Machine Learning Techniques Applied to COVID-19 Prediction: A Systematic Literature Review
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
- (1)
- Used at least one ML technique to predict COVID-19 transmission trends.
- (2)
- Page length 8+.
- (3)
- Reported predictive performance metrics of ML models.
- (4)
- Experimentation on different datasets related to COVID-19.
- (5)
- Limited to journal articles.
- (1)
- Full text unavailable.
- (2)
- Not relevant to COVID-19 prevalence or trend projections.
- (3)
- No practical theoretical research (e.g., survey and review papers).
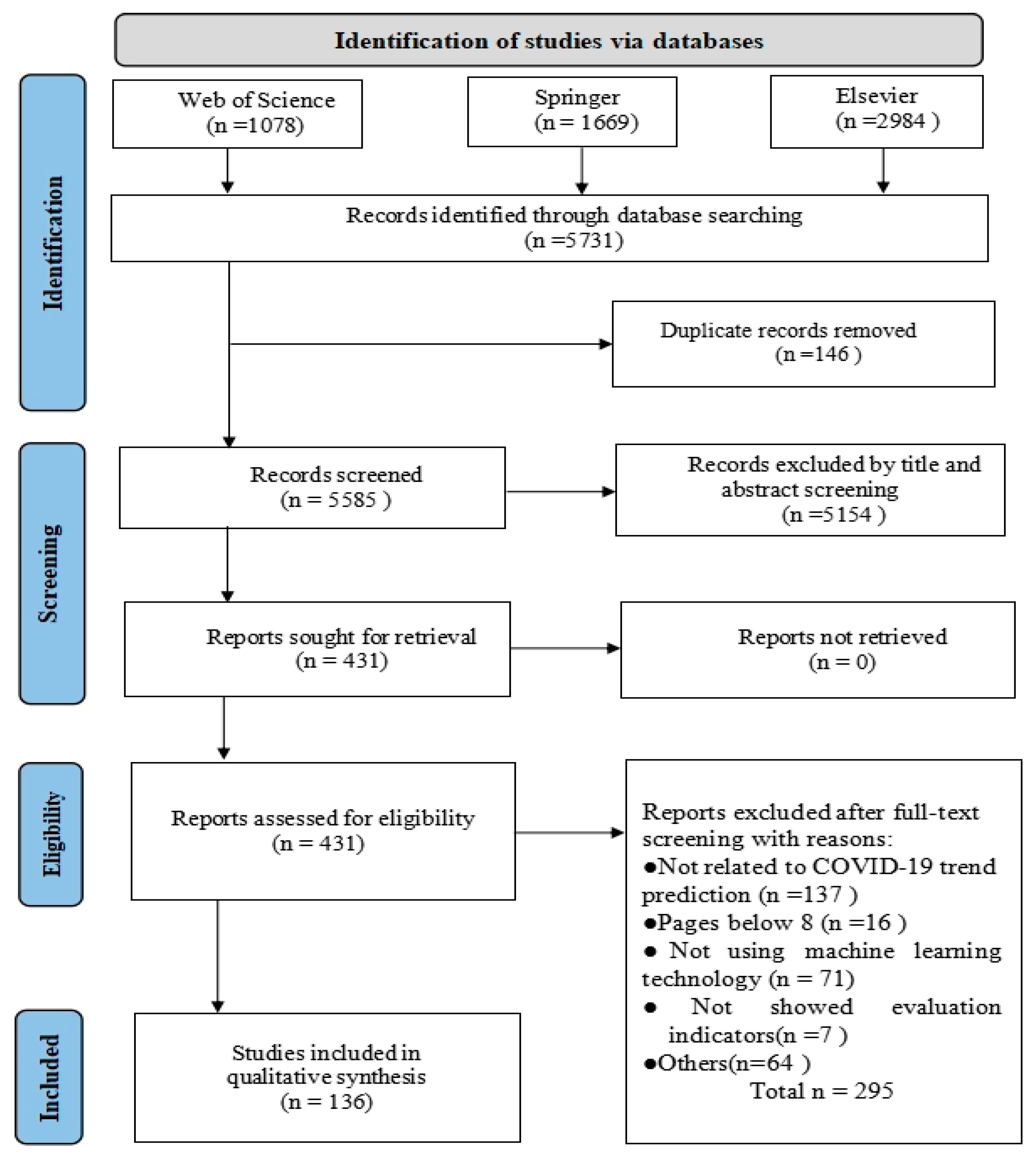
2.3. Study Selection
2.4. Study Risk of Bias Assessment
2.5. Data Synthesis
3. Results
3.1. Quality Assessment
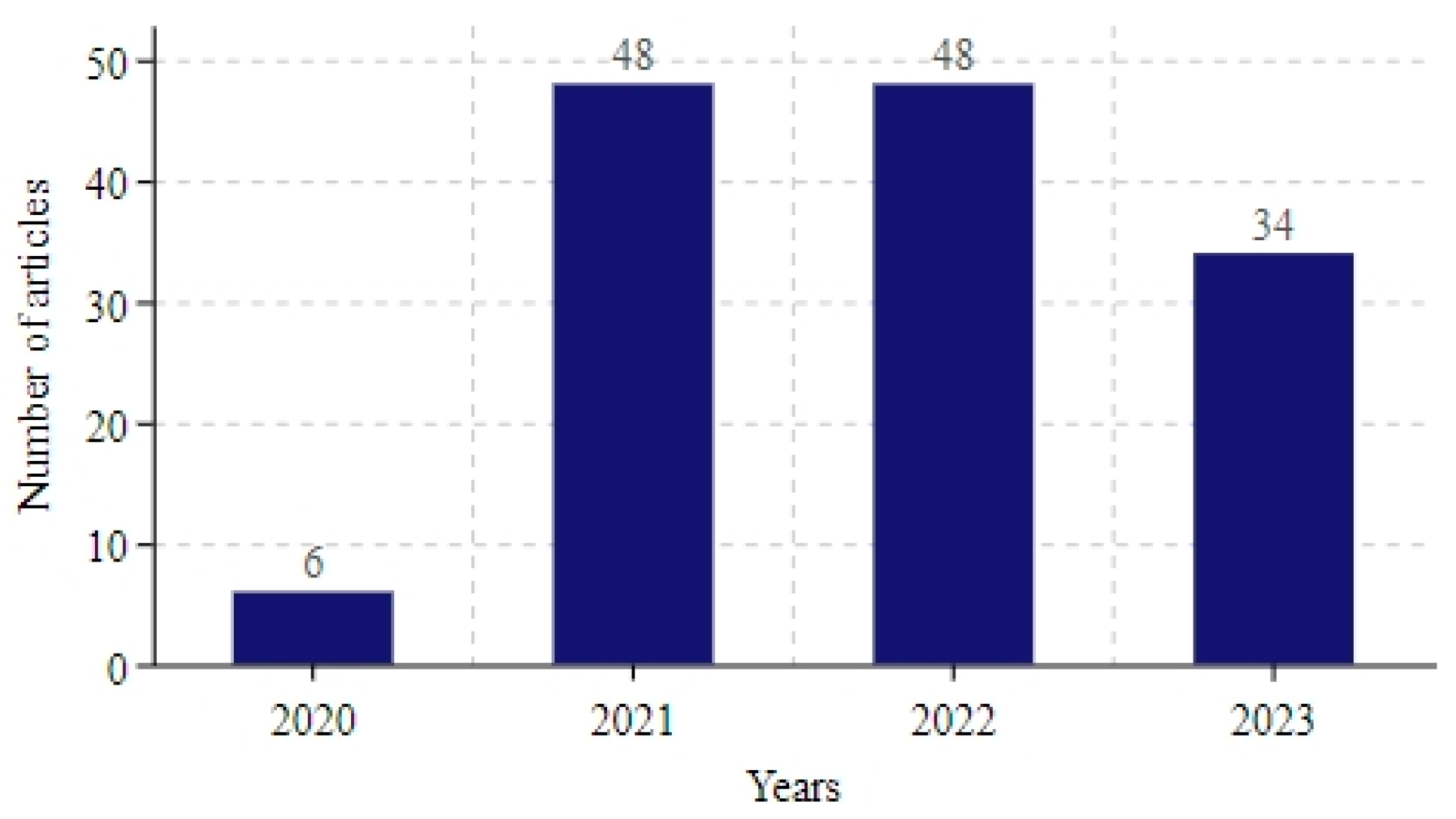
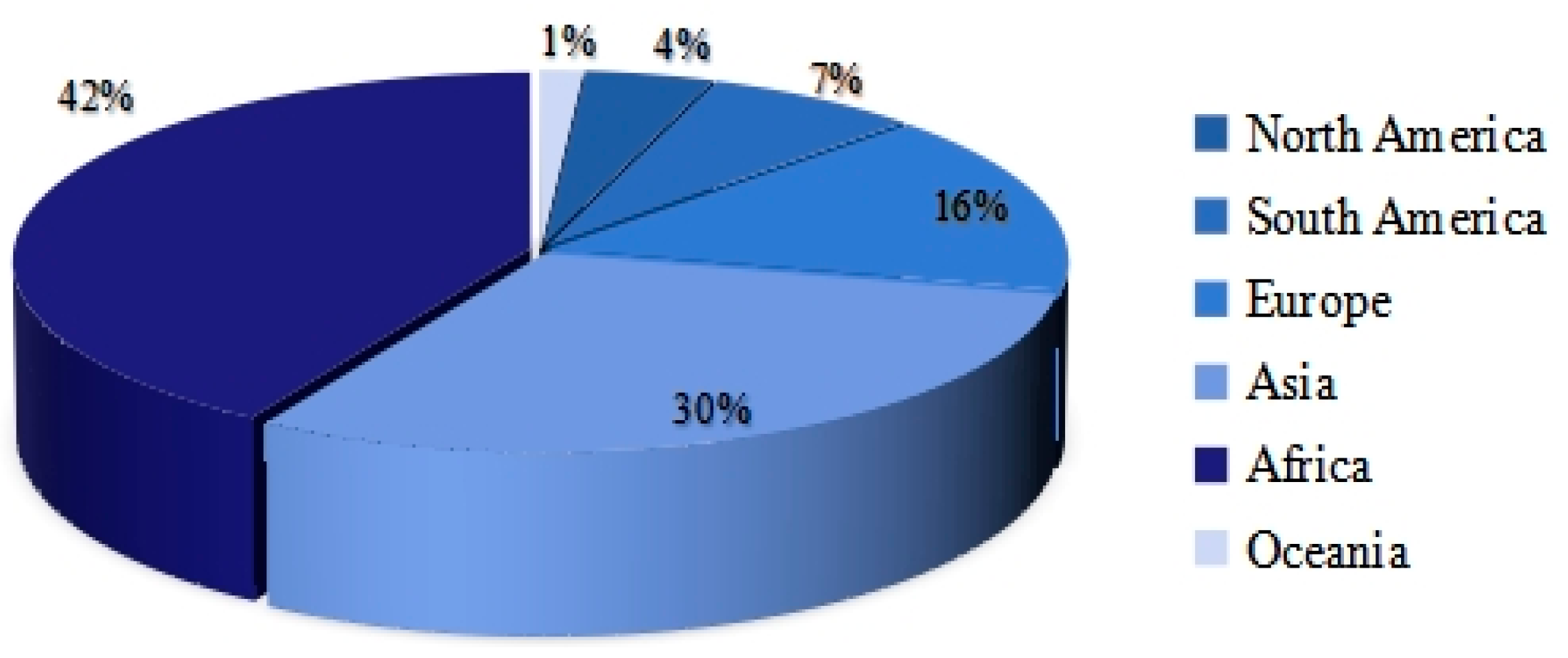
3.2. Bibliometric Analysis
3.3. Basic Content of COVID-19 Prediction
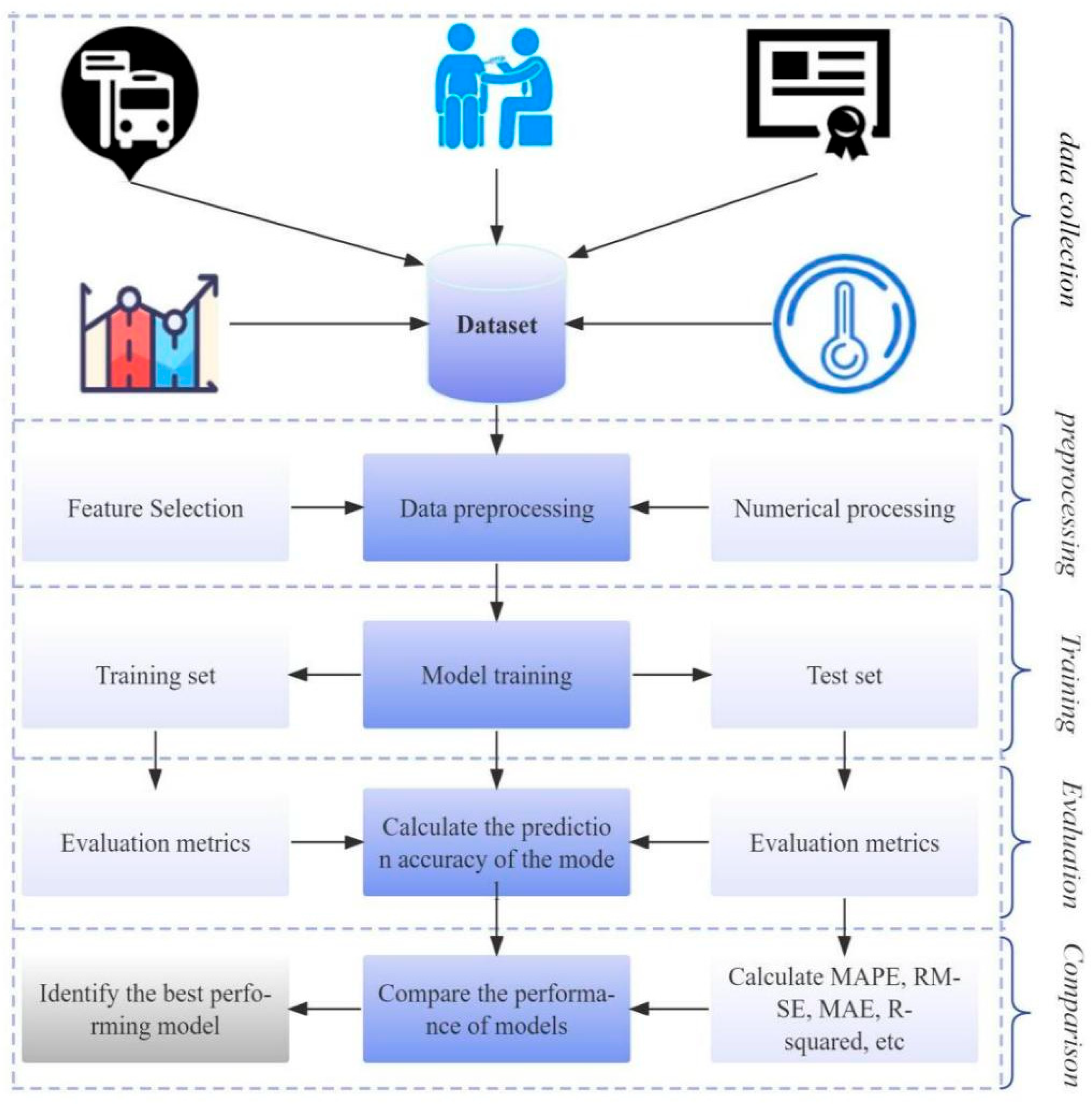
3.3.1. Dataset
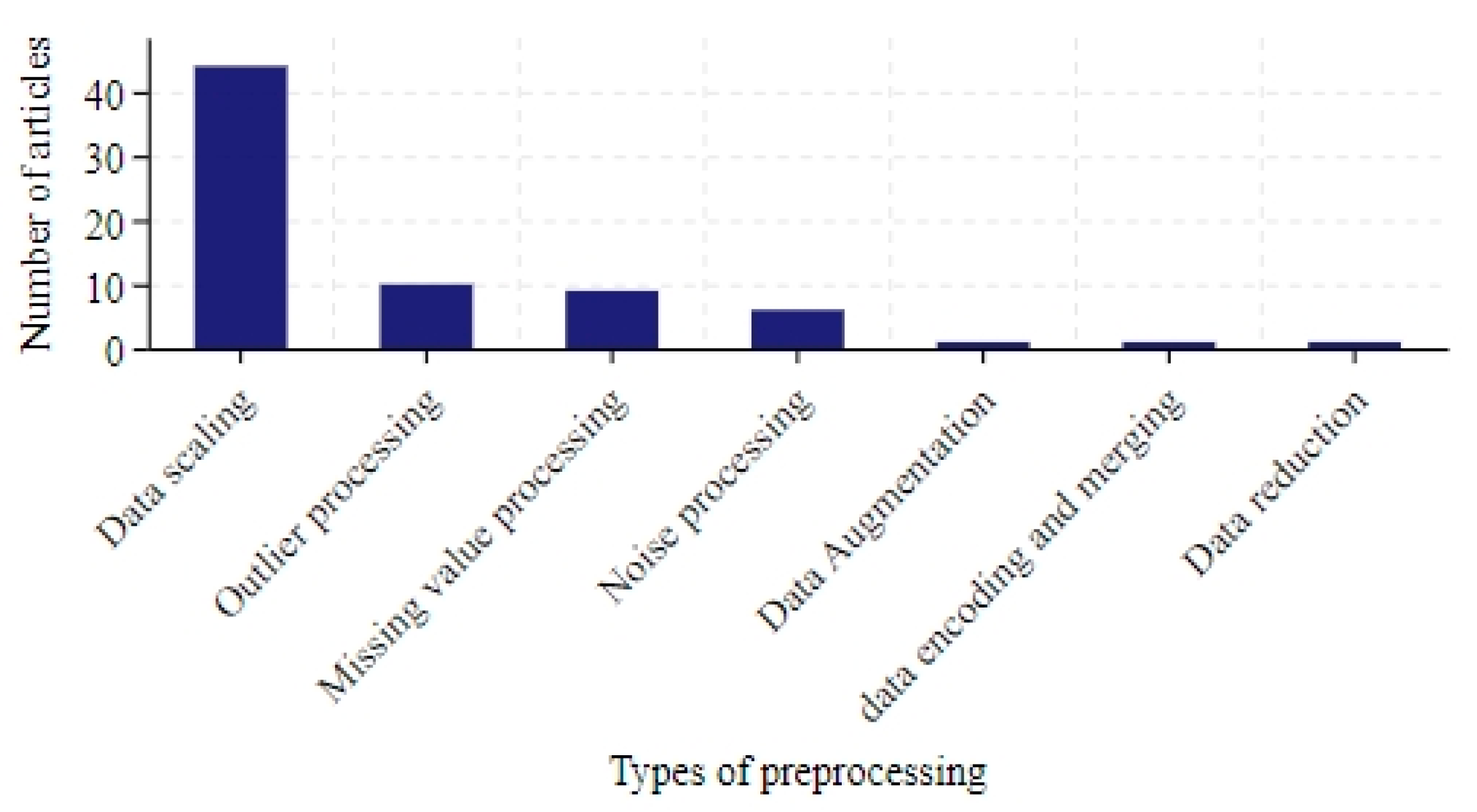
3.3.2. Data Preprocessing
3.3.3. Evaluation Indicator
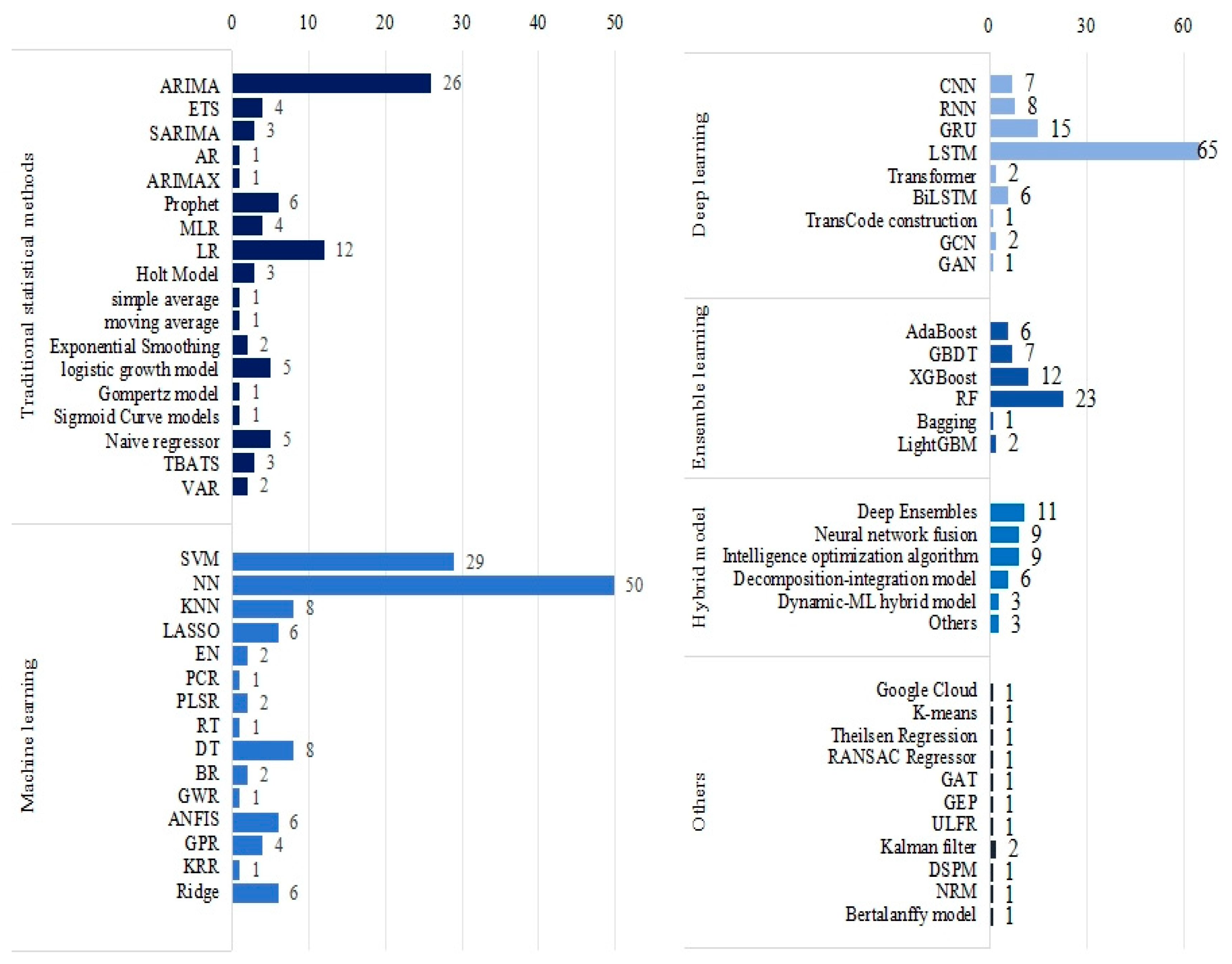
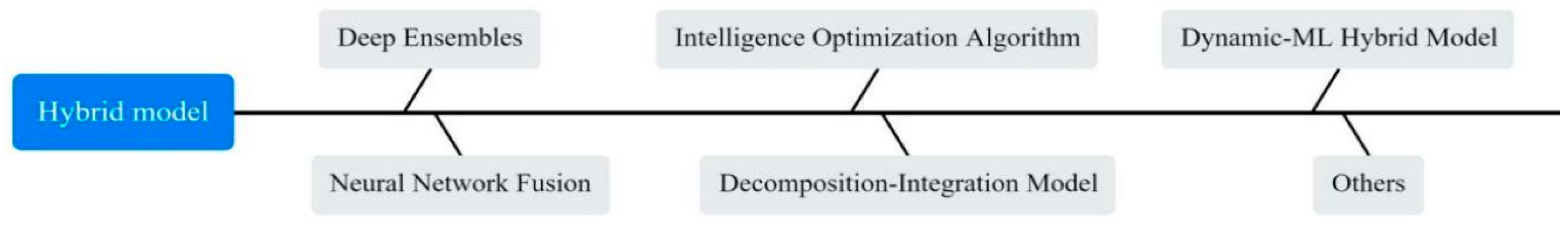
3.4. Machine Learning Models for COVID-19 Prediction
3.4.1. Meta-Heuristic Algorithmic Optimization Models
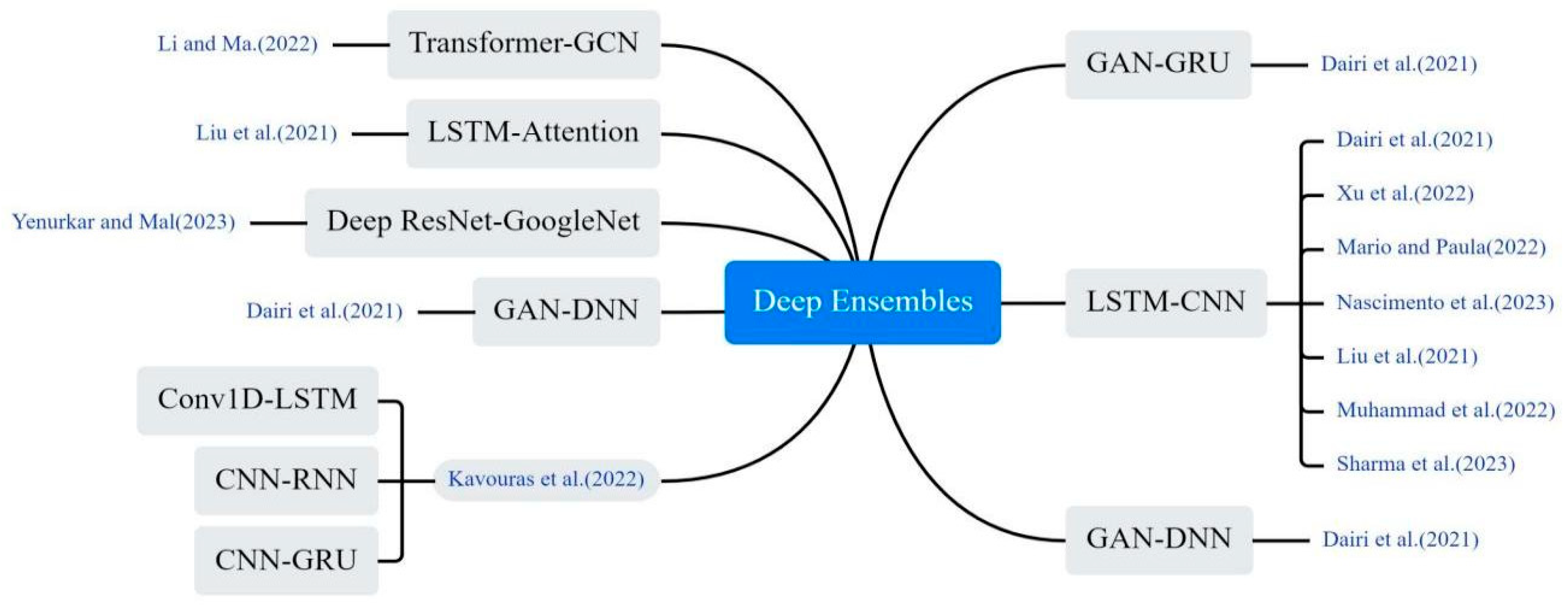
3.4.2. Deep Ensembles Models
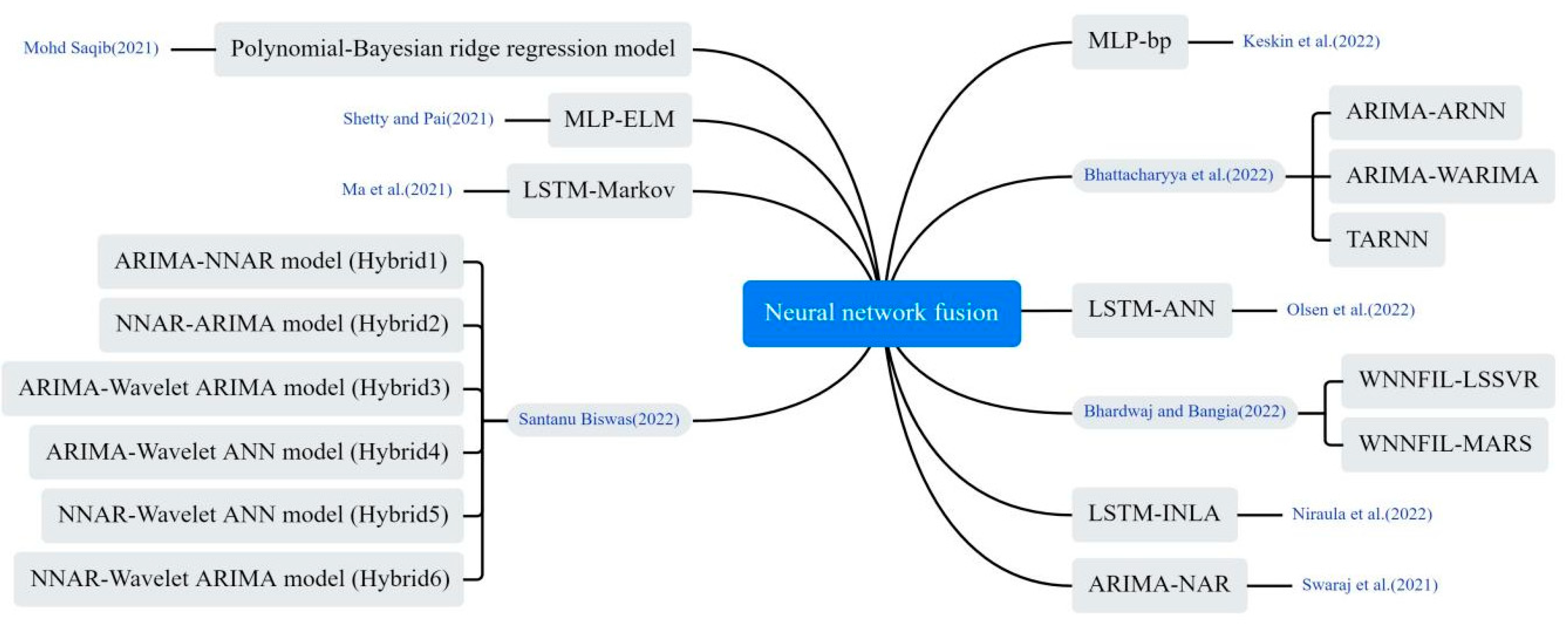
3.4.3. Neural Network Fusion Models
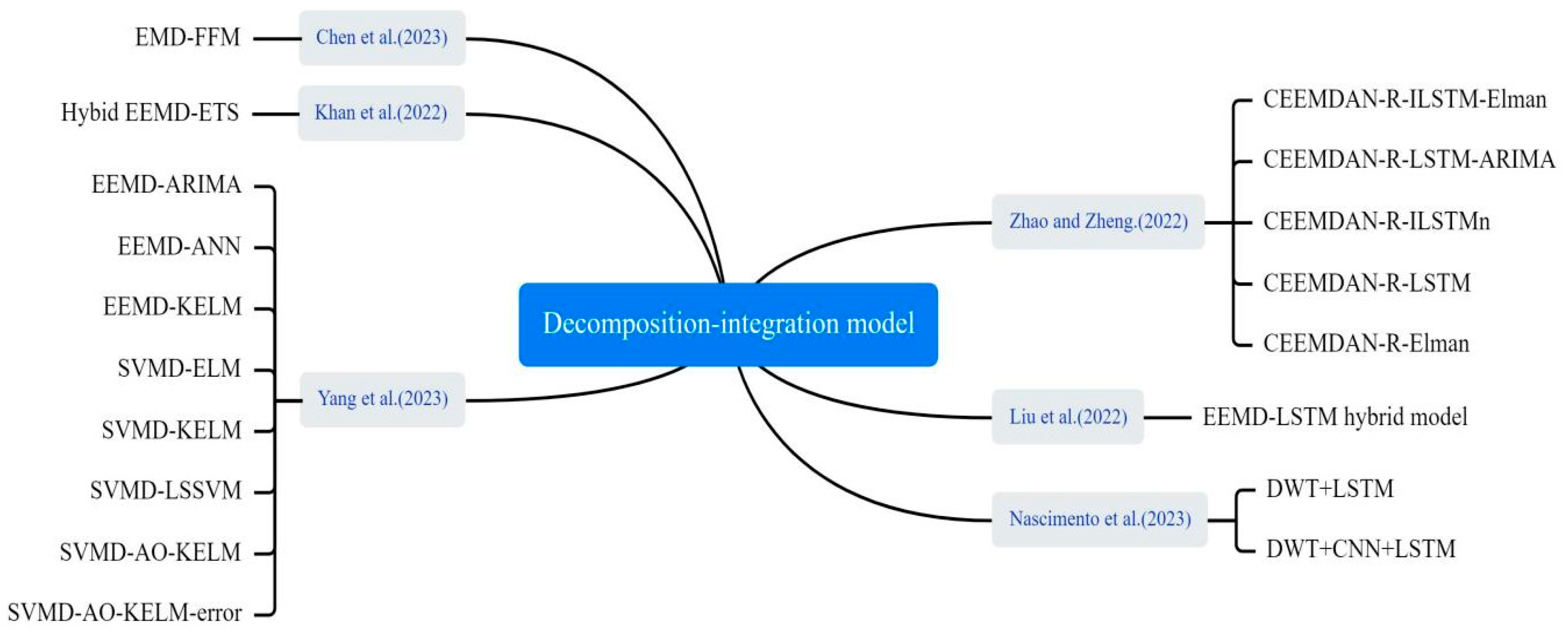
3.4.4. Decomposition–Integration Models
3.4.5. Dynamic–ML Hybrid Model
3.4.6. Other Models
3.5. Performance Comparison Between Machine Learning Models and Other Models
4. Conclusions
- The spread of infectious diseases is influenced by a variety of factors, including historical cases, meteorological conditions, and socio-economic factors such as population movements. Consideration of these influences in COVID-19 projections helps to more fully understand and predict trends and impacts of outbreak spread.
- Data scaling, outlier processing, missing value processing and noise processing are commonly used in data preprocessing methods.
- LSTM and SVM are the most commonly used ML models. The prediction accuracy of the model can be effectively improved by various hybrid strategies, such as heuristic algorithms, decomposition–reconstruction methods, and hybrid dynamics models.
- ML models typically have higher predictive accuracy than non-ML models.
- Despite the better performance of machine learning in COVID-19 prediction, it still has some limitations. Interpretability may limit the practical application of machine learning in infectious diseases.
5. Limitations and Future Challenges
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ML | Machine Learning |
| ARIMA | Auto Regressive Integrated Moving Average |
| SVM | Support Vector Machines |
| CNN | Convolutional Neural Networks |
| LSTM | Long Short-Term Memory Networks |
| WHO | World Health Organization |
| DL | Deep Learning |
| EHR | Electronic Health Records |
| MAPE | Mean Absolute Percentage Error |
| RMSE | Root Mean Square Error |
| MAE | Mean Absolute Error |
| MSE | Mean Squared Error |
| NN | Neural Networks |
| RF | Random Forest |
| GRU | Gated Recurrent |
| LR | Linear Regression |
| FNN | Feed Forward Neural Network |
| MLP | Multi-layer Perceptron |
| RNN | Recurrent Neural Networks |
| ICU | Intensive Care Units |
| DT | Decision Tree |
| GA | Genetic Algorithm |
| PSO | Particle Swarm Optimization |
| DE | Differential Evolution |
| FA | Firefly Algorithm |
| HS | Harmony Search |
| TLBO | Teaching–Learning-Based Optimization |
| BA | Bees Algorithm |
| mBA | mutation-based Bees Algorithm |
| LsOA | Lioness Optimization Algorithm |
| HBA | Honey Badger Algorithm |
| ABC | Artificial Bee Colony |
| CS | Cuckoo Search Algorithm |
| BBO | Biogeography-Based Optimization |
| IBAS | Improved Beetle Antennae Search Algorithm |
| SSA | Sparrow Search Algorithm |
| ANFIS | Adaptive Neuro-Fuzzy Inference System |
| GCN | Graph Convolutional Network |
| ANN | Artificial Neural Network |
| GEP | Gene Expression Programming |
| ELM | Extreme Learning Machine |
| LSSVR | Least Squares Support Vector Machine |
| WNN | Wavelet Neural Network |
| MARS | Multivariate Adaptive Regression Spline |
| LMBP | Levenberg–Marquardt Back Propagation |
| NAR | Nonlinear Auto Regressive |
| CEEMD | Complete Ensemble Empirical Mode Decomposition |
| VMD | Variational Mode Decomposition |
| AO-KELM | Adaptive Optimization-based Kernel Extreme Learning Machine |
| GLM | General Linear Model |
| NLP | Natural Language Processing |
| LIME | Local Interpretable Model-agnostic Explanations |
| SHAP | Shapley Additive Explanations |
Appendix A
| No. | QA1 | QA2 | QA3 | QA4 | QA5 | QA6 | QA7 | QA8 | QA9 | QA10 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 15 |
| 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 18 |
| 3 | 2 | 2 | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 0 | 13 |
| 4 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 17 |
| 5 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 18 |
| 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 7 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 17 |
| 8 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 9 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 0 | 15 |
| 10 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 11 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 14 |
