Artificial Intelligence Approaches for Geographic Atrophy Segmentation: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
1.1. Age-Related Macular Degeneration—Geographic Atrophy Definition
1.2. Geographic Atrophy Segmentation
1.3. Artificial Intelligence
2. Materials and Methods
2.1. Research Question
2.2. Eligibility Criteria
2.3. Search Strategy and Study Selection
2.4. Quality Assessment
2.5. Data Extraction
2.6. Statistical Analysis
3. Results
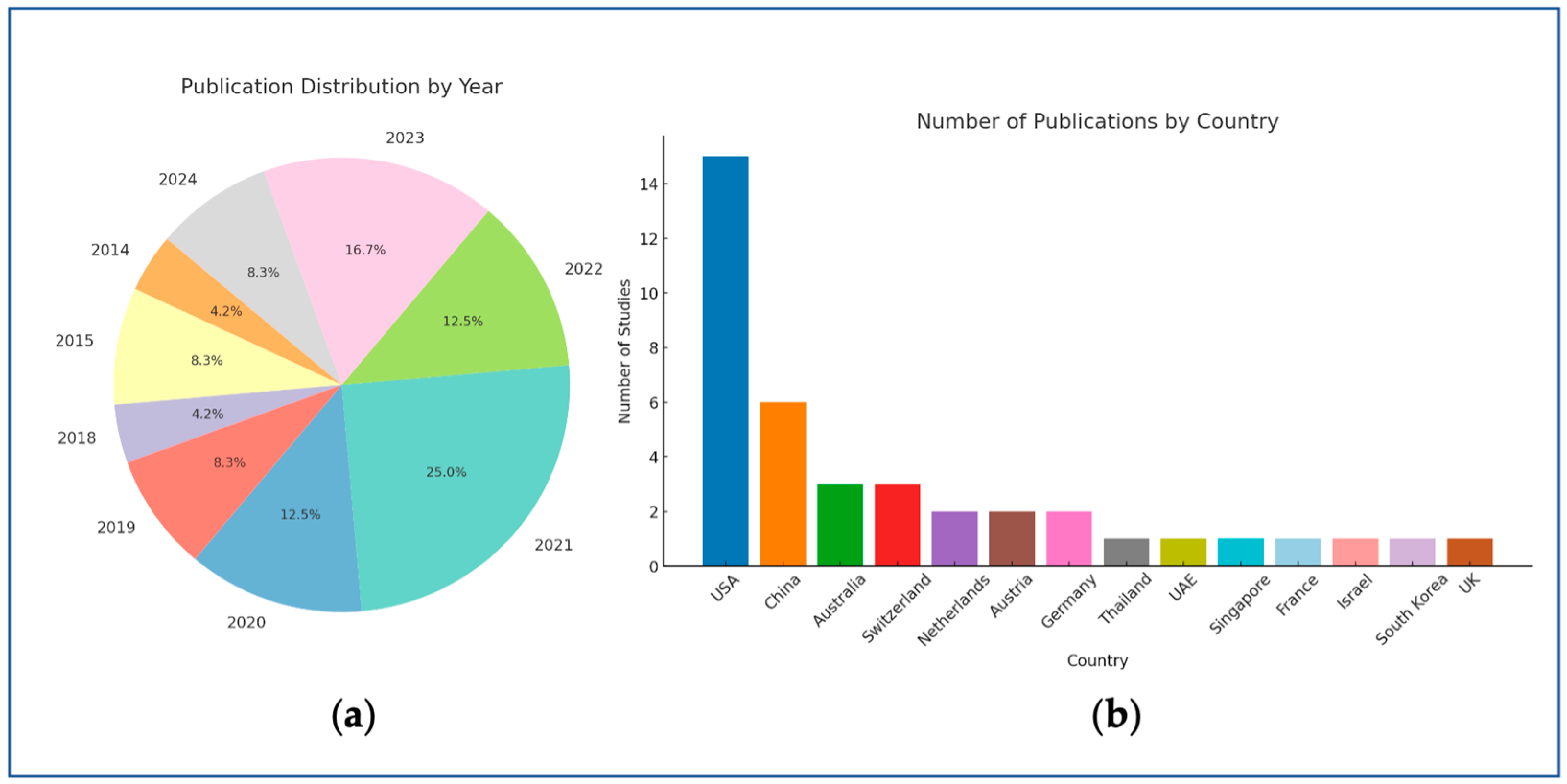
3.1. Study Selection
3.2. Risk of Bias, Applicability Concerns, and Reporting Quality
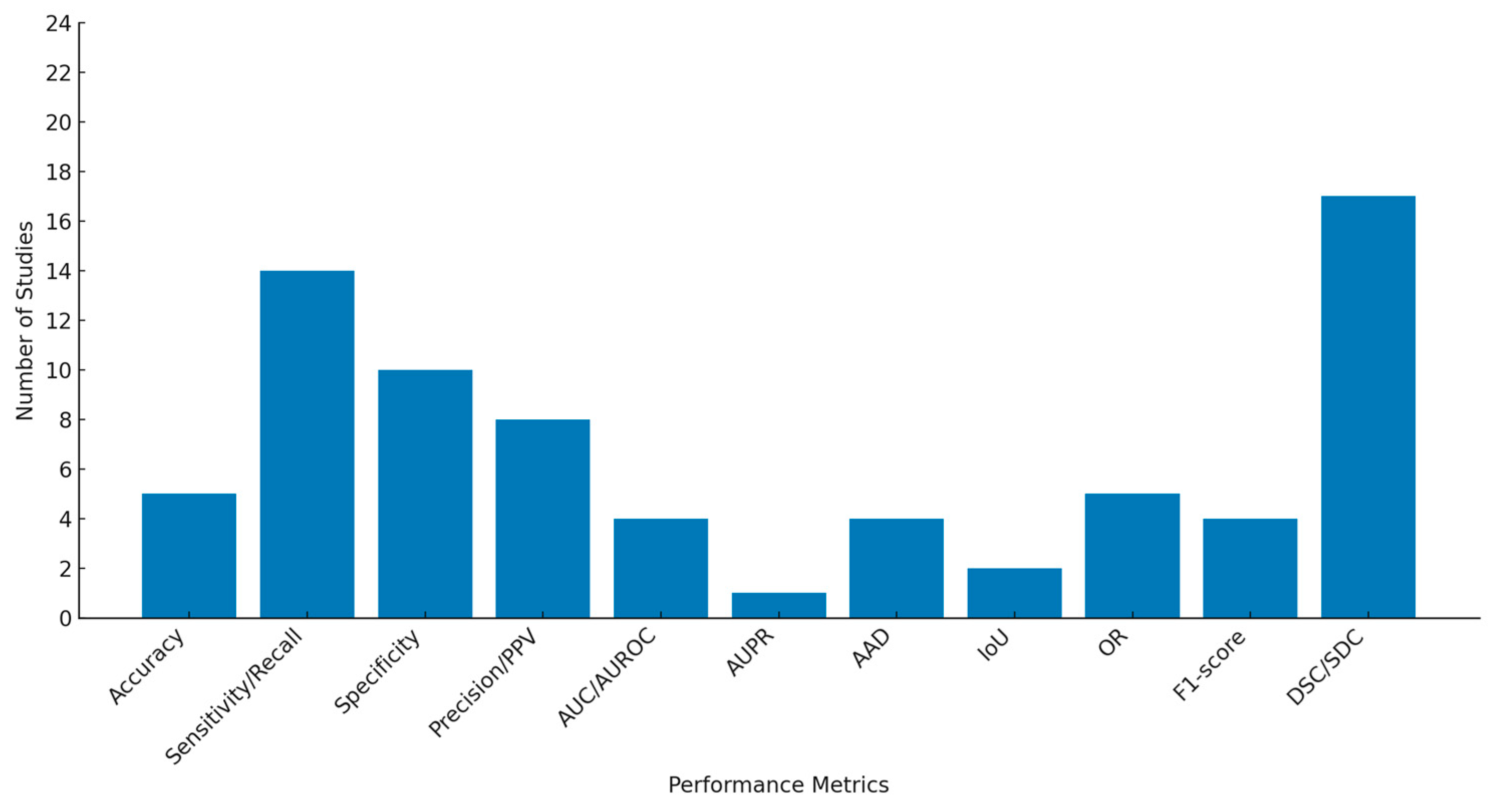
3.3. Characteristics of Included Studies
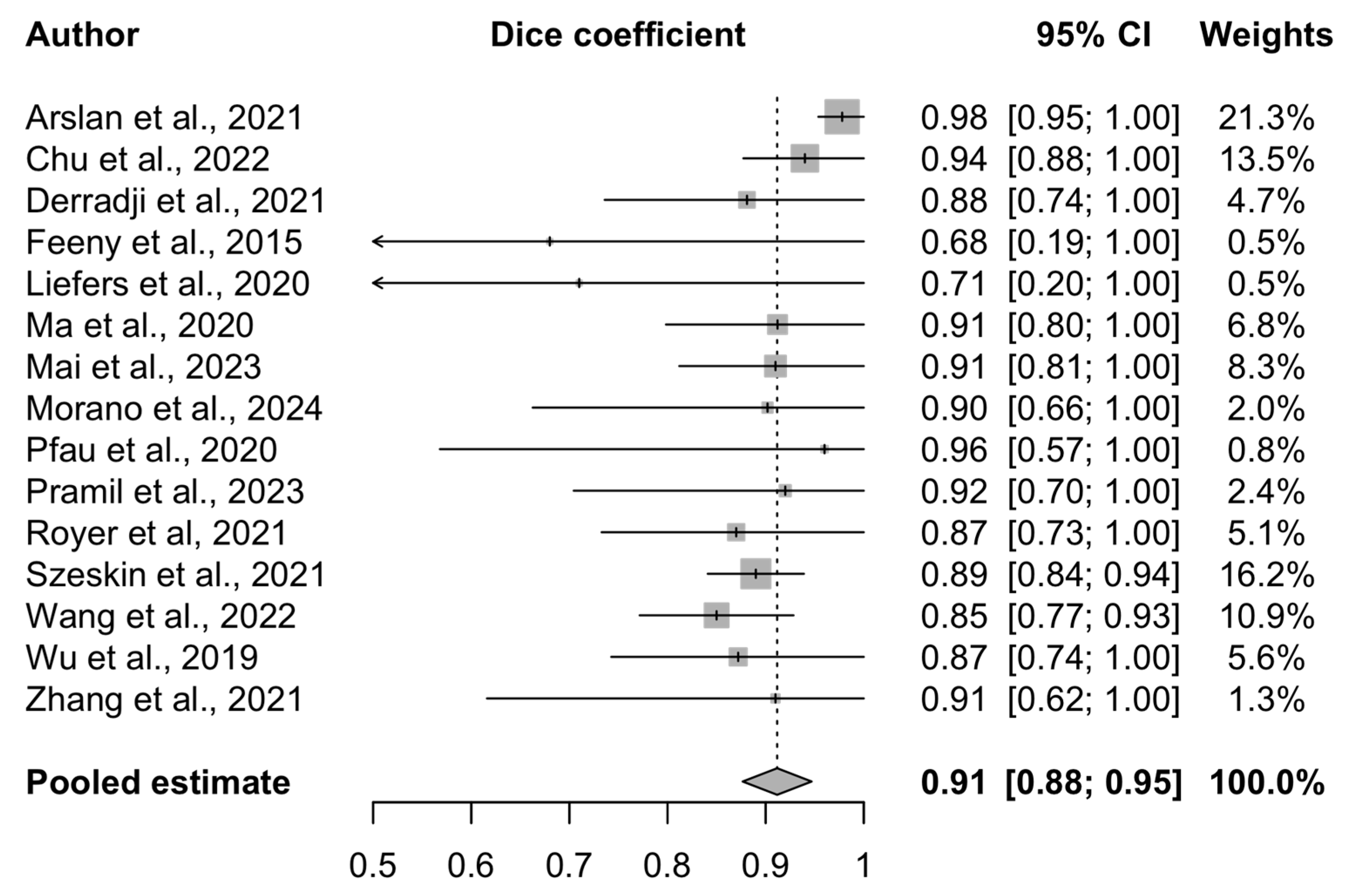
3.4. Meta-Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAD | Absolute Area Difference |
| AI | Artificial Intelligence |
| AIA | AI Act |
| AMD | Age-related Macular Degeneration |
| AUC | Area Under the Curve |
| AUPR | Area Under the Precision-Recall Curve |
| AUROC | Area Under the Receiver Operating Characteristic Curve |
| BCVA | Best-Corrected Visual Acuity |
| CAD | Computer-Aided Diagnosis |
| CAM | Class Activation Mapping |
| CAM* | Classification of Atrophy Meetings |
| CFP | Color Fundus Photography |
| CI | Confidence Interval |
| CLAIM | Checklist for Artificial Intelligence in Medical Imaging |
| CNN | Convolutional Neural Network |
| cORA | Complete Outer Retinal Atrophy |
| cRORA | Complete Retinal Pigment Epithelium and Outer Retinal Atrophy |
| cSLO | Confocal Scanning Laser Ophthalmoscopy |
| DL | Deep Learning |
| DSC | Dice Similarity Coefficient |
| DSS | Decision Support System |
| EHDS | European Health Data Space |
| FA | Fluorescein Angiography |
| FAF | Fundus Autofluorescence |
| GA | Geographic Atrophy |
| GDPR | General Data Protection Regulation |
| GT | Ground Truth |
| HIPAA | Health Insurance Portability and Accountability Act |
| ICTRP | International Clinical Trials Registry Platform |
| iORA | Incomplete Outer Retinal Atrophy |
| IoU | Intersection over Union |
| iRORA | Incomplete Retinal Pigment Epithelium and Outer Retinal Atrophy |
| k-NN | k-Nearest Neighbor |
| ML | Machine Learning |
| NIR | Near-Infrared Reflectance |
| OCT | Optical Coherence Tomography |
| OR | Overlap Ratio |
| PICOS | Population Intervention Comparator Outcome Study Design |
| PPV | Positive Predictive Value |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| QUADAS-2 | Quality Assessment of the Diagnostic Accuracy Studies-2 |
| RCT | Randomized Controlled Trial |
| ROI | Region of Interest |
| RORA | Retinal Pigment Epithelium and Outer Retinal Atrophy |
| RPE | Retinal Pigment Epithelium |
| SD | Standard Deviation |
| SD-OCT | Spectral-Domain Optical Coherence Tomography |
| SS-OCT | Swept-Source Optical Coherence Tomography |
| WHO | World Health Organization |
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef] [PubMed]
- Holz, F.G.; Strauss, E.C.; Schmitz-Valckenberg, S.; van Lookeren Campagne, M. Geographic Atrophy. Ophthalmology 2014, 121, 1079–1091. [Google Scholar] [CrossRef]
- Keenan, T.D.; Agrón, E.; Domalpally, A.; Clemons, T.E.; van Asten, F.; Wong, W.T.; Danis, R.G.; Sadda, S.; Rosenfeld, P.J.; Klein, M.L.; et al. Progression of Geographic Atrophy in Age-related Macular Degeneration. Ophthalmology 2018, 125, 1913–1928. [Google Scholar] [CrossRef]
- Schmitz-Valckenberg, S.; Sadda, S.; Staurenghi, G.; Chew, E.Y.; Fleckenstein, M.; Holz, F.G.; Group, C.A.M. GEOGRAPHIC ATROPHY: Semantic Considerations and Literature Review. Retina 2016, 36, 2250–2264. [Google Scholar] [CrossRef]
- Bird, A.C.; Bressler, N.M.; Bressler, S.B.; Chisholm, I.H.; Coscas, G.; Davis, M.D.; de Jong, P.T.; Klaver, C.C.; Klein, B.E.; Klein, R.; et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv. Ophthalmol. 1995, 39, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sadda, S.R.; Guymer, R.; Holz, F.G.; Schmitz-Valckenberg, S.; Curcio, C.A.; Bird, A.C.; Blodi, B.A.; Bottoni, F.; Chakravarthy, U.; Chew, E.Y.; et al. Consensus Definition for Atrophy Associated with Age-Related Macular Degeneration on OCT: Classification of Atrophy Report 3. Ophthalmology 2018, 125, 537–548. [Google Scholar] [CrossRef]
- Kaiser, P.K.; Karpecki, P.M.; Regillo, C.D.; Baumal, C.; Ryan, E.H.; Ip, M.S.; Modi, Y.; Yeu, E.; Nijm, L.; Farid, M.; et al. Geographic Atrophy Management Consensus (GA-MAC): A Delphi panel study on identification, diagnosis and treatment. BMJ Open Ophthalmol. 2023, 8. [Google Scholar] [CrossRef]
