Identification of Potential Therapeutic Targets for Sensorineural Hearing Loss and Evaluation of Drug Development Potential Using Mendelian Randomization Analysis
Abstract
1. Introduction
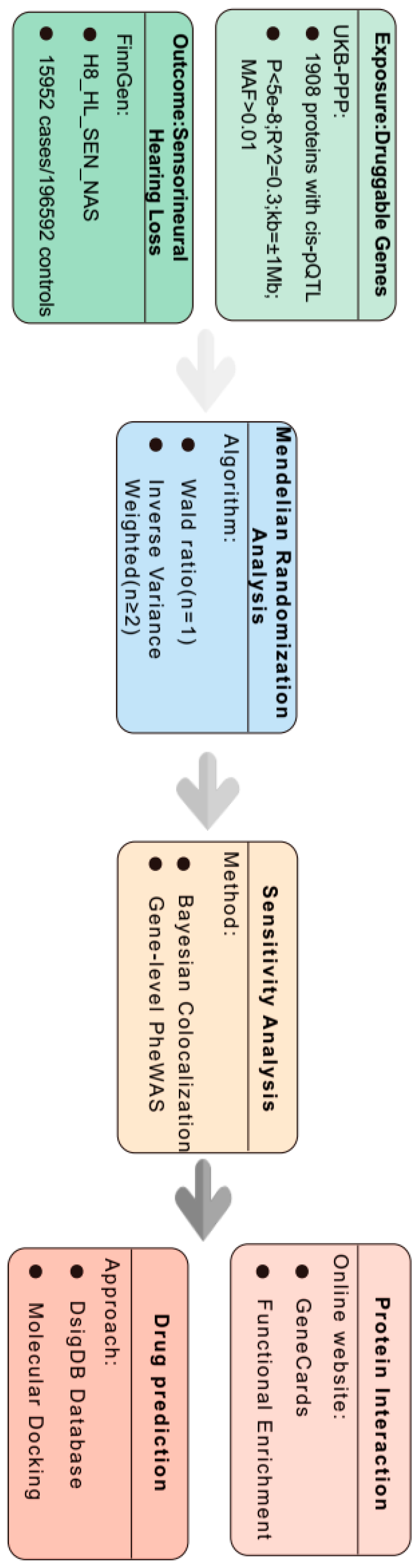
2. Materials and Methods
2.1. Plasma Protein Quantitative Trait Loci (pQTL)
2.2. SNHL GWAS Dataset
2.3. Mendelian Randomization Analysis
2.4. Colocalization Analysis
2.5. Phenotype-Wide Association Analysis
2.6. Functional Enrichment Analysis
2.7. Prediction of Candidate Drugs
2.8. Molecular Docking
3. Results
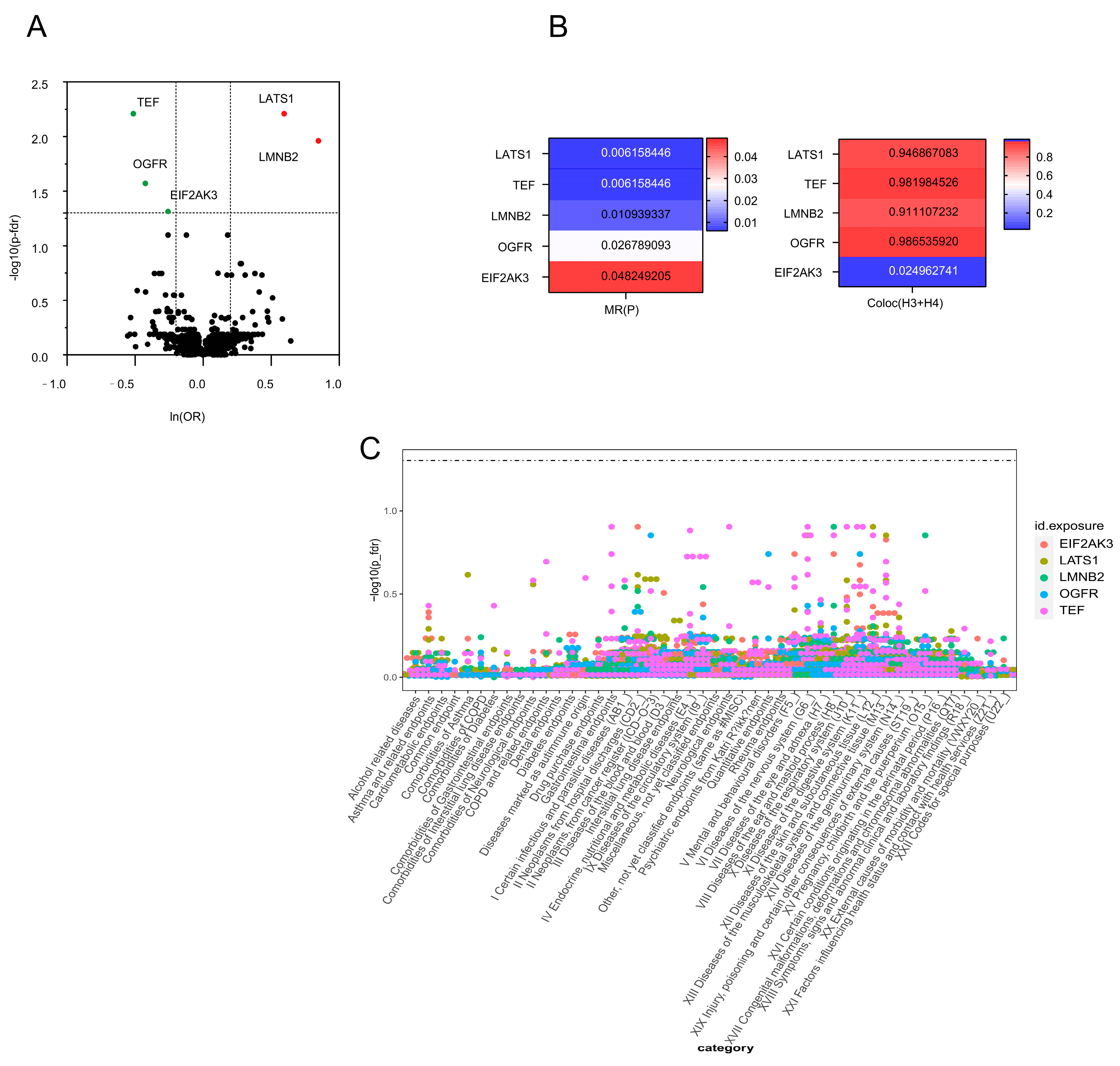
3.1. We Found1908 Plasma Proteins Associated with SNHL in the MR Analysis
3.2. Sensitivity Analysis for Five Plasma Proteins Associated with SNHL
3.3. Phenotype-Wide MR Analysis of Four Plasma Proteins in SNHL
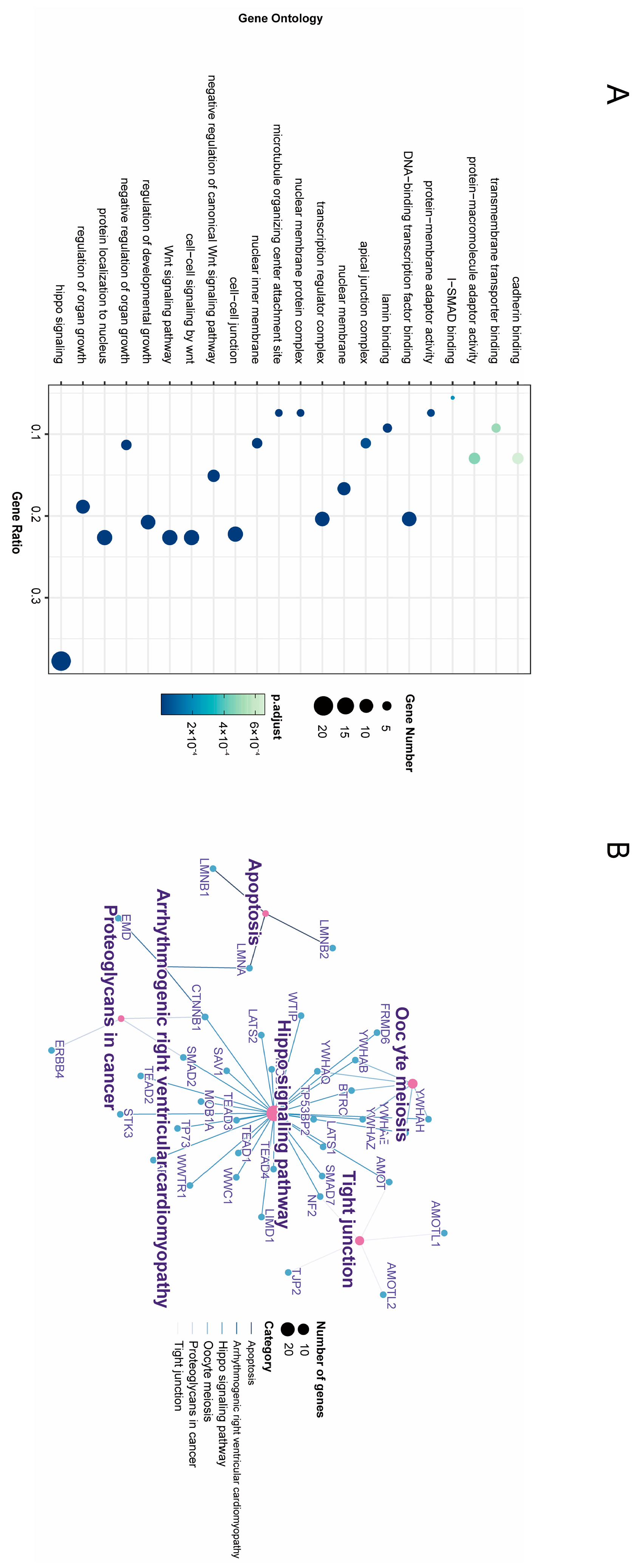
3.4. Enrichment Analysis of GO Annotation and KEGG Pathway
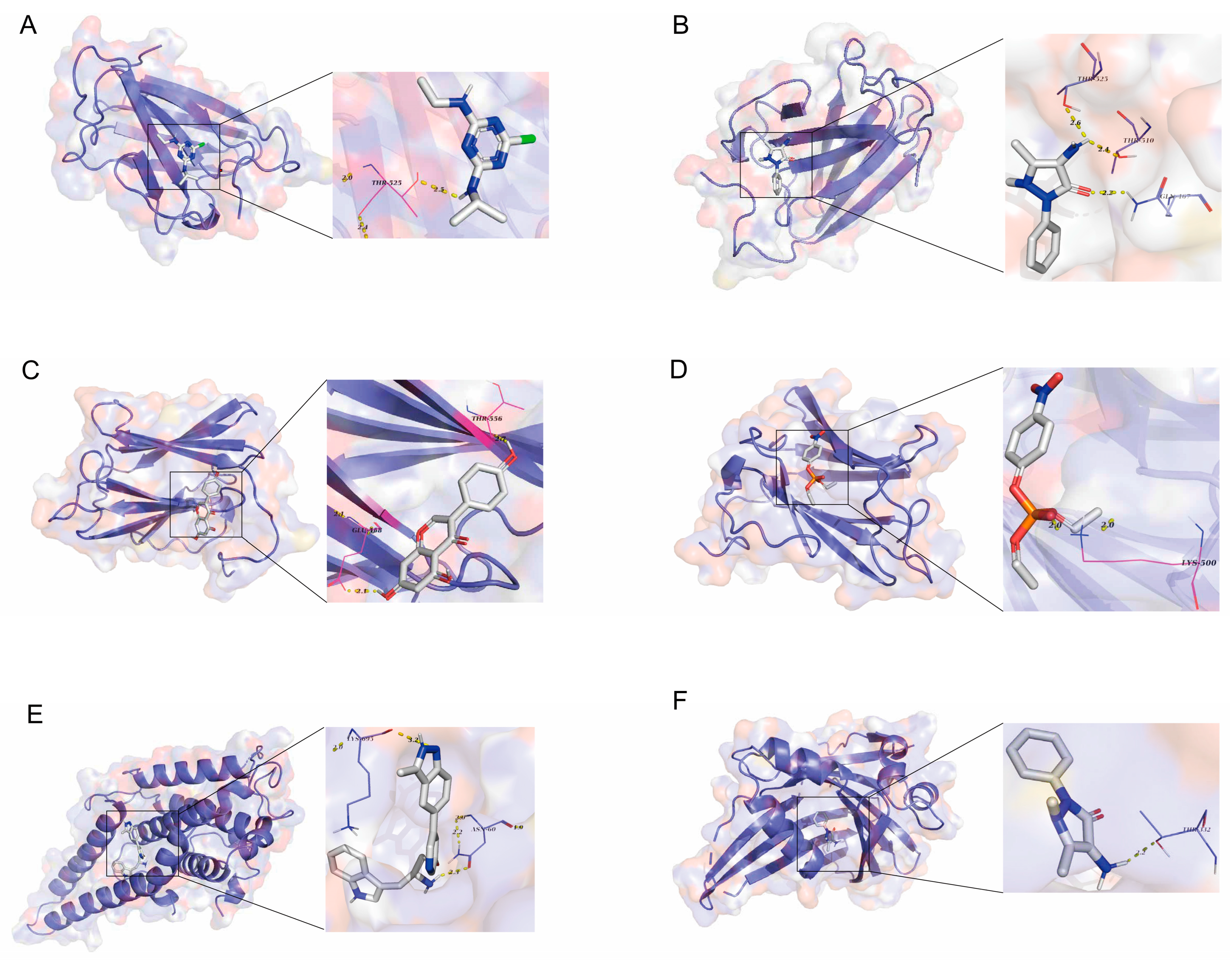
3.5. Screening of Potential Therapeutic Drugs
3.6. Validation of Candidate Plasma Proteins in Treating SNHL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, X.; Wan, P.; Li, P.; Wang, J.; Guo, S.; Zhang, Y.; An, Y.; Ye, C.; Liu, Z.; Gao, J.; et al. Mechanism and Prevention of Ototoxicity Induced by Aminoglycosides. Front. Cell. Neurosci. 2021, 15, 692762. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Boroujeni, M.; Zahedi-Amiri, A.; Coombs, K.M. Embryonic Origins of Virus-Induced Hearing Loss: Overview of Molecular Etiology. Viruses 2021, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Crowson, M.G.; Hertzano, R.; Tucci, D.L. Emerging Therapies for Sensorineural Hearing Loss. Otol. Neurotol. Off. Publ. Am. Otol. Soc. Am. Neurotol. Soc. Eur. Acad. Otol. Neurotol. 2017, 38, 792–803. [Google Scholar] [CrossRef]