| 12 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 18 |
| 13 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 18 |
| 14 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 17 |
| 15 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 16 |
| 16 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 16 |
| 17 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 0 | 12 |
| 18 | 2 | 2 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 0 | 10 |
| 19 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 20 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 21 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 22 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 15 |
| 23 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | 2 | 0 | 15 |
| 24 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 25 | 1 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 15 |
| 26 | 2 | 2 | 1 | 0 | 1 | 2 | 2 | 2 | 2 | 0 | 14 |
| 27 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 19 |
| 28 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 29 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 30 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 31 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 32 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 33 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 17 |
| 34 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 0 | 13 |
| 35 | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 12 |
| 36 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 37 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 18 |
| 38 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 0 | 15 |
| 39 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 19 |
| 40 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 12 |
| 41 | 2 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 42 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 43 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 13 |
| 44 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 45 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 13 |
| 46 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 47 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 17 |
| 48 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 16 |
| 49 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 50 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 15 |
| 51 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 52 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 18 |
| 53 | 2 | 1 | 1 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 11 |
| 54 | 2 | 2 | 1 | 2 | 0 | 2 | 1 | 2 | 2 | 0 | 14 |
| 55 | 2 | 2 | 1 | 0 | 0 | 2 | 2 | 2 | 1 | 0 | 12 |
| 56 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 57 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 16 |
| 58 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 59 | 2 | 2 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 60 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 2 | 1 | 0 | 12 |
| 61 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 16 |
| 62 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 63 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 64 | 2 | 2 | 1 | 0 | 1 | 2 | 2 | 2 | 2 | 0 | 14 |
| 65 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 0 | 12 |
| 66 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 0 | 14 |
| 67 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 68 | 2 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 11 |
| 69 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 70 | 2 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 11 |
| 71 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 12 |
| 72 | 2 | 2 | 1 | 1 | 1 | 2 | 0 | 2 | 2 | 2 | 15 |
| 73 | 2 | 2 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 74 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 75 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 14 |
| 76 | 2 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 77 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 1 | 1 | 2 | 12 |
| 78 | 2 | 1 | 0 | 1 | 0 | 2 | 2 | 2 | 1 | 0 | 11 |
| 79 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 80 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 17 |
| 81 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 13 |
| 82 | 2 | 2 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 83 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 0 | 15 |
| 84 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 1 | 2 | 17 |
| 85 | 2 | 1 | 1 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 10 |
| 86 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 87 | 2 | 2 | 2 | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 12 |
| 88 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 16 |
| 89 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 90 | 2 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 91 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 16 |
| 92 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 1 | 16 |
| 93 | 2 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 11 |
| 94 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 0 | 17 |
| 95 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 18 |
| 96 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 1 | 0 | 13 |
| 97 | 2 | 1 | 1 | 1 | 0 | 2 | 0 | 2 | 2 | 0 | 11 |