- Shen, L.L.; Sun, M.; Ahluwalia, A.; Park, M.M.; Young, B.K.; Del Priore, L.V. Local Progression Kinetics of Geographic Atrophy Depends Upon the Border Location. Investig. Ophthalmol. Vis. Sci. 2021, 62, 28. [Google Scholar] [CrossRef] [PubMed]
- Schaal, K.B.; Rosenfeld, P.J.; Gregori, G.; Yehoshua, Z.; Feuer, W.J. Anatomic Clinical Trial Endpoints for Nonexudative Age-Related Macular Degeneration. Ophthalmology 2016, 123, 1060–1079. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef]
- Mharib, A.M.; Ramli, A.R.; Mashohor, S.; Mahmood, R.B. Survey on liver CT image segmentation methods. Artif. Intell. Rev. 2011, 37, 83–95. [Google Scholar] [CrossRef]
- Li, J.; Udupa, J.K.; Tong, Y.; Wang, L.; Torigian, D.A. Segmentation evaluation with sparse ground truth data: Simulating true segmentations as perfect/imperfect as those generated by humans. Med Image Anal. 2021, 69, 101980. [Google Scholar] [CrossRef] [PubMed]
- Philip, M.M.; Watts, J.; Moeini, S.N.M.; Musheb, M.; McKiddie, F.; Welch, A.; Nath, M. Comparison of semi-automatic and manual segmentation methods for tumor delineation on head and neck squamous cell carcinoma (HNSCC) positron emission tomography (PET) images. Phys. Med. Biol. 2024, 69, 095005. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, Y.; Boufama, B. Biomedical Image Segmentation: A Survey. SN Comput. Sci. 2021, 2, 310. [Google Scholar] [CrossRef]
- Arslan, J.; Samarasinghe, G.; Benke, K.K.; Sowmya, A.; Wu, Z.; Guymer, R.H.; Baird, P.N. Artificial Intelligence Algorithms for Analysis of Geographic Atrophy: A Review and Evaluation. Transl. Vis. Sci. Technol. 2020, 9, 57. [Google Scholar] [CrossRef]
- Arabahmadi, M.; Farahbakhsh, R.; Rezazadeh, J. Deep Learning for Smart Healthcare-A Survey on Brain Tumor Detection from Medical Imaging. Sensors 2022, 22, 1960. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef]
- Paul, S.K.; Pan, I.; Sobol, W.M. A Systematic Review of Deep Learning Applications for Optical Coherence Tomography in Age-Related Macular Degeneration. Retina 2022, 42, 1417–1424. [Google Scholar] [CrossRef]
- Riedl, S.; Vogl, W.D.; Mai, J.; Reiter, G.S.; Lachinov, D.; Grechenig, C.; McKeown, A.; Scheibler, L.; Bogunovic, H.; Schmidt-Erfurth, U. The Effect of Pegcetacoplan Treatment on Photoreceptor Maintenance in Geographic Atrophy Monitored by Artificial Intelligence-Based OCT Analysis. Ophthalmol. Retin. 2022, 6, 1009–1018. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Enzendorfer, M.L.; Schmidt-Erfurth, U. Artificial intelligence for geographic atrophy: Pearls and pitfalls. Curr. Opin. Ophthalmol. 2024, 35, 455–462. [Google Scholar] [CrossRef]
- Xu, Y.; Goodacre, R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. J. Anal. Test 2018, 2, 249–262. [Google Scholar] [CrossRef]
- Vlăsceanu, G.V.; Tarbă, N.; Voncilă, M.L.; Boiangiu, C.A. Selecting the Right Metric: A Detailed Study on Image Segmentation Evaluation. BRAIN Broad Res. Artif. Intell. Neurosci. 2024, 15, 295–318. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.; Pereira, S.; Goncalves, L.; Ferreira, M.; Silva, C.A. Multi-surface segmentation of OCT images with AMD using sparse high order potentials. Biomed. Opt. Express 2017, 8, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Freyer, N.; Gross, D.; Lipprandt, M. The ethical requirement of explainability for AI-DSS in healthcare: A systematic review of reasons. BMC Med. Ethics 2024, 25, 104. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Methley, A.M.; Campbell, S.; Chew-Graham, C.; McNally, R.; Cheraghi-Sohi, S. PICO, PICOS and SPIDER: A comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv. Res. 2014, 14, 579. [Google Scholar] [CrossRef]
- Bramer, W.M.; Milic, J.; Mast, F. Reviewing retrieved references for inclusion in systematic reviews using EndNote. J. Med. Libr. Assoc. 2017, 105, 84–87. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Tejani, A.S.; Klontzas, M.E.; Gatti, A.A.; Mongan, J.T.; Moy, L.; Park, S.H.; Kahn, C.E., Jr.; Panel, C.U. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): 2024 Update. Radiol. Artif. Intell. 2024, 6, e240300. [Google Scholar] [CrossRef] [PubMed]
- Arslan, J.; Samarasinghe, G.; Sowmya, A.; Benke, K.K.; Hodgson, L.A.B.; Guymer, R.H.; Baird, P.N. Deep Learning Applied to Automated Segmentation of Geographic Atrophy in Fundus Autofluorescence Images. Transl. Vis. Sci. Technol. 2021, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Wang, L.; Zhou, X.; Shi, Y.; Cheng, Y.; Laiginhas, R.; Zhou, H.; Shen, M.; Zhang, Q.; de Sisternes, L.; et al. Automatic geographic atrophy segmentation using optical attenuation in OCT scans with deep learning. Biomed. Opt. Express 2022, 13, 1328–1343. [Google Scholar] [CrossRef]