- Zhang, L.; Du, Z.; He, L.; Liang, W.; Liu, K.; Gong, S. ROS-Induced Oxidative Damage and Mitochondrial Dysfunction Mediated by Inhibition of SIRT3 in Cultured Cochlear Cells. Neural Plast. 2022, 2022, 5567174. [Google Scholar] [CrossRef]
- Tanna, R.J.; Lin, J.W.; De Jesus, O. Sensorineural Hearing Loss. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Toroslu, T.; Erdoğan, H.; Çağlar, Ö.; Güçlü, O.; Dereköy, F.S. Comparison of Different Treatment Methods for Idiopathic Sudden Sensorineural Hearing Loss. Turk. Arch. Otorhinolaryngol. 2018, 56, 226–232. [Google Scholar] [CrossRef]
- Evans, D.S. Target Discovery for Drug Development Using Mendelian Randomization. Methods Mol. Biol. 2022, 2547, 1–20. [Google Scholar] [CrossRef]
- Burgess, S.; Mason, A.M.; Grant, A.J.; Slob, E.A.W.; Gkatzionis, A.; Zuber, V.; Patel, A.; Tian, H.; Liu, C.; Haynes, W.G.; et al. Using genetic association data to guide drug discovery and development: Review of methods and applications. Am. J. Hum. Genet. 2023, 110, 195–214. [Google Scholar] [CrossRef]
- Gill, D.; Georgakis, M.K.; Walker, V.M.; Schmidt, A.F.; Gkatzionis, A.; Freitag, D.F.; Finan, C.; Hingorani, A.D.; Howson, J.M.M.; Burgess, S.; et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021, 6, 16. [Google Scholar] [CrossRef]
- Sun, B.B.; Chiou, J.; Traylor, M.; Benner, C.; Hsu, Y.H.; Richardson, T.G.; Surendran, P.; Mahajan, A.; Robins, C.; Vasquez-Grinnell, S.G.; et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 2023, 622, 329–338. [Google Scholar] [CrossRef]