| 98 | 2 | 2 | 1 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 12 |
| 99 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 0 | 16 |
| 100 | 2 | 2 | 1 | 1 | 2 | 2 | 0 | 2 | 2 | 1 | 15 |
| 101 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 102 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 17 |
| 103 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 19 |
| 104 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 1 | 1 | 2 | 14 |
| 105 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 106 | 2 | 1 | 0 | 0 | 0 | 2 | 2 | 1 | 1 | 0 | 9 |
| 107 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 108 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 16 |
| 109 | 2 | 1 | 1 | 0 | 0 | 2 | 2 | 1 | 1 | 0 | 10 |
| 110 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 111 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | 2 | 1 | 0 | 14 |
| 112 | 2 | 2 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | 10 |
| 113 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 14 |
| 114 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 17 |
| 115 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 18 |
| 116 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 17 |
| 117 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 118 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 14 |
| 119 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 16 |
| 120 | 2 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 121 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 12 |
| 122 | 2 | 1 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 12 |
| 123 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 17 |
| 124 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 16 |
| 125 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 15 |
| 126 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 0 | 16 |
| 127 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 0 | 15 |
| 128 | 2 | 2 | 1 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 12 |
| 129 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 14 |
| 130 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 17 |
| 131 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 1 | 0 | 13 |
| 132 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 15 |
| 133 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 134 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 15 |
| 135 | 2 | 2 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 13 |
| 136 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 14 |
| Project | Attribute Name | Motivation |
|---|---|---|
| Publication characteristics | Title | Literature title |
| Authors | The author of the document | |
| Keywords | Key words of literature | |
| Journal | Published journals | |
| Country | The country of study | |
| Year | Year of publication | |
| Data | Data sources | Where did the data information come from? |
| Number of samples | The total number of samples in the dataset | |
| Input features | Variables in data sets used to train ML models | |
| Target variable | Variables that the model tries to predict or explain | |
| Feature Selection | Select the most relevant feature from all available features | |
| Data Correction | The process of cleaning and transforming the original data | |
| Data Split | Divide the data set into training set and test set according to a certain proportion | |
| Methods | Models | ML methods used in literature |
| Intelligence Algorithms | Intelligent optimization algorithm used in literature | |
| Performance | Index | Evaluation index of model performance used in the literature |
| Best model | The model with the best prediction performance |
References
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Albrecht, J.K. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar] [CrossRef] [PubMed]
- van den Brand, J.M.; Stittelaar, K.J.; van Amerongen, G.; Rimmelzwaan, G.F.; Simon, J.; de Wit, E.; Osterhaus, A.D. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J. Infect. Dis. 2010, 201, 993–999. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, J.M.; Dong, S.J.; Li, J.; Ji, J.S. The Ebola epidemic is ongoing in West Africa and responses from China are positive. Mil. Med. Res. 2015, 2, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamaguchi, F.; Suzuki, A.; Hashiguchi, M.; Kondo, E.; Maeda, A.; Yokoe, T.; Sasaki, J.; Shikama, Y.; Hayashi, M.; Kobayashi, S.; et al. Combination of rRT-PCR and Clinical Features to Predict Coronavirus Disease 2019 for Nosocomial Infection Control. Infect. Drug Resist. 2024, 17, 161–170. [Google Scholar] [CrossRef]
- Hu, X.; Flahault, A.; Temerev, A.; Rozanova, L. The Progression of COVID-19 and the Government Response in China. Int. J. Environ. Res. Public Health. 2021, 18, 3002. [Google Scholar] [CrossRef]
- Cenggoro, T.W.; Pardamean, B. A systematic literature review of machine learning application in COVID-19 medical image classification. Procedia Comput. Sci. 2023, 216, 749–756. [Google Scholar]
- Kamran, F.; Tang, S.; Otles, E.; McEvoy, D.S.; Saleh, S.N.; Gong, J.; Wiens, J. Early identification of patients admitted to hospital for covid-19 at risk of clinical deterioration: Model development and multisite external validation study. BMJ 2022, 376, e068576. [Google Scholar] [CrossRef]