- Derradji, Y.; Mosinska, A.; Apostolopoulos, S.; Ciller, C.; De Zanet, S.; Mantel, I. Fully-automated atrophy segmentation in dry age-related macular degeneration in optical coherence tomography. Sci. Rep. 2021, 11, 21893. [Google Scholar] [CrossRef]
- Hassan, B.; Qin, S.; Hassan, T.; Ahmed, R.; Werghi, N. Joint Segmentation and Quantification of Chorioretinal Biomarkers in Optical Coherence Tomography Scans: A Deep Learning Approach. IEEE Trans. Instrum. Meas. 2021, 70, 2508817. [Google Scholar] [CrossRef]
- Hu, Z.; Medioni, G.G.; Hernandez, M.; Sadda, S.R. Automated segmentation of geographic atrophy in fundus autofluorescence images using supervised pixel classification. J. Med. Imaging 2015, 2, 014501. [Google Scholar] [CrossRef]
- Kalra, G.; Cetin, H.; Whitney, J.; Yordi, S.; Cakir, Y.; McConville, C.; Whitmore, V.; Bonnay, M.; Lunasco, L.; Sassine, A.; et al. Machine Learning-Based Automated Detection and Quantification of Geographic Atrophy and Hypertransmission Defects Using Spectral Domain Optical Coherence Tomography. J. Pers. Med. 2022, 13, 37. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Q.; Zhang, Y.; Wang, M.; Tang, J. TSSK-Net: Weakly supervised biomarker localization and segmentation with image-level annotation in retinal OCT images. Comput. Biol. Med. 2023, 153, 106467. [Google Scholar] [CrossRef]
- Mai, J.; Lachinov, D.; Riedl, S.; Reiter, G.S.; Vogl, W.D.; Bogunovic, H.; Schmidt-Erfurth, U. Clinical validation for automated geographic atrophy monitoring on OCT under complement inhibitory treatment. Sci. Rep. 2023, 13, 7028. [Google Scholar] [CrossRef]
- Morano, J.; Aresta, G.; Grechenig, C.; Schmidt-Erfurth, U.; Bogunovic, H. Deep Multimodal Fusion of Data With Heterogeneous Dimensionality via Projective Networks. IEEE J. Biomed. Health Inform. 2024, 28, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Royer, C.; Sublime, J.; Rossant, F.; Paques, M. Unsupervised Approaches for the Segmentation of Dry ARMD Lesions in Eye Fundus cSLO Images. J. Imaging 2021, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Szeskin, A.; Yehuda, R.; Shmueli, O.; Levy, J.; Joskowicz, L. A column-based deep learning method for the detection and quantification of atrophy associated with AMD in OCT scans. Med. Image Anal. 2021, 72, 102130. [Google Scholar] [CrossRef]
- Wang, Z.; Sadda, S.R.; Lee, A.; Hu, Z.J. Automated segmentation and feature discovery of age-related macular degeneration and Stargardt disease via self-attended neural networks. Sci. Rep. 2022, 12, 14565. [Google Scholar] [CrossRef]
- Wu, M.; Cai, X.; Chen, Q.; Ji, Z.; Niu, S.; Leng, T.; Rubin, D.L.; Park, H. Geographic atrophy segmentation in SD-OCT images using synthesized fundus autofluorescence imaging. Comput. Methods Programs Biomed. 2019, 182, 105101. [Google Scholar] [CrossRef]
- Ji, Z.; Chen, Q.; Niu, S.; Leng, T.; Rubin, D.L. Beyond Retinal Layers: A Deep Voting Model for Automated Geographic Atrophy Segmentation in SD-OCT Images. Transl. Vis. Sci. Technol. 2018, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ji, Z.; Niu, S.; Leng, T.; Rubin, D.L.; Chen, Q. MS-CAM: Multi-Scale Class Activation Maps for Weakly-Supervised Segmentation of Geographic Atrophy Lesions in SD-OCT Images. IEEE J. Biomed. Health Inform. 2020, 24, 3443–3455. [Google Scholar] [CrossRef]
- Xu, R.; Niu, S.; Chen, Q.; Ji, Z.; Rubin, D.; Chen, Y. Automated geographic atrophy segmentation for SD-OCT images based on two-stage learning model. Comput. Biol. Med. 2019, 105, 102–111. [Google Scholar] [CrossRef]
- Domalpally, A.; Slater, R.; Linderman, R.E.; Balaji, R.; Bogost, J.; Voland, R.; Pak, J.; Blodi, B.A.; Channa, R.; Fong, D.; et al. Strong versus Weak Data Labeling for Artificial Intelligence Algorithms in the Measurement of Geographic Atrophy. Ophthalmol. Sci. 2024, 4, 100477. [Google Scholar] [CrossRef]
- Feeny, A.K.; Tadarati, M.; Freund, D.E.; Bressler, N.M.; Burlina, P. Automated segmentation of geographic atrophy of the retinal epithelium via random forests in AREDS color fundus images. Comput. Biol. Med. 2015, 65, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Liefers, B.; Colijn, J.M.; Gonzalez-Gonzalo, C.; Verzijden, T.; Wang, J.J.; Joachim, N.; Mitchell, P.; Hoyng, C.B.; van Ginneken, B.; Klaver, C.C.W.; et al. A Deep Learning Model for Segmentation of Geographic Atrophy to Study Its Long-Term Natural History. Ophthalmology 2020, 127, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Pfau, M.; von der Emde, L.; de Sisternes, L.; Hallak, J.A.; Leng, T.; Schmitz-Valckenberg, S.; Holz, F.G.; Fleckenstein, M.; Rubin, D.L. Progression of Photoreceptor Degeneration in Geographic Atrophy Secondary to Age-related Macular Degeneration. JAMA Ophthalmol. 2020, 138, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, D.J.; Sunness, J.S.; Malviya, P.; Applegate, C.; Hager, G.D.; Handa, J.T. Automated image alignment and segmentation to follow progression of geographic atrophy in age-related macular degeneration. Retina 2014, 34, 1296–1307. [Google Scholar] [CrossRef]
