Validation (95% CI) (n = 36) | External Test (95% CI) (n = 50) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SENS | SPEC | ACC | F1 | AUC | P | SENS | SPEC | ACC | F1 | AUC | P | SENS | SPEC | ACC | F1 | AUC | P | |
| Intratumoral (ITR) | 0.756 (0.727–0.781) | 0.844 (0.816–0.865) | 0.830 (0.787–0.855) | 0.641 (0.596–0.678) | 0.816 (0.772–0.845) | ** | 0.857 (0.836–0.883) | 0.814 (0.786–0.845) | 0.843 (0.826–0.867) | 0.683 (0.647–0.728) | 0.858 (0.832–0.874) | * | 0.723 (0.701–0.757) | 0.818 (0.798–0.843) | 0.810 (0.786–0.845) | 0.540 (0.508–0.587) | 0.803 (0.783–0.822) | ** |
| Peritumoral (PTR) | 0.644 (0.618–0.661) | 0.685 (0.657–0.713) | 0.672 (0.658–0.695) | 0.450 (0.420–0.477) | 0.664 (0.647–0.708) | *** | 0.665 (0.642–0.686) | 0.723 (0.691–0.754) | 0.684 (0.658–0.692) | 0.487 (0.458–0.518) | 0.692 (0.667–0.732) | *** | 0.642 (0.627–0.678) | 0.682 (0.550–0.724) | 0.674 (0.647–0.688) | 0.388 (0.314–0.434) | 0.658 (0.631–0.692) | *** |
| Periprostatic (PPR) | 0.682 (0.657–0.714) | 0.744 (0.716–0.768) | 0.735 (0.713–0.767) | 0.511 (0.477–0.547) | 0.699 (0.671–0.737) | *** | 0.756 (0.730–0.774) | 0.699 (0.651–0.720) | 0.715 (0.678–0.747) | 0.538 (0.495–0.562) | 0.731 (0.704–0.762) | *** | 0.664 (0.647–0.694) | 0.741 (0.713–0.758) | 0.702 (0.676–0.735) | 0.439 (0.410–0.468) | 0.684 (0.654–0.723) | *** |
| Combined | 0.788 (0.759–0.806) | 0.859 (0.834–0.887) | 0.852 (0.817–0.866) | 0.675 (0.632–0.719) | 0.835 (0.812–0.855) | - | 0.872 (0.858–0.884) | 0.848 (0.825–0.867) | 0.866 (0.828–0.897) | 0.725 (0.695–0.753) | 0.875 (0.851–0.940) | - | 0.781 (0.764–0.798) | 0.847 (0.819–0.863) | 0.819 (0.786–0.855) | 0.604 (0.563–0.634) | 0.825 (0.808–0.852) | - |
| Training (95% CI) (n = 146) | Validation (95% CI) (n = 36) | External Test (95% CI) (n = 50) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SENS | SPEC | ACC | F1 | AUC | SENS | SPEC | ACC | F1 | AUC | SENS | SPEC | ACC | F1 | AUC | |
| MRI-based model | 0.788 (0.759–0.806) | 0.859 (0.834–0.887) | 0.852 (0.817–0.866) | 0.675 (0.632–0.719) | 0.835 (0.812–0.855) | 0.872 (0.858–0.884) | 0.848 (0.825–0.867) | 0.866 (0.828–0.897) | 0.725 (0.695–0.753) | 0.875 (0.851–0.940) | 0.781 (0.764–0.798) | 0.847 (0.819–0.863) | 0.819 (0.786–0.855) | 0.604 (0.563–0.634) | 0.825 (0.808–0.852) |
| Clinical model | 0.675 (0.624–0.747) | 0.776 (0.643–0.813) | 0.753 (0.724–0.787) | 0.531 (0.415–0.605) | 0.725 (0.664–0.753) | 0.828 (0.694–0.907) | 0.723 (0.684–0.807) | 0.758 (0.725–0.798) | 0.592 (0.496–0.702) | 0.745 (0.721–0.796) | 0.579 (0.546–0.633) | 0.672 (0.634–0.681) | 0.637 (0.611–0.679) | 0.351 (0.315–0.383) | 0.612 (0.574–0.646) |
| Integrated model | 0.846 (0.781–0.883) | 0.868 (0.769–0.903) | 0.855 (0.827–0.881) | 0.718 (0.584–0.782) | 0.861 (0.835–0.872) | 0.851 (0.837–0.916) | 0.882 (0.854–0.923) | 0.867 (0.824–0.883) | 0.752 (0.713–0.838) | 0.857 (0.836–0.894) | 0.774 (0.732–0.819) | 0.866 (0.824–0.893) | 0.842 (0.815–0.863) | 0.625 (0.551–0.688) | 0.835 (0.818–0.869) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, T.; Zhou, M.; Shao, Y.; Chen, X.; Zhao, Y.; Feng, Q. MRMS-CNNFormer: A Novel Framework for Predicting the Biochemical Recurrence of Prostate Cancer on Multi-Sequence MRI. Bioengineering 2025, 12, 538. https://doi.org/10.3390/bioengineering12050538
Lian T, Zhou M, Shao Y, Chen X, Zhao Y, Feng Q. MRMS-CNNFormer: A Novel Framework for Predicting the Biochemical Recurrence of Prostate Cancer on Multi-Sequence MRI. Bioengineering. 2025; 12(5):538. https://doi.org/10.3390/bioengineering12050538
Chicago/Turabian StyleLian, Tao, Mengting Zhou, Yangyang Shao, Xiaqing Chen, Yinghua Zhao, and Qianjin Feng. 2025. "MRMS-CNNFormer: A Novel Framework for Predicting the Biochemical Recurrence of Prostate Cancer on Multi-Sequence MRI" Bioengineering 12, no. 5: 538. https://doi.org/10.3390/bioengineering12050538
APA StyleLian, T., Zhou, M., Shao, Y., Chen, X., Zhao, Y., & Feng, Q. (2025). MRMS-CNNFormer: A Novel Framework for Predicting the Biochemical Recurrence of Prostate Cancer on Multi-Sequence MRI. Bioengineering, 12(5), 538. https://doi.org/10.3390/bioengineering12050538