- Sharin, S.N.; Radzali, M.K.; Sani, M.S.A. A network analysis and support vector regression approaches for visualising and predicting the COVID-19 outbreak in Malaysia. Healthc. Anal. 2022, 2, 100080. [Google Scholar] [CrossRef]
- Oyewola, D.O.; Dada, E.G.; Misra, S. Machine learning for optimizing daily COVID-19 vaccine dissemination to combat the pandemic. Health Technol. 2022, 12, 1277–1293. [Google Scholar] [CrossRef]
- Jacob, J. Utilisation of deep learning for COVID-19 diagnosis: A review. Clin. Radiol. 2023, 78, 150–157. [Google Scholar]
- Chen, W.; Sá, R.C.; Bai, Y.; Napel, S.; Gevaert, O.; Lauderdale, D.S.; Giger, M.L. Machine learning with multimodal data for COVID-19. Heliyon 2023, 9, e17934. [Google Scholar] [CrossRef] [PubMed]
- Sailunaz, K.; Özyer, T.; Rokne, J.; Alhajj, R. A survey of machine learning-based methods for COVID-19 medical image analysis. Med. Biol. Eng. Comput. 2023, 61, 1257–1297. [Google Scholar] [CrossRef]
- Abbasi Habashi, S.; Koyuncu, M.; Alizadehsani, R. A survey of COVID-19 diagnosis using routine blood tests with the aid of artificial intelligence techniques. Diagnostics 2023, 13, 1749. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ayus, I.; Gupta, D. A comprehensive review of COVID-19 detection with machine learning and deep learning techniques. Health Technol. 2023, 13, 679–692. [Google Scholar] [CrossRef]
- Soda, P.; D’Amico, N.C.; Tessadori, J.; Valbusa, G.; Guarrasi, V.; Bortolotto, C.; Akbar, M.U.; Sicilia, R.; Cordelli, E.; Fazzini, D.; et al. AIforCOVID: Predicting the clinical outcomes in patients with COVID-19 applying AI to chest-X-rays. An Italian multicentre study. Med. Image Anal. 2021, 74, 102216. [Google Scholar] [CrossRef]
- Prinzi, F.; Militello, C.; Scichilone, N.; Gaglio, S.; Vitabile, S. Explainable machine-learning models for COVID-19 prognosis prediction using clinical, laboratory and radiomic features. IEEE Access 2023, 11, 121492–121510. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, S.; Li, L.; Wu, Q.; Qian, W.; Hu, Y.; Li, L.; Zhou, X.; Ma, H.; Li, H.; et al. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics 2020, 10, 7231–7244. [Google Scholar] [CrossRef]
- Wang, D.; Huang, C.; Bao, S.; Fan, T.; Sun, Z.; Wang, Y.; Jiang, H.; Wang, S. Study on the prognosis predictive model of COVID-19 patients based on CT radiomics. Sci. Rep. 2021, 11, 11591. [Google Scholar] [CrossRef]
- Signoroni, A.; Savardi, M.; Benini, S.; Adami, N.; Leonardi, R.; Gibellini, P.; Vaccher, F.; Ravanelli, M.; Borghesi, A.; Maroldi, R.; et al. BS-Net: Learning COVID-19 pneumonia severity on a large chest X-ray dataset. Med. Image Anal. 2021, 71, 102046. [Google Scholar] [CrossRef]
- Kitchenham, B.; Brereton, P. A systematic review of systematic review process research in software engineering. Inf. Softw. Technol. 2013, 55, 2049–2075. [Google Scholar] [CrossRef]
- Kumar, Y.; Koul, A.; Kaur, S.; Hu, Y.C. Machine learning and deep learning based time series prediction and forecasting of ten nations’ COVID-19 pandemic. SN Comput. Sci. 2022, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, C.; Rong, Y.; Pang, C.P.; Chen, X.; Chen, H. Real-time prediction of the daily incidence of COVID-19 in 215 countries and territories using machine learning: Model development and validation. J. Med. Internet Res. 2021, 23, e24285. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, K. A Hybrid Model Based on Improved Transformer and Graph Convolutional Network for COVID-19 Forecasting. Int. J. Environ. Res. Public Health 2022, 19, 12528. [Google Scholar] [CrossRef] [PubMed]
- Ilu, S.Y.; Prasad, R. Improved autoregressive integrated moving average model for COVID-19 prediction by using statistical significance and clustering techniques. Heliyon 2023, 9, e13483. [Google Scholar] [CrossRef]
- Zheng, N.; Du, S.; Wang, J.; Zhang, H.; Cui, W.; Kang, Z.; Yang, T.; Lou, B.; Chi, Y.; Long, H.; et al. Predicting COVID-19 in China using hybrid AI model. IEEE Trans. Cybern. 2020, 50, 2891–2904. [Google Scholar] [CrossRef]
- Kuo, C.P.; Fu, J.S. Evaluating the impact of mobility on COVID-19 pandemic with machine learning hybrid predictions. Sci. Total Environ. 2021, 758, 144151. [Google Scholar] [CrossRef]
- Shrivastav, L.K.; Jha, S.K. A gradient boosting machine learning approach in modeling the impact of temperature and humidity on the transmission rate of COVID-19 in India. Appl. Intell. 2021, 51, 2727–2739. [Google Scholar] [CrossRef]
- Olsen, F.; Schillaci, C.; Ibrahim, M.; Lipani, A. Borough-level COVID-19 forecasting in London using deep learning techniques and a novel MSE-Moran’s I loss function. Results Phys. 2022, 35, 105374. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, Y.K.; Mishra, A.K. Analysis and prediction of COVID-19 multivariate data using deep ensemble learning methods. Int. J. Environ. Res. Public Health 2023, 20, 5943. [Google Scholar] [CrossRef]
- De Ruvo, S.; Pio, G.; Vessio, G.; Volpe, V. Forecasting and what-if analysis of new positive COVID-19 cases during the first three waves in Italy. Med. Biol. Eng. Comput. 2023, 61, 2051–2066. [Google Scholar] [CrossRef]
- Xu, L.; Magar, R.; Farimani, A.B. Forecasting COVID-19 new cases using deep learning methods. Comput. Biol. Med. 2022, 144, 105342. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Gomes, D.C.; de Oliveira Serra, G.L. Machine learning model for computational tracking and forecasting the COVID-19 dynamic propagation. IEEE J. Biomed. Health Inform. 2021, 25, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Saqib, M. Forecasting COVID-19 outbreak progression using hybrid polynomial-Bayesian ridge regression model. Appl. Intell. 2021, 51, 2703–2713. [Google Scholar] [CrossRef] [PubMed]