- Spaide, T.; Jiang, J.; Patil, J.; Anegondi, N.; Steffen, V.; Kawczynski, M.G.; Newton, E.M.; Rabe, C.; Gao, S.S.; Lee, A.Y.; et al. Geographic Atrophy Segmentation Using Multimodal Deep Learning. Transl. Vis. Sci. Technol. 2023, 12, 10. [Google Scholar] [CrossRef]
- Zhang, G.; Fu, D.J.; Liefers, B.; Faes, L.; Glinton, S.; Wagner, S.; Struyven, R.; Pontikos, N.; Keane, P.A.; Balaskas, K. Clinically relevant deep learning for detection and quantification of geographic atrophy from optical coherence tomography: A model development and external validation study. Lancet Digit. Health 2021, 3, e665–e675. [Google Scholar] [CrossRef]
- Pramil, V.; de Sisternes, L.; Omlor, L.; Lewis, W.; Sheikh, H.; Chu, Z.; Manivannan, N.; Durbin, M.; Wang, R.K.; Rosenfeld, P.J.; et al. A Deep Learning Model for Automated Segmentation of Geographic Atrophy Imaged Using Swept-Source OCT. Ophthalmol. Retin. 2023, 7, 127–141. [Google Scholar] [CrossRef]
- Kocak, B.; Keles, A.; Akinci D’Antonoli, T. Self-reporting with checklists in artificial intelligence research on medical imaging: A systematic review based on citations of CLAIM. Eur. Radiol. 2024, 34, 2805–2815. [Google Scholar] [CrossRef]
- Chen, Q.; de Sisternes, L.; Leng, T.; Zheng, L.; Kutzscher, L.; Rubin, D.L. Semi-automatic geographic atrophy segmentation for SD-OCT images. Biomed. Opt. Express 2013, 4, 2729–2750. [Google Scholar] [CrossRef]
- Muller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef]
- Chandra, S.; Rasheed, R.; Sen, P.; Menon, D.; Sivaprasad, S. Inter-rater reliability for diagnosis of geographic atrophy using spectral domain OCT in age-related macular degeneration. Eye 2022, 36, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Asgari Taghanaki, S.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 2020, 54, 137–178. [Google Scholar] [CrossRef]
- Piffer, S.; Ubaldi, L.; Tangaro, S.; Retico, A.; Talamonti, C. Tackling the small data problem in medical image classification with artificial intelligence: A systematic review. Prog. Biomed. Eng. 2024, 6, 032001. [Google Scholar] [CrossRef] [PubMed]
- Dow, E.R.; Keenan, T.D.L.; Lad, E.M.; Lee, A.Y.; Lee, C.S.; Loewenstein, A.; Eydelman, M.B.; Chew, E.Y.; Keane, P.A.; Lim, J.I.; et al. From Data to Deployment: The Collaborative Community on Ophthalmic Imaging Roadmap for Artificial Intelligence in Age-Related Macular Degeneration. Ophthalmology 2022, 129, e43–e59. [Google Scholar] [CrossRef]
- Yang, F.; Zamzmi, G.; Angara, S.; Rajaraman, S.; Aquilina, A.; Xue, Z.; Jaeger, S.; Papagiannakis, E.; Antani, S.K. Assessing Inter-Annotator Agreement for Medical Image Segmentation. IEEE Access 2023, 11, 21300–21312. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef]
- Eche, T.; Schwartz, L.H.; Mokrane, F.Z.; Dercle, L. Toward Generalizability in the Deployment of Artificial Intelligence in Radiology: Role of Computation Stress Testing to Overcome Underspecification. Radiol. Artif. Intell. 2021, 3, e210097. [Google Scholar] [CrossRef]
- Balki, I.; Amirabadi, A.; Levman, J.; Martel, A.L.; Emersic, Z.; Meden, B.; Garcia-Pedrero, A.; Ramirez, S.C.; Kong, D.; Moody, A.R.; et al. Sample-Size Determination Methodologies for Machine Learning in Medical Imaging Research: A Systematic Review. Can. Assoc. Radiol. J. 2019, 70, 344–353. [Google Scholar] [CrossRef]
- Teo, K.Y.C.; Fujimoto, S.; Sadda, S.R.; Kokame, G.; Gomi, F.; Kim, J.E.; Cheng, M.F.S.; Corradetti, G.; Amornpetchsathaporn, A.; Chainakul, M.; et al. Geographic Atrophy Phenotypes in Subjects of Different Ethnicity: Asia-Pacific Ocular Imaging Society Work Group Report 3. Ophthalmol. Retin. 2023, 7, 593–604. [Google Scholar] [CrossRef]
- European Parliament. EU AI Act: First Regulation on Artificial Intelligence (n.d.); European Parliament: Strasbourg, France, 2023; Available online: https://www.europarl.europa.eu/topics/en/article/20230601STO93804/eu-ai-act-first-regulation-on-artificial-intelligence (accessed on 31 March 2025).