- Lin, J.; Zhou, J.; Xu, Y. Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis. Brain 2023, 146, 3364–3372. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Sipilä, T.P.; Kristiansson, K.; Donner, K.M.; Reeve, M.P.; Laivuori, H.; Aavikko, M.; Kaunisto, M.A.; et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023, 613, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Howard, D.M.; Adams, M.J.; Hill, W.D.; Clarke, T.K.; Deary, I.J.; Whalley, H.C.; McIntosh, A.M. A phenome-wide association and Mendelian Randomisation study of polygenic risk for depression in UK Biobank. Nat. Commun. 2020, 11, 2301. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Deng, Y.T.; Ou, Y.N.; Wu, B.S.; Yang, Y.X.; Jiang, Y.; Huang, Y.Y.; Liu, Y.; Tan, L.; Dong, Q.; Suckling, J.; et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol. Psychiatry 2022, 27, 2849–2857. [Google Scholar] [CrossRef]
- Feng, R.; Lu, M.; Xu, J.; Zhang, F.; Yang, M.; Luo, P.; Xu, K.; Xu, P. Pulmonary embolism and 529 human blood metabolites: Genetic correlation and two-sample Mendelian randomization study. BMC Genom. Data 2022, 23, 69. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Su, W.M.; Gu, X.J.; Dou, M.; Duan, Q.Q.; Jiang, Z.; Yin, K.F.; Cai, W.C.; Cao, B.; Wang, Y.; Chen, Y.P. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2023, 94, 954–961. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar] [CrossRef] [PubMed]
- Kim, S. Getting the most out of PubChem for virtual screening. Expert Opin. Drug Discov. 2016, 11, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef]
- Hsin, K.Y.; Ghosh, S.; Kitano, H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS ONE 2013, 8, e83922. [Google Scholar] [CrossRef]