- Davahli, M.R.; Karwowski, W.; Fiok, K. Optimizing COVID-19 vaccine distribution across the United States using deterministic and stochastic recurrent neural networks. PLoS ONE 2021, 16, e0253925. [Google Scholar] [CrossRef]
- Rashed, E.A.; Hirata, A. One-year lesson: Machine learning prediction of COVID-19 positive cases with meteorological data and mobility estimate in Japan. Int. J. Environ. Res. Public Health 2021, 18, 5736. [Google Scholar] [CrossRef]
- Didi, Y.; Walha, A.; Ben Halima, M.; Wali, A. COVID-19 Outbreak Forecasting Based on Vaccine Rates and Tweets Classification. Comput. Intell. Neurosci. 2022, 2022, 4535541. [Google Scholar] [CrossRef]
- Yu, C.S.; Chang, S.S.; Chang, T.H.; Wu, J.L.; Lin, Y.J.; Chien, H.F.; Chen, R.J. A COVID-19 pandemic artificial intelligence–based system with deep learning forecasting and automatic statistical data acquisition: Development and implementation study. J. Med. Internet Res. 2021, 23, e27806. [Google Scholar] [CrossRef]
- Kavouras, I.; Kaselimi, M.; Protopapadakis, E.; Bakalos, N.; Doulamis, N.; Doulamis, A. COVID-19 spatio-temporal evolution using deep learning at a European level. Sensors 2022, 22, 3658. [Google Scholar] [CrossRef]
- Yeung, A.Y.; Roewer-Despres, F.; Rosella, L.; Rudzicz, F. Machine learning–based prediction of growth in confirmed COVID-19 infection cases in 114 countries using metrics of nonpharmaceutical interventions and cultural dimensions: Model development and validation. J. Med. Internet Res. 2021, 23, e26628. [Google Scholar] [CrossRef]
- Saif, S.; Das, P.; Biswas, S. A hybrid model based on mba-anfis for COVID-19 confirmed cases prediction and forecast. J. Inst. Eng. Ser. B 2021, 102, 1123–1136. [Google Scholar] [CrossRef]
- Li, D.; Ren, X.; Su, Y. Predicting COVID-19 using lioness optimization algorithm and graph convolution network. Soft Comput. 2023, 27, 5437–5501. [Google Scholar] [CrossRef] [PubMed]
- Shaibani, M.J.; Emamgholipour, S.; Moazeni, S.S. Investigation of robustness of hybrid artificial neural network with artificial bee colony and re y algorithm in predicting COVID-19 new cases: Case study of Iran. Stoch. Environ. Res. Risk Assess. 2022, 36, 2461–2476. [Google Scholar] [CrossRef] [PubMed]
- Qasem, S.N. A novel honey badger algorithm with multilayer perceptron for predicting COVID-19 time series data. J. Supercomput. 2024, 80, 3943–3969. [Google Scholar] [CrossRef]
- Shetty, R.P.; Pai, P.S. Forecasting of COVID 19 cases in Karnataka state using artificial neural network (ANN). J. Inst. Eng. Ser. B 2021, 102, 1201–1211. [Google Scholar] [CrossRef]
- Singh, K.K.; Kumar, S.; Dixit, P.; Bajpai, M.K. Kalman filter based short term prediction model for COVID-19 spread. Appl. Intell. 2020, 51, 2714–2726. [Google Scholar] [CrossRef]
- Sarmiento Varón, L.; González-Puelma, J.; Medina-Ortiz, D.; Aldridge, J.; Alvarez-Saravia, D.; Uribe-Paredes, R.; Navarrete, M.A. The role of machine learning in health policies during the COVID-19 pandemic and in long COVID management. Front. Public Health 2023, 11, 1140353. [Google Scholar] [CrossRef]
- Muñoz-Organero, M.; Queipo-Álvarez, P. Deep spatiotemporal model for COVID-19 forecasting. Sensors 2022, 22, 3519. [Google Scholar] [CrossRef]
- Sperandio Nascimento, E.G.; Ortiz, J.; Furtado, A.N.; Frias, D. Using discrete wavelet transform for optimizing COVID-19 new cases and deaths prediction worldwide with deep neural networks. PLoS ONE 2023, 18, e0282621. [Google Scholar] [CrossRef]
- Muhammad, L.J.; Haruna, A.A.; Sharif, U.S.; Mohammed, M.B. CNN-LSTM deep learning based forecasting model for COVID-19 infection cases in Nigeria, South Africa and Botswana. Health Technol. 2022, 12, 1259–1276. [Google Scholar] [CrossRef]
- Liu, Q.; Fung, D.L.; Lac, L.; Hu, P. A novel matrix profile-guided attention LSTM model for forecasting COVID-19 cases in USA. Front. Public Health 2021, 9, 741030. [Google Scholar] [CrossRef]
- Yenurkar, G.; Mal, S. Future forecasting prediction of Covid-19 using hybrid deep learning algorithm. Multimed. Tools Appl. 2023, 82, 22497–22523. [Google Scholar] [CrossRef]
- Dairi, A.; Harrou, F.; Zeroual, A.; Hittawe, M.M.; Sun, Y. Comparative study of machine learning methods for COVID-19 transmission forecasting. J. Biomed. Inform. 2021, 118, 103791. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zheng, X.; Wang, P.; Liu, H.; Zhang, C. The prediction and analysis of COVID-19 epidemic trend by combining LSTM and Markov method. Sci. Rep. 2021, 11, 17421. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, R.; Bangia, A. Hybridized wavelet neuronal learning-based modelling to predict novel COVID-19 effects in India and USA. Eur. Phys. J. Spec. Top. 2022, 231, 3471–3488. [Google Scholar] [CrossRef] [PubMed]
- Niraula, P.; Mateu, J.; Chaudhuri, S. A Bayesian machine learning approach for spatio-temporal prediction of COVID-19 cases. Stoch. Environ. Res. Risk Assess. 2022, 36, 2265–2283. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Chakraborty, T.; Rai, S.N. Stochastic forecasting of COVID-19 daily new cases across countries with a novel hybrid time series model. Nonlinear Dyn. 2022, 107, 3025–3040. [Google Scholar] [CrossRef] [PubMed]