- Aboy, M.; Minssen, T.; Vayena, E. Navigating the EU AI Act: Implications for regulated digital medical products. NPJ Digit. Med. 2024, 7, 237. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H. Secondary Use of Health Data for Medical AI: A Cross-Regional Examination of Taiwan and the EU. Asian Bioeth. Rev. 2024, 16, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Kondylakis, H.; Catalan, R.; Alabart, S.M.; Barelle, C.; Bizopoulos, P.; Bobowicz, M.; Bona, J.; Fotiadis, D.I.; Garcia, T.; Gomez, I.; et al. Documenting the de-identification process of clinical and imaging data for AI for health imaging projects. Insights Imaging 2024, 15, 130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11045, pp. 3–11. [Google Scholar]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; She, W.; Xie, M. Biomedical Image Segmentation Using Denoising Diffusion Probabilistic Models: A Comprehensive Review and Analysis. Appl. Sci. 2024, 14, 632. [Google Scholar] [CrossRef]



| PICOS | Description |
|---|---|
| Population | Adult patients with geographic atrophy due to advanced dry age-related macular degeneration. |
| Intervention | Artificial intelligence algorithms for the segmentation of geographic atrophy. Various AI tools trained to automatically delineate geographic atrophy lesions using different retinal imaging modalities (CFP, OCT, FAF, etc.). |
| Comparator | Alternative segmentation methods: manual segmentation performed by human experts, validated semi-automated segmentation techniques, or other software-assisted methods. |
| Outcomes | Accuracy and efficacy of artificial intelligence techniques for segmenting geographic atrophy as measured through performance metrics (sensitivity, specificity, AUC, F1 score, IoU, DSC, etc.). |
| Study Design | Studies included observational studies, randomized clinical trials, and registry/database studies. |
| Additional Details | Language: English. Time frame: from inception to 23 March 2024. Location: worldwide. |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Observational studies, randomized clinical trials, registry/database studies. | Systematic reviews, meta-analyses, narrative reviews, scoping reviews, opinion pieces, surveys, editorials, commentary letters, case reports, book chapters, conference abstracts, proceedings, or presentations. |
| Studies using artificial intelligence techniques. | Studies using traditional image processing techniques. |
| Studies including eyes of adult patients. | Studies including pediatric patients, geographic atrophy due to neovascular age-related macular degeneration, or non-human studies. |
| Studies addressing geographic atrophy lesion segmentation with the use of artificial intelligence segmentation techniques. | Studies addressing solely geographic atrophy diagnosis, classification, progression, future prediction, treatment response, area quantification, or retinal layer-by-layer segmentation. |
| Studies reporting at least one of the following metrics: accuracy, sensitivity/recall, specificity, precision/PPV, F1 score, AUC/AUROC, AUPR, IoU/OR, DSC/SDC, or AAD. | Studies without clear documentation of the purpose, dataset, data sources, patient distribution, methods, results, or conclusions. |
| Articles published in English language. | Ineligible population and ineligible outcome. |
| Articles with full text unavailable. |
| Category | Description |
|---|---|
| Study information |
|
| Dataset and annotation methods |
|
| AI model characteristics and validation strategy |
|
| Model performance evaluation |
|
| Author (Year) | Country | Diseases | CAM* | Dataset | Imaging Modality | Total pts/Eyes | Total Sample Size | GA Sample Size (% of Dataset) | Ground Truth |
|---|---|---|---|---|---|---|---|---|---|
| Arslan (2021) [34] | Australia | GA secondary to dry AMD | No | Institutional dataset (Centre for Eye Research Australia) and private dataset | FAF (Heidelberg Spectralis) | 51/99 | 702 FAF images | 702 FAF images (100% of dataset) | Manual annotation of FAF by principal grader, additional grading of subset by senior grader for GT evaluation, and RegionFinder software outputs for comparison. |
| Chu (2022) [35] | USA | GA and Healthy | Yes | Institutional dataset (part of the University of Miami) | SS-OCT (PLEX Elite 9000 Carl Zeiss) | 115/140 | 184 OCT scans | 124 OCT scans (67.3% of dataset) | Manual annotation by 2 independent graders reaching consensus on the en face subRPE OCT images (with senior adjudicator). |
| Derradji (2021) [36] | Switzerland | Atrophic AMD with RORA and Healthy | Yes | Institutional dataset (JG Eye Hospital Medical Retinal Department) | SD-OCT (Heidelberg Spectralis) | 57/62 | 62 OCT volumes (3595 B-scans) | 2085 B-scans with RORA (58% of dataset) | Manual annotation by 2 expert graders (senior and junior). |
| Domalpally (2024) [50] | USA | GA | No | AREDS2 subset | FAF (Heidelberg Spectralis HRA) | 271/362 | 601 FAF images | 601 FAF images (100% of dataset) | Manual GA demarcation by certified graders. |
| Feeny (2015) [51] | USA Thailand | GA | No | AREDS dbGAP subset | CFP | NR/55 | 143 CFP images | 143 CFP images (100% of dataset) | Manual delineation by 1 retina specialist and review/correction by retina specialist team. |
| Hassan (2021) [37] | China, UAE | 11 CRBMs from a pool of datasets containing AMD, ME, CSCR, MH, DR, CNV, and Healthy | No | 7 OCT datasets spread over 4 DBs (Isfahan University of Medical Sciences, Duke University, University of Waterloo, and Guangzhou Medical University) | OCT (Spectralis, Cirrus, Topcon, and Bioptigen) | 6953/NR | 7000 OCT scans | 500 OCT scans (7.1% of dataset) | Manual pixel-wise annotations of 11 biomarkers by 3 retina specialists (1 grader per scan). |
| Hu (2015) [38] | USA | GA | No | Institutional FAF dataset (University of Southern California Retina Clinics) | FAF (Heidelberg cSLO Spectralis HRA + OCT) | 16/16 | 16 FAF images | 16 FAF images (100% of dataset) | Manual delineation by certified grader. |
| Ji (2018) [47] | China, USA | Advanced non- exudative AMD with GA | No | 2 datasets previously described and used [59] | SD-OCT (Cirrus Carl Zeiss) FAF (for GT annotations) | 62/66 | 51 SD-OCT cube scans (DS1); 54 SD-OCT cube scans (DS2) and corresponding FAF images | 105 SD-OCT cube scans (100% of DS1 and DS2) | DS1: manual outline average of 2 independent experts from 2 sessions, based on B-scans. DS2: manual outline based on FAF images and manual registering to projection images. |
| Kalra (2022) [39] | USA | Nonexudative AMD with or without GA (including iRORA and cRORA) | Yes | Institutional OCT dataset (Cole Eye Institute of the Cleveland Clinic) | SD-OCT (Heidelberg Spectralis HRA + OCT and Cirrus HD-OCT Zeiss) | 341/NR | 100.266 SD-OCT B-scans and 900 en face SD-OCT images | NR | B-scan model: Retinal layer segmentation masks by previously validated ML model, corrected by expert reader, reviewed by senior expert, and adjudicated by analysis director. Binary GA masks in areas of outer layer attenuation overlap. En face model: generated from assembled B-scan masks. |