- Zheng, J.; Haberland, V.; Baird, D.; Walker, V.; Haycock, P.C.; Hurle, M.R.; Gutteridge, A.; Erola, P.; Liu, Y.; Luo, S.; et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020, 52, 1122–1131. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Kelley, M.W. Cell adhesion molecules during inner ear and hair cell development, including notch and its ligands. Curr. Top. Dev. Biol. 2003, 57, 321–356. [Google Scholar] [CrossRef]
- Li, J.Z.; Fan, B.Y.; Sun, T.; Wang, X.X.; Li, J.J.; Zhang, J.P.; Gu, G.J.; Shen, W.Y.; Liu, D.R.; Wei, Z.J.; et al. Bioinformatics analysis of ferroptosis in spinal cord injury. Neural Regen. Res. 2023, 18, 626–633. [Google Scholar] [CrossRef]
- Wang, M.; Dong, Y.; Gao, S.; Zhong, Z.; Cheng, C.; Qiang, R.; Zhang, Y.; Shi, X.; Qian, X.; Gao, X.; et al. Hippo/YAP signaling pathway protects against neomycin-induced hair cell damage in the mouse cochlea. Cell. Mol. Life Sci. CMLS 2022, 79, 79. [Google Scholar] [CrossRef]
- Nishiyama, T.; Fujioka, M.; Saegusa, C.; Oishi, N.; Harada, T.; Hosoya, M.; Saya, H.; Ogawa, K. Deficiency of large tumor suppressor kinase 1 causes congenital hearing loss associated with cochlear abnormalities in mice. Biochem. Biophys. Res. Commun. 2021, 534, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Nadar-Ponniah, P.T.; Taiber, S.; Caspi, M.; Koffler-Brill, T.; Dror, A.A.; Siman-Tov, R.; Rubinstein, M.; Padmanabhan, K.; Luxenburg, C.; Lang, R.A.; et al. Striatin Is Required for Hearing and Affects Inner Hair Cells and Ribbon Synapses. Front. Cell Dev. Biol. 2020, 8, 615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, H.; Salvi, R.; Ding, D. Paraquat initially damages cochlear support cells leading to anoikis-like hair cell death. Hear. Res. 2018, 364, 129–141. [Google Scholar] [CrossRef]