- Keskin, G.A.; Doğruparmak, Ş.Ç.; Ergün, K. Estimation of COVID-19 patient numbers using artificial neural networks based on air pollutant concentration levels. Environ. Sci. Pollut. Res. 2022, 29, 68269–68279. [Google Scholar] [CrossRef]
- Biswas, S. Forecasting and comparative analysis of Covid-19 cases in India and US. Eur. Phys. J. Spec. Top. 2022, 231, 3537–3544. [Google Scholar] [CrossRef]
- Swaraj, A.; Verma, K.; Kaur, A.; Singh, G.; Kumar, A.; de Sales, L.M. Implementation of stacking based ARIMA model for prediction of Covid-19 cases in India. J. Biomed. Inform. 2021, 121, 103887. [Google Scholar] [CrossRef]
- Zhao, Q.; Zheng, Z. Computational and Mathematical Methods in Medicine Prediction of COVID-19 in BRICS Countries: An Integrated Deep Learning Model of CEEMDAN-R-ILSTM-Elman. Comput. Math. Methods Med. 2022, 2022, 1566727. [Google Scholar] [CrossRef]
- Liu, S.; Wan, Y.; Yang, W.; Tan, A.; Jian, J.; Lei, X. A hybrid model for coronavirus disease 2019 forecasting based on ensemble empirical mode decomposition and deep learning. Int. J. Environ. Res. Public Health 2022, 20, 617. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, H.; Li, G. A novel prediction model based on decomposition-integration and error correction for COVID-19 daily confirmed and death cases. Comput. Biol. Med. 2023, 156, 106674. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.M.; Ali, M.; Iqbal, N.; Khalil, U.; Aljohani, H.M.; Alharthi, A.S.; Afify, A.Z. Short-term prediction of COVID-19 using novel hybrid ensemble empirical mode decomposition and error trend seasonal model. Front. Public Health 2022, 10, 922795. [Google Scholar] [CrossRef]
- Chen, B.L.; Shen, Y.Y.; Zhu, G.C.; Yu, Y.T.; Ji, M. An empirical mode decomposition fuzzy forecast model for COVID-19. Neural Process. Lett. 2023, 55, 2369–2390. [Google Scholar] [CrossRef]
- Liu, X.D.; Wang, W.; Yang, Y.; Hou, B.H.; Olasehinde, T.S.; Feng, N.; Dong, X.P. Nesting the SIRV model with NAR, LSTM and statistical methods to fit and predict COVID-19 epidemic trend in Africa. BMC Public Health 2023, 23, 138. [Google Scholar] [CrossRef]
- Feng, L.; Chen, Z.; Lay, H.A., Jr.; Furati, K.; Khaliq, A. Data driven time-varying SEIR-LSTM/GRU algorithms to track the spread of COVID-19. Math. Biosci. Eng. 2022, 19, 8935–8962. [Google Scholar] [CrossRef]
- Farooq, J.; Bazaz, M.A. A deep learning algorithm for modeling and forecasting of COVID-19 in five worst affected states of India. Alex. Eng. J. 2021, 60, 587–596. [Google Scholar] [CrossRef]
- Zheng, H.L.; An, S.Y.; Qiao, B.J.; Guan, P.; Huang, D.S.; Wu, W. A data-driven interpretable ensemble framework based on tree models for forecasting the occurrence of COVID-19 in the USA. Environ. Sci. Pollut. Res. 2023, 30, 13648–13659. [Google Scholar] [CrossRef]
- Chakraborty, D.; Goswami, D.; Ghosh, S.; Ghosh, A.; Chan, J.H.; Wang, L. Transfer-recursive-ensemble learning for multi-day COVID-19 prediction in India using recurrent neural networks. Sci. Rep. 2023, 13, 6795. [Google Scholar] [CrossRef]
- Safari, A.; Hosseini, R.; Mazinani, M. A novel deep interval type-2 fuzzy LSTM (DIT2FLSTM) model applied to COVID-19 pandemic time-series prediction. J. Biomed. Inform. 2021, 123, 103920. [Google Scholar] [CrossRef]
- Silva, C.D.; Lima, C.D.; Silva, A.D.; Silva, E.L.; Marques, G.S.; Araújo, L.J.B.; Júnior, L.A.A.; Souza, S.B.J.; Santana, M.D.; Gomes, J.C.; et al. COVID-19 Dynamic Monitoring and Real-Time Spatio-Temporal Forecasting. Front. Public Health 2021, 9, 10–3389. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Sharma, R.; Qian, C.; Glass, L.M.; Spaeder, J.; Romberg, J.; Sun, J.; Xiao, C. STAN: Spatio-temporal attention network for pandemic prediction using real-world evidence. J. Am. Med. Inform. Assoc. 2021, 28, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Malki, Z.; Atlam, E.S.; Ewis, A.; Dagnew, G.; Ghoneim, O.A.; Mohamed, A.A.; Abdel-Daim, M.M.; Gad, I. The COVID-19 pandemic: Prediction study based on machine learning models. Environ. Sci. Pollut. Res. 2021, 28, 40496–40506. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, V.; Pal, S. Application of machine learning time series analysis for prediction COVID-19 pandemic. Res. Biomed. Eng. 2020, 38, 35–47. [Google Scholar] [CrossRef]
- Khakharia, A.; Shah, V.; Jain, S.; Shah, J.; Tiwari, A.; Daphal, P.; Warang, M.; Mehendale, N. Outbreak prediction of COVID-19 for dense and populated countries using machine learning. Ann. Data Sci. 2021, 8, 1–19. [Google Scholar] [CrossRef]
- Rahman, M.S.; Chowdhury, A.H. A data-driven eXtreme gradient boosting machine learning model to predict COVID-19 transmission with meteorological drivers. PLoS ONE 2022, 17, e0273319. [Google Scholar] [CrossRef]
- Said, A.B.; Erradi, A.; Aly, H.A.; Mohamed, A. Predicting COVID-19 cases using bidirectional LSTM on multivariate time series. Environ. Sci. Pollut. Res. 2021, 28, 56043–56052. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Vo, N.Q.; Nguyen, T.T.; Nguyen-An, K.; Nguyen, Q.H.; Tran, D.N.; Quan, T.T. BeCaked: An explainable artificial intelligence model for COVID-19 forecasting. Sci. Rep. 2022, 12, 7969. [Google Scholar] [CrossRef]
- Gomez-Cravioto, D.A.; Diaz-Ramos, R.E.; Cantu-Ortiz, F.J.; Ceballos, H.G. Data analysis and forecasting of the COVID-19 spread: A comparison of recurrent neural networks and time series models. Cogn. Comput. 2024, 16, 1794–1805. [Google Scholar] [CrossRef]