| Liefers (2020) [52] | The Netherlands, Australia, Singapore, Switzerland | AMD with GA | No | BMES and RS cohorts I, II, and III | CFI (Zeiss, Canon CF-60 Dsi, Topcon TRV-50VT/TRC 50EX, and Sony DXC-950P) | 175/238 | 409 CFI images | 409 CFI images (100% of dataset) | Manual delineation by 4 expert graders for BMES and at least 2 expert graders for RS using in-house software and final consensus grading. Consultation with additional multimodal data when available. |
| Liu (2023) [40] | China, USA | GA, Healthy, and other CRBMs (IRF/DME, DRUSEN, CNV, PED, HF, and SRF) | No | Private retinal biomarker dataset from Wuhan Aier Eye Hospital and Kermany’s public dataset | OCT Triton DRI for DS1, NR for DS2) | NR/NR | 30.850 B-scans | 700 B-scans (2.27% of dataset) | Previously published manual annotations for IRF/DRUSEN and ophthalmologist annotations for CNV. Annotation methods for GA are not reported. |
| Ma (2020) [48] | China, USA | Advanced non- exudative AMD with GA | No | 2 datasets previously described and used [59] | SD-OCT (Cirrus Carl Zeiss) FAF (for GT annotations) | 62/66 | 105 SD-OCT cube scans | 105 SD-OCT cube scans (100% of dataset) | DS1: manual outline by 2 independent experts from 2 sessions based on B-scans. DS2: manual outline by 1 expert based on FAF images and mapping to projection image. |
| Mai (2023) [41] | Austria | GA | No | Institutional dataset (Medical University of Vienna) | SD-OCT (Heidelberg Spectralis) FAF (for GT annotations) | 100/184 | 967 OCT volumes | 967 OCT volumes (100% of dataset) | Manual annotation of FAF by 2 trained graders using OCTAVO software, with automatic NIR registration and final 2D en face OCT annotations. |
| Morano (2024) [42] | Austria | GA | No | Institutional dataset (Medical University of Vienna) | OCT, SLO, and FAF (Heidelberg Spectralis) | 100/184 | 967 OCT volumes, including OCT B-Scans, SLO, and FAF images | 967 OCT volumes, including OCT B-Scans, SLO, and FAF images (100% of dataset) | En face masks annotated by 1 retinal expert on FAF, additional annotations on 35 OCT samples by retinal experts, with automatic co-registration of OCT-SLO and FAF-SLO. |
| Pfau (2020) [53] | USA, Germany | GA and Healthy | No | Directional spread in GA 2 study dataset | FAF, IR, and SD-OCT (Spectralis HRA + OCT2) | 182/251 | UC | UC | B-scan and en face manual segmentation by 2 readers on test set (3 B-scans per patient). |
| Pramil (2023) [57] | USA, Australia | GA, Early AMD, Intermediate AMD, and Healthy | No | Study OCT dataset (“SWAGGER” cohort) and institutional dataset from New England Eye Center (Tufts Medical Center) | SS-OCT (PLEX Elite 9000 Carl Zeiss) | 138/198 | 351 OCT scans | 273 OCT scans (77.7% of dataset) | Plex Elite Review software layer segmentation, then manual annotations on generated en face images by 1 expert grader for development set and 2 expert graders during 2 separate sessions for test set (ophthalmology research fellows), with grader training and annotation verification by retina specialist. |
| Ramsey (2014) [54] | USA | GA | No | Fenretinide study subset | FAF and CFP | 10/NR | 26 FAF-CFP image pairs | 26 FAF-CFP image pairs (100% of dataset) | Manual delineation by expert grader. |
| Royer (2021) [43] | France | GA | No | Institutional dataset (Clinical Imaging Center, Quinze-Vingts Hospital) | cSLO | 13/NR | 328 cSLO images | 328 cSLO images (100% of dataset) | GA delineation by ophthalmologists. |
| Spaide (2023) [55] | USA, Germany | GA | No | Proxima A and Proxima B study datasets | FAF and NIR (Spectralis cSLO) | 337/337 | 1437 FAF-NIR image pairs | 1437 FAF-NIR image pairs (100% of dataset) | Semi-automatic delineation on FAF by trained graders using RegionFinder software. DS1 later assessed by 2 junior graders with senior adjudicator. DS2 later assessed by junior and senior grader with optional second senior grader involvement. Total of 384 images used for GT. |
| Szeskin (2021) [44] | Israel | cRORA and Macular atrophy (cRORA, iRORA, cORA, and iORA) | Yes | 2 institutional datasets (Hadassah University Medical Center) | OCT and IR (Heidelberg Spectralis) | 34/NR | 106 OCT scans + IR (5.207 slices) (DS1); 19 OCT scans + IR (829 slices) (DS2) | 2952 slices (56.70% of DS1); 829 slices (100% of DS2) | DS1: multistep manual annotation of OCT by 2 technical co-authors and 2 ophthalmologists (of which one was senior reviewer). DS2: Manual delineation on IR image by medical student and projection onto OCT. Both DSs used in-house OCT-E GUI software. |
| Wang (2022) [45] | USA | AMD GA and Stargardt atrophy | No | In-house datasets | FAF (Spectralis HRA + OCT) | 217/296 | 180 FAF images (AMD GA DS); 412 FAF images (Stargardt DS) | 90 FAF images (50% of AMD GA dataset) (15.2% of both datasets) | Manual delineation/labeling of FAF by certified grader using RegionFinder software and reviewed by senior grader with senior adjudicator. |
| Wu (2019) [46] | China, USA, South Korea | Advanced non- neovascular AMD with GA | No | In-house dataset | SD-OCT (Cirrus Carl Zeiss) FAF (Heidelberg Spectralis) | 56/NR | 56 SD-OCT volumes | 56 SD-OCT volumes (100% of dataset) | Manual segmentation of FAF images and manual registration on en face OCT by fellowship-trained retinal specialist. |
| Xu (2019) [49] | China, USA | GA | No | 2 datasets previously described and used [59] | SD-OCT (Carl Zeiss) | 64/NR | 55 SD-OCT cubes (DS1); 56 SD-OCT cubes with corresponding FAF images (DS2) | 111 SD-OCT cubes (100% of DS1 and DS2) | DS1: manual outline of OCT cubes by 2 ophthalmologists during 2 sessions. DS2: manual outline based on FAF image by ophthalmologist and registered to projection image. |
| Zhang (2021) [56] | UK, the Netherlands, Switzerland | GA (including cRORA and iRORA) | Yes | FILLY dataset | OCT (Heidelberg Spectralis OCT + HRA) | 200/399 | 984 OCT volumes (5049 B-scans) | 984 OCT volumes (100% of dataset) | Manual annotation of 5 B-scans per OCT volume by 3 expert graders. |
| Author (Year) | Segmentation Technique | Internal Validation Method | Training Set | Validation Set | Internal Test Set | External Test Set |
|---|---|---|---|---|---|---|
| Arslan (2021) [34] | Pixel prediction and classification—binary classification problem. | 5-fold cross-validation | 5 sets of 140 or 142 FAF images each | 5 sets of 140 or 142 FAF images each | 5 sets of 140 or 142 FAF images each | NR |
| Chu (2022) [35] | 2 models (trained on OAC false-color en face images or OCT subRPE en face images). | train–test split | 80% of all eyes (89 eyes—133 OCT scans) | 20% of training set (23 eyes—23 OCT scans) | 20% of all eyes (28 eyes—28 OCT scans) | NR |