- Rudolf, M.A.; Andreeva, A.; Kozlowski, M.M.; Kim, C.E.; Moskowitz, B.A.; Anaya-Rocha, A.; Kelley, M.W.; Corwin, J.T. YAP Mediates Hair Cell Regeneration in Balance Organs of Chickens, But LATS Kinases Suppress Its Activity in Mice. J. Neurosci. Off. J. Soc. Neurosci. 2020, 40, 3915–3932. [Google Scholar] [CrossRef]
- Kasturirangan, S.; Mehdi, B.; Chadee, D.N. LATS1 Regulates Mixed-Lineage Kinase 3 (MLK3) Subcellular Localization and MLK3-Mediated Invasion in Ovarian Epithelial Cells. Mol. Cell. Biol. 2021, 41, e0007821. [Google Scholar] [CrossRef]
- Wang, J.; Ma, M.; Liu, X. Lamins B2 Promotes Esophageal Cancer by Stimulating Proliferation and Inhibiting Apoptosis. Ann. Clin. Lab. Sci. 2022, 52, 202–212. [Google Scholar]
- Dong, C.H.; Jiang, T.; Yin, H.; Song, H.; Zhang, Y.; Geng, H.; Shi, P.C.; Xu, Y.X.; Gao, H.; Liu, L.Y.; et al. LMNB2 promotes the progression of colorectal cancer by silencing p21 expression. Cell Death Dis. 2021, 12, 331. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, M.; Luo, Y.; Li, Y.; Xu, Y.; Wang, N. Comprehensive analysis of LMNB2 in pan-cancer and identification of its biological role in sarcoma. Aging 2024, 16. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Yu, Z.; Li, H.; Jin, X. The role of lamin B2 in human diseases. Gene 2023, 870, 147423. [Google Scholar] [CrossRef]
- Zagon, I.S.; Verderame, M.F.; McLaughlin, P.J. The biology of the opioid growth factor receptor (OGFr). Brain Res. Brain Res. Rev. 2002, 38, 351–376. [Google Scholar] [CrossRef]