- Rguibi, M.A.; Moussa, N.; Madani, A.; Aaroud, A.; Zine-Dine, K. Forecasting covid-19 transmission with arima and lstm techniques in morocco. SN Comput. Sci. 2022, 3, 133. [Google Scholar] [CrossRef]
- Ketu, S.; Mishra, P.K. Enhanced Gaussian process regression-based forecasting model for COVID-19 outbreak and significance of IoT for its detection. Appl. Intell. 2021, 51, 1492–1512. [Google Scholar] [CrossRef] [PubMed]
- Sardar, I.; Akbar, M.A.; Leiva, V.; Alsanad, A.; Mishra, P. Machine learning and automatic ARIMA/Prophet models-based forecasting of COVID-19: Methodology, evaluation, and case study in SAARC countries. Stoch. Environ. Res. Risk Assess. 2023, 37, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Vig, V.; Kaur, A. Time series forecasting and mathematical modeling of COVID-19 pandemic in India: A developing country struggling to cope up. Int. J. Syst. Assur. Eng. Manag. 2022, 13, 2920–2933. [Google Scholar] [CrossRef]
- Islam, A.R.M.T.; Elbeltagi, A.; Mallick, J.; Fattah, M.A.; Roy, M.C.; Pal, S.C.; Shahjaman, M.; Patwary, M.A. Application of optimal subset regression and stacking hybrid models to estimate COVID-19 cases in Dhaka, Bangladesh. Theor. Appl. Climatol. 2023, 154, 797–814. [Google Scholar] [CrossRef]
- Pourroostaei Ardakani, S.; Xia, T.; Cheshmehzangi, A.; Zhang, Z. An urban-level prediction of lockdown measures impact on the prevalence of the COVID-19 pandemic. Genus 2022, 78, 28. [Google Scholar] [CrossRef]
- Rakhshan, S.A.; Nejad, M.S.; Zaj, M.; Ghane, F.H. Global analysis and prediction scenario of infectious outbreaks by recurrent dynamic model and machine learning models: A case study on COVID-19. Comput. Biol. Med. 2023, 158, 106817. [Google Scholar] [CrossRef]
- Kumar, R.; Al-Turjman, F.; Srinivas, L.N.B.; Braveen, M.; Ramakrishnan, J. ANFIS for prediction of epidemic peak and infected cases for COVID-19 in India. Neural Comput. Appl. 2023, 35, 7207–7220. [Google Scholar] [CrossRef]
- Banerjee, S.; Lian, Y. Data driven covid-19 spread prediction based on mobility and mask mandate information. Appl. Intell. 2022, 52, 1969–1978. [Google Scholar] [CrossRef]
- Bej, A.; Maulik, U.; Sarkar, A. Time-Series prediction for the epidemic trends of COVID-19 using Conditional Generative adversarial Networks Regression on country-wise case studies. SN Comput. Sci. 2022, 3, 352. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, R.; Algarni, F.; Kumar, I.; Choudhary, A.; Srivastava, A. Performance evaluation of regression models for COVID-19: A statistical and predictive perspective. Ain Shams Eng. J. 2022, 13, 101574. [Google Scholar] [CrossRef]
- Busari, S.I.; Samson, T.K. Modelling and forecasting new cases of Covid-19 in Nigeria: Comparison of regression, ARIMA and machine learning models. Sci. Afr. 2022, 18, e01404. [Google Scholar] [CrossRef] [PubMed]
- Kao, I.H.; Perng, J.W. Early prediction of coronavirus disease epidemic severity in the contiguous United States based on deep learning. Results Phys. 2021, 25, 104287. [Google Scholar] [CrossRef] [PubMed]
- Aljaaf, A.J.; Mohsin, T.M.; Al-Jumeily, D.; Alloghani, M. A fusion of data science and feed-forward neural network-based modelling of COVID-19 outbreak forecasting in IRAQ. J. Biomed. Inform. 2021, 118, 103766. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, J.; Elavarasan, R.M.; Pugazhendhi, R.; Shafiullah, G.M.; Ganesan, S.; Jeysree, A.K.; Khan, I.A.; Hossain, E. Forecasting of COVID-19 cases using deep learning models: Is it reliable and practically significant? Results Phys. 2021, 21, 103817. [Google Scholar] [CrossRef]
- ArunKumar, K.E.; Kalaga, D.V.; Kumar, C.M.S.; Kawaji, M.; Brenza, T.M. Comparative analysis of Gated Recurrent Units (GRU), long Short-Term memory (LSTM) cells, autoregressive Integrated moving average (ARIMA), seasonal autoregressive Integrated moving average (SARIMA) for forecasting COVID-19 trends. Alex. Eng. J. 2022, 61, 7585–7603. [Google Scholar] [CrossRef]
- Lounis, M.; Torrealba-Rodriguez, O.; Conde-Gutiérrez, R.A. Predictive models for COVID-19 cases, deaths and recoveries in Algeria. Results Phys. 2021, 30, 104845. [Google Scholar] [CrossRef]
| Infectious Disease | Influence |
|---|---|
| SARS | In 2002–2003, over 8000 people were infected, resulting in approximately 800 deaths and a mortality rate of around 10%. Most cases are concentrated in China, Hong Kong, Taiwan, Canada, the United States, and other places. |
| H1N1 | Over 1 billion people were infected, with an estimated death toll of 200,000 to 300,000, spreading globally. |
| MERS | Approximately 2500 people were infected and 900 people died, with a mortality rate of about 30%. It mainly spreads in the Middle East and also spreads to Asia, Europe, and the United States. |
| Ebola | Nearly 30,000 people were infected and approximately 11,000 people died. Mainly occurring in West Africa, the most severe outbreaks occurred in Liberia, Guinea, and Sierra Leone. |
| COVID-19 | More than 700 million people have been infected and over 6 million have died (as of 2023), and the COVID-19 pandemic has rapidly spread to almost every country worldwide |
| ID | Research Question |
|---|---|
| Q1 | What type of data is used in the study? |
| Q2 | How to handle incomplete, inaccurate, or noisy data? |
| Q3 | Which ML methods are applied to COVID-19 trend prediction? |
| Q4 | How to measure the prediction accuracy of ML technology? |