| Derradji (2021) [36] | CNN trained using single 2D B-scan as input and producing corresponding 2D RORA probability mask as output. | 5-fold cross-validation on the merged training and validation sets | 2301 OCT B-scans | 256 OCT B-scans | 1038 OCT B-scans | NR |
| Domalpally (2024) [50] | Weakly labeled model for GA measurement and strongly labeled model for GA measurement and pixel-level segmentation. | 5-fold cross-validation | 80% of dataset (~481 FAF images) for each iteration | 20% of dataset (~120 FAF images) for each iteration | NR | 156 FAF images from GSK BAM114341 study dataset |
| Feeny (2015) [51] | 52 features computed per pixel for binary classification (GA vs. not-GA). | Leave-one-out cross-validation | 142 CFP images for each iteration | NR | 1 CFP image for each iteration | NR |
| Hassan (2021) [37] | Asymmetric encoder–decoder structure for joint segmentation and quantification of 11 CRBMs with preprocessing, feature map utilization, and postprocessing stages. | Hold-out validation | 4200 OCT B-scans (352 for GA) from University of Waterloo DB and Guangzhou Medical University DB | 1400 OCT B-scans in total (75 for GA) | 1400 OCT B-scans in total (73 for GA) from Isfahan University of Medical Sciences DB and Duke University DB | NR |
| Hu (2015) [38] | Supervised pixel classification employing image texture features. | 8-fold cross-validation | 8 rotating FAF image sets | NR | 8 rotating FAF image sets | NR |
| Ji (2018) [47] | Deep voting model with 5 layers. A-scans labeled with 1024 features fed into the network and a soft-max classifier determined pixel-level labels. 10 trained models. No retinal layer segmentation. | Hold-out validation | 10.000 GA-positive OCT A-scans and 10.000 GA-negative OCT A-scans for each dataset (~5% and ~9% of total data) | NR | NR | NR |
| Kalra (2022) [39] | 2 models (B-scan and en face) for binary detection of GA presence and pixel-wise lesion segmentation. | Hold-out validation | 80% of total patients | 10% of total patients | Unseen 10% of patients | NR |
| Liefers (2020) [52] | Ensemble of 20 models obtained during 5-fold cross-validation with average of pixel-wise predictions used to form a single binary image. | 5-fold cross-validation | ~254 CFI images for each model (60% of data) | NR | NR | 50 random CFIs from AREDS subset |
| Liu (2023) [40] | 3 stages: a supervised, contrastive learning-based pre-training; a fine-tuning module combining two loss functions; and a knowledge distillation-based teacher–student network for anomaly localization. | Hold-out validation | 500 GA OCT B-scans | 100 GA OCT B-scans | 100 GA OCT B-scans | NR |
| Ma (2020) [48] | Stage 1: B-scan denoising, RPE segmentation, flattening. Stage 2: B-scans input into weakly supervised network to generate attention maps. Stage 3: Segmentation masks by graph-based algorithm using positive/negative seeds. | 5-fold cross-validation | NR | NR | NR | NR |
| Mai (2023) [41] | Patch-based training, 3D to 2D image segmentation. | 5-fold cross-validation | ~695 OCT volumes (90% of development set) | ~77 OCT volumes (10% of development set) | ~193 OCT volumes | 226 OCT volumes from FILLY dataset |
| Morano (2024) [42] | Multimodal Late Fusion and Multiscale Fusion approaches with 2 branches to extract and project features from different modalities and dimensions (3D OCT and 2D FAF or SLO images) onto a common feature subspace, enabling their joint use for segmentation tasks. | Hold-out validation | ~580 OCT volumes (B-scans, SLO, and FAF) (60% of DS) | ~97 OCT volumes (B-scans, SLO, and FAF) (fixed 10% of DS) | ~290 OCT volumes (B-scans, SLO, and FAF) (fixed 30% of DS) | NR |
| Pfau (2020) [53] | 1st CNN for 6-layer retinal segmentation, and multimodal input stacking into 2nd CNN for en face GA segmentation. | Hold-out validation | 135 eyes for en face GA segmentation | 45 eyes for en face GA segmentation | 75 OCT B-scans of 25 patients with GA | NR |
| Pramil (2023) [57] | Encoding of OCT-derived GA features onto a pseudocolor image using RGB channels for RPE loss, hypertransmission, and retinal thinning, respectively. | 5-fold cross-validation | 126 OCT scans | ~25 OCT scans (~20% of training set) | 225 OCT scans | NR |
| Ramsey (2014) [54] | Image registration, digital vessel subtraction, user-defined ROI, soft FCM segmentation, user selection of relevant topographies, and final GA quantification. | NR | NR | NR | NR | NR |
| Royer (2021) [43] | Unsupervised fully convolutional autoencoder trained on 2 loss functions (reconstruction error and soft N-cut loss) and 3 classes to segment GA by maximizing pixel intensity and spatial dissimilarity. | 8 different random combinations of 12 series for training and 6 for testing | NR | NR | NR | NR |
| Spaide (2023) [55] | Single encoder–decoder architecture for pixel prediction and classification (UNet), two encoder branches to encode FAF and NIR images separately, and one joint decoder to decode the embeddings (Ynet). | Hold-out validation | 748 FAF-NIR image pairs from DS2 | 192 FAF-NIR image pairs from DS2 | 497 FAF-NIR image pairs from DS1 | NR |
| Szeskin (2021) [44] | CNN that classified light-scattering patterns in 2D and 3D columns of vertical pixel-wide vectors (A-scans) on atrophic OCT B-scans, utilizing the BCE and F1 loss functions without layer segmentation. | 4-fold cross-validation | 93 OCT scans for cRORA; 10 OCT scans for macular atrophy for training and cross-validation | 93 OCT scans for cRORA; 10 OCT scans for macular atrophy for training and cross-validation | 12 OCT scans for cRORA; 9 OCT scans for macular atrophy for testing | NR |
| Wang (2022) [45] | Integrated soft-labeled self-attended deep CNN system and binary pixel classification producing feature maps. | 8-fold cross-validation | 70 AMD FAF images (in each fold) | 10 fixed AMD FAF images | 10 AMD FAF images (in each fold) | NR |
| Wu (2019) [46] | FAF image synthesis from generated en face OCT by GA RA-CGAN; image fusion net and segmentation net refined with SFCM. | Leave-four-out cross-validation | NR | NR | NR | NR |
| Xu (2019) [49] | Offline learning phase to capture common features from training samples and self-learning phase to identify discriminative features and reduce FPs. Fusion of both outputs for final segmentation. | Independent dataset validation | Random 100.000 axial data with GA and 100.000 without GA for each DS | NR | 111 3D OCT cubes in total from both DSs | NR |
| Zhang (2021) [56] | 2 approaches: direct GA segmentation or individual segmentation of overlapping features (RPE loss, photoreceptor degeneration, and hypertransmission). | Hold-out validation | 582 OCT volumes (3024 B-scans) (60% of dataset) | 191 OCT volumes (958 B-scans) (20% of dataset) | 211 OCT volumes (1067 B-scans) (20% of dataset) | 192 OCT volumes (884 B-scans) from Moorfields Eye Hospital institutional dataset |
| Author (Year) | AI Type | AI Algorithm | XAI | Performance Metrics (Internal Testing) | Performance Metrics (External Testing) |
|---|---|---|---|---|---|