| Q5 | What are the main challenges and limitations of ML in COVID-19 prediction? |
| Digital Databases | Search Query |
|---|---|
| Web of Science | (“Machine learning” OR “AI” OR “Deep learning”) AND (COVID-19) AND (“case” OR “trend” OR “outbreak” OR “transmissions” OR “Spread”) AND (“Prediction” OR “Forecasting”) |
| Elsevier | (“Machine learning” OR “AI” OR “Deep learning”) AND (COVID-19) AND (“case” OR “trend” OR “outbreak” OR “transmissions” OR “Spread”) AND (“Prediction” OR “Forecasting”) |
| Springer | (Machine learning OR AI OR Deep learning) AND COVID-19 AND (case OR trend OR outbreak OR transmissions OR Spread) AND (Prediction OR Forecasting) |
| No. | Assessment Questions |
|---|---|
| AQ1 | Are the aims of the research clearly defined? |
| AQ2 | Is the topic of the article associated with the review? |
| AQ3 | Are data sources provided in the article? |
| AQ4 | Is the description of the data set clear in this article (data size, data splitting)? |
| AQ5 | Are there any data preprocessing methods in the article? |
| AQ6 | Are the research methods accurately described in the article? |
| AQ7 | Did the study compare the proposed method with other methods? |
| AQ8 | Is predictive performance measured and reported? |
| AQ9 | Are the findings/results clearly reported? |
| AQ10 | Are the limitations of research analyzed explicitly? |
| Quality Level | n | % |
|---|---|---|
| Very high (17 ≤ score ≤ 20) | 30 | 22 |
| High (14 ≤ score ≤ 16) | 65 | 48 |
| Medium (11 ≤ score ≤ 13) | 36 | 26 |
| Low (0 ≤ score ≤ 10) | 5 | 4 |
| Total | 136 | 100 |
| Metrics | Formula | n | % |
|---|---|---|---|
| RMSE | 93 | 24.2 | |
| MAE | 62 | 16.1 | |
| MAPE | 50 | 13 | |
| R-square | 47 | 12.2 | |
| MSE | 27 | 7 | |
| Accuracy | 13 | 3.4 | |
| R | 6 | 1.6 | |
| Code openness | Y/N | 23 | 16.9 |
| Data Availability | Y/N | 69 | 50.7 |
| Others | 79 | 22.5 |
| ML | Non-ML | Ref. | Best Model |
|---|---|---|---|
| RF, DT, KNR, Lasso, BR, KRR, Ransac Regressor, XGBoost, Elastic, Stacked LSTM, Stacked GRU | LR, Theilsen Regression, Holt Model | [21] | RF, Prophet, Stacked LSTM |
| Transformer-GCN, Transformer, LSTM, GRU | ARIMA, SARIMA | [23] | Transformer-GCN |
| XGboost, LSTM, NAIVEBAYESI | ARIMAI | [24] | ARIMAI |
| LASSO, LSTM, Interval type-2 fuzzy Kalman filter | ARIMA | [32] | Interval type-2 fuzzy Kalman filter |
| LSTM, Hybrid polynomial–Bayesian ridge regression model | ARIMA | [33] | Hybrid polynomial–Bayesian ridge regression model |
| Deterministic LSTM model, stochastic LSTM/MDN | LR | [34] | LSTM |
| LSTM | Google Cloud | [35] | LSTM |
| FNN, MLP, LSTM | ARIMA | [37] | LSTM |
| GAN-GRU, LSTM-CNN, RBM, GAN-DNN, CNN, LSTM, SVM | LR | [46] | LSTM-CNN |
| CEEMDAN-R-ILSTM-Elman | CEEMDAN-R-LSTM-ARIMA | [60] | CEEMDAN-R-ILSTM-Elman |
| SVR, MLP, RF | LR | [71] | LR |
| GRU, ColaGNN, CovidGNN, STAN-PC, STAN-Graph, STAN | SIR, SEIR | [72] | STAN |
| DT, RF, DL | ARIMA | [73] | DT |
| Naive method | Simple average, Moving average, Single exponential smoothing, Holt linear trend method, Holt–Winters method, ARIMA | [74] | Naive method |
| SVR, BR, RF, HW, XGBoost | ARMA, ARIMA, LR | [75] | ARIMA |
| XGBoost | ARIMAX | [76] | XGBoost |
| K-means, LSTM, Bi-LSTM | ARIMA, SMA-6, D-EXP-MA | [77] | Bi-LSTM |
| DTR, BeCaked, Ridge, SVR, LASSO, BR, RF | ARIMA | [78] | BeCaked |
| LSTM | Polynomial, VAR, LR, Sigmoid Curve models, Logistic model | [79] | LSTM |
| LSTM | ARIMA | [80] | LSTM |
| RF, SVR, LSTM, MTGP | LR | [81] | MTGP |
| RF, XGBoost | ARIMA, Prophet, GLMNet | [82] | ARIMA |
| RF | SMOreg, ARIMA, lBk, Gaussian Process, LR | [83] | ARIMA |
| SVM | AR, M5P, RSS | [84] | SVM |
| RF | KNN | [85] | RF |
| MLP, RBF, LSTM, ANFIS, GRNN | SEIRS | [86] | ANFIS, RBF |
| ANFIS | MLR | [87] | ANFIS |
| LSTM | ARIMA | [88] | LSTM |
| Ridge regression, ElasticNet, CGAN | Logistic, Lasso | [89] | CGAN |
| Ridge regression, Polynomial ridge regression, SVR | Polynomial regression, LR | [90] | Polynomial ridge |
| Fine Tree, Bagged Trees, Exponential GPR, Medium Tree, Boosted Trees, Trilayered Neural Network, Wide N.N., Matern 5/2 GPR, Squared exponential GPR, Rational Quadratic GPR | LR, Quadratic, Cubic, Inverse, ARIMA | [91] | Fine tree |
| AL-CNN | CAE | [92] | AL-CNN |
| FFNN | ETS, ARIMA | [93] | FFNN |
| LSTM, SLSTM | ARIMA, prophet | [94] | SLSTM, LSTM |
| GRU, LSTM | ARIMA, SARIMA | [95] | LSTM, GRU |
| ANNi | Gompertz model, Logistic, Bertalanffy model | [96] | ANNi |
| Total number (ML vs. Non-ML) | 31:5 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Cheng, R.; Xu, T.; Tan, X.; Bai, Y. Machine Learning Techniques Applied to COVID-19 Prediction: A Systematic Literature Review. Bioengineering 2025, 12, 514. https://doi.org/10.3390/bioengineering12050514
Cheng Y, Cheng R, Xu T, Tan X, Bai Y. Machine Learning Techniques Applied to COVID-19 Prediction: A Systematic Literature Review. Bioengineering. 2025; 12(5):514. https://doi.org/10.3390/bioengineering12050514
Chicago/Turabian StyleCheng, Yunyun, Rong Cheng, Ting Xu, Xiuhui Tan, and Yanping Bai. 2025. "Machine Learning Techniques Applied to COVID-19 Prediction: A Systematic Literature Review" Bioengineering 12, no. 5: 514. https://doi.org/10.3390/bioengineering12050514
APA StyleCheng, Y., Cheng, R., Xu, T., Tan, X., & Bai, Y. (2025). Machine Learning Techniques Applied to COVID-19 Prediction: A Systematic Literature Review. Bioengineering, 12(5), 514. https://doi.org/10.3390/bioengineering12050514