| Arslan (2021) [34] | DL | U-Net | NR | Accuracy: 0.9774; Sensitivity: 0.9903; Specificity: 0.7498; Precision: 0.9837; DSC: 0.9780 | NR |
| Chu (2022) [35] | DL | U-Net | NR | OAC composite model: GA Identification Sensitivity: 100% GA Identification Specificity: 100%; DSC: 0.940 OCT en face subRPE model: GA Identification Sensitivity: 100% GA Identification Specificity: 100%; DSC: 0.889 | NR |
| Derradji (2021) [36] | DL | U-Net style network with EfficientNet-b3 architecture | NR | Model with prior layer: Sensitivity (grader 1): 0.850; Sensitivity (grader 2): 0.915 Precision (grader 1): 0.928; Precision (grader 2): 0.799 DSC (grader 1): 0.881; DSC (grader 2): 0.844 Model without prior layer: Sensitivity (grader 1): 0.765; Sensitivity (grader 2): 0.845 Precision (grader 1): 0.955; Precision (grader 2): 0.845 DSC (grader 1): 0.841; DSC (grader 2): 0.831 | NR |
| Domalpally (2024) [50] | DL | EfficientNet-B5 (weakly labeled model) Feature Pyramid Network with EfficientNet-B5 encoder (strongly labeled model) | Saliency maps | Strongly labeled model: DSC: 0.885 | Strongly labeled model: DSC: 0.918 |
| Feeny (2015) [51] | ML | Random forest classifier | NR | Sensitivity: 0.65; Specificity: 0.99; PPV: 0.82; DSC: 0.68 | NR |
| Hassan (2021) [37] | DL | RASP-Net | NR | GA detection: Sensitivity: 0.904; Specificity: 0.930; Precision: 0.853 GA segmentation: IoU: 0.606; DSC: 0.755 | NR |
| Hu (2015) [38] | ML | k-NN | NR | Accuracy: 0.94; Sensitivity: 0.87; Specificity: 0.96; PPV: 0.80; OR: 0.72 | NR |
| Ji (2018) [47] | DL | Deep voting model (sparse autoencoders) | NR | DS1: AAD (mm2): 0.67; AAD (%): 11.49; OR: 0.8694 DS2: AAD (mm2): 0.34; AAD (%): 8.30; OR: 0.8166 | NR |
| Kalra (2022) [39] | DL | U-Net | NR | B-scan model: Detection Accuracy: 0.91; Pixel-wise Accuracy: 0.94 Detection Sensitivity: 0.86; Pixel-wise Sensitivity: 0.90 Detection Specificity: 0.94; Pixel-wise Specificity: 0.90 Detection f-score: 0.87; Pixel-wise f-score: 0.71 En face model: Detection Accuracy: 0.82; Pixel-wise Accuracy: 0.96 Detection Sensitivity: 0.97; Pixel-wise Sensitivity: 0.95 Detection Specificity: 0.42; Pixel-wise Specificity: 0.93 Detection f-score: 0.88; Pixel-wise f-score: 0.82 | NR |
| Liefers (2020) [52] | DL | Deep encoder–decoder structure with residual blocks and shortcut connections | NR | DSC: 0.72 | DSC: 0.66 (total cases); DSC: 0.71 (pure GA cases) |
| Liu (2023) [40] | DL | TSSK-Net with ResNet-18 backbone | CAM-based techniques (Grad-CAM, LayerCAM, and MS-CAM) and knowledge distillation-based anomaly localization (heatmaps) | GA Segmentation: AUC: 0.9704; DSC: 0.4504 | NR |
| Ma (2020) [48] | DL | VGG16 backbone | Saliency maps (MS-CAM) | DS1: AAD (mm2): 0.63; AAD (%): 12.87; AUC: 0.9417; OR: 0.8430; DSC: 0.9121 DS2: AAD (mm2): 0.78; AAD (%): 14.99; AUC: 0.9326; OR: 0.7441; DSC: 0.847 | NR |
| Mai (2023) [41] | DL | DL model based on U-Net architecture | NR | Recall: 0.87; Precision: 0.87; DSC (mean): 0.86; DSC (median): 0.90 | Recall: 0.93; Precision: 0.90; DSC: 0.91; DSC (mean): 0.91; DSC (median): 0.93 |
| Morano (2024) [42] | DL | FCNN | NR | 100% of training data Multiscale OCT + FAF: AUROC: 0.996; AUPR: 0.9895; DSC: 0.9019 Multiscale OCT + SLO: AUROC: 0.9939; AUPR: 0.984; DSC: 0.8915 Late OCT + FAF: AUROC: 0.9948; AUPR: 0.9859; DSC: 0.8991 Late OCT + SLO: AUROC: 0.9933; AUPR: 0.9822; DSC: 0.8908 | NR |
| Pfau (2020) [53] | DL | Deeplabv3 model with ResNet-50 backbone | NR | DSC: 0.96 | NR |
| Pramil (2023) [57] | DL | U-Net | NR | GA detection: Sensitivity: 0.95; Specificity: 0.91 GA segmentation: SDC (grader 1): 0.92; SDC (grader 2): 0.91; SDC (after BM correction): 0.97 | NR |
| Ramsey (2014) [54] | ML | FCM | NR | Sensitivity (FAF): 0.94; Sensitivity (CFP): 0.47 Specificity (FAF): 0.98; Specificity (CFP): 0.98 | NR |
| Royer (2021) [43] | DL | W-net (autoencoder) | NR | Sensitivity: 0.85; Precision: 0.90; F1 score: 0.87 | NR |
| Spaide (2023) [55] | DL | Y-Net and U-Net | NR | DSC: 0.90–0.92 | NR |
| Szeskin (2021) [44] | DL | Custom column-based CNN | NR | cRORA: Mean Detection Recall: 0.67; Mean Detection Precision: 0.70; (F1-3D) avg. Precision: 0.68; AUC (F1-3D): 0.937; F1 score: 0.78 Macular atrophy: Mean Detection Recall: 0.91; Mean Detection precision: 0.72; F1-3D avg. Precision: 0.83; AUC: 0.970; F1 score: 0.89 | NR |
| Wang (2022) [45] | DL | Self-attended U-Net | Attention modules | Baseline: Accuracy: 0.98; Sensitivity: 0.85; Specificity: 0.99; IoU: 0.77; DSC: 0.85 Month 12: Accuracy: 0.95; Sensitivity: 0.75; Specificity: 0.98; IoU: 0.68; DSC: 0.78 | NR |
| Wu (2019) [46] | DL | U-Net | Probability map | AAD: 11.0; OR: 0.779; DSC: 0.87 | NR |
| Xu (2019) [49] | DL | Stacked sparse autoencoder | NR | DS1 with training data: AAD (mm2): 0.18; AAD (%): 3.67; OR: 0.9985 DS1 without training data: AAD (mm2): 0.21; AAD (%): 4.79; OR: 0.9979 DS2 with training data: AAD (mm2): 0.48; AAD (%): 11.09; OR: 0.8455 DS2 without training data: AAD (mm2): 0.4418; AAD (%): 11.09; OR: 0.8448 | NR |
| Zhang (2021) [56] | DL | Modified U-Net | NR | GA approach 1: DSC median: 0.84; DSC mean: 0.75 GA approach 2: DSC median: 0.83; DSC mean: 0.75 | GA approach 1: Accuracy: 0.91; Sensitivity: 0.99; Specificity: 0.54; F1 score: 0.94; DSC median: 0.96; DSC mean: 0.91 GA approach 2: Accuracy: 0.94; Sensitivity: 0.98; Specificity: 0.76; F1 score: 0.96; DSC median: 0.95; DSC mean: 0.87 iRORA/cRORA approach 1: Accuracy: 0.89; Sensitivity: 0.90; Specificity: 0.68; F1 score: 0.94 iRORA/cRORA approach 2: Accuracy: 0.87; Sensitivity: 0.90; Specificity: 0.68; F1 score: 0.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatzara, A.; Maliagkani, E.; Mitsopoulou, D.; Katsimpris, A.; Apostolopoulos, I.D.; Papageorgiou, E.; Georgalas, I. Artificial Intelligence Approaches for Geographic Atrophy Segmentation: A Systematic Review and Meta-Analysis. Bioengineering 2025, 12, 475. https://doi.org/10.3390/bioengineering12050475
Chatzara A, Maliagkani E, Mitsopoulou D, Katsimpris A, Apostolopoulos ID, Papageorgiou E, Georgalas I. Artificial Intelligence Approaches for Geographic Atrophy Segmentation: A Systematic Review and Meta-Analysis. Bioengineering. 2025; 12(5):475. https://doi.org/10.3390/bioengineering12050475
Chicago/Turabian StyleChatzara, Aikaterini, Eirini Maliagkani, Dimitra Mitsopoulou, Andreas Katsimpris, Ioannis D. Apostolopoulos, Elpiniki Papageorgiou, and Ilias Georgalas. 2025. "Artificial Intelligence Approaches for Geographic Atrophy Segmentation: A Systematic Review and Meta-Analysis" Bioengineering 12, no. 5: 475. https://doi.org/10.3390/bioengineering12050475
APA StyleChatzara, A., Maliagkani, E., Mitsopoulou, D., Katsimpris, A., Apostolopoulos, I. D., Papageorgiou, E., & Georgalas, I. (2025). Artificial Intelligence Approaches for Geographic Atrophy Segmentation: A Systematic Review and Meta-Analysis. Bioengineering, 12(5), 475. https://doi.org/10.3390/bioengineering12050475