- Titunick, M.B.; Lewis, G.S.; Cain, J.D.; Zagon, I.S.; McLaughlin, P.J. Blockade of the OGF-OGFr pathway in diabetic bone. Connect. Tissue Res. 2019, 60, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Hankins, G.R.; Harris, R.T. The Opioid Growth Factor in Growth Regulation and Immune Responses in Cancer. Adv. Neurobiol. 2024, 35, 45–85. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, L.; Zhou, Z.; Wang, M.; Liu, R.; Wang, L.; Jiang, Q.; Song, L. An opioid growth factor receptor (OGFR) for [Met5]-enkephalin in Chlamys farreri. Fish Shellfish Immunol. 2013, 34, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Urosevic, M.; Oberholzer, P.A.; Maier, T.; Hafner, J.; Laine, E.; Slade, H.; Benninghoff, B.; Burg, G.; Dummer, R. Imiquimod treatment induces expression of opioid growth factor receptor: A novel tumor antigen induced by interferon-alpha? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 4959–4970. [Google Scholar] [CrossRef]
- Gachon, F.; Fonjallaz, P.; Damiola, F.; Gos, P.; Kodama, T.; Zakany, J.; Duboule, D.; Petit, B.; Tafti, M.; Schibler, U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004, 18, 1397–1412. [Google Scholar] [CrossRef]
- Chopra, A.; Tye, S.J.; Lee, K.H.; Sampson, S.; Matsumoto, J.; Adams, A.; Klassen, B.; Stead, M.; Fields, J.A.; Frye, M.A. Underlying neurobiology and clinical correlates of mania status after subthalamic nucleus deep brain stimulation in Parkinson’s disease: A review of the literature. J. Neuropsychiatry Clin. Neurosci. 2012, 24, 102–110. [Google Scholar] [CrossRef]
- Li, W.K.; Li, H.; Lu, Y.F.; Li, Y.Y.; Fu, Z.D.; Liu, J. Atorvastatin alters the expression of genes related to bile acid metabolism and circadian clock in livers of mice. PeerJ 2017, 5, e3348. [Google Scholar] [CrossRef]
- Wu, J.; Ye, J.; Kong, W.; Zhang, S.; Zheng, Y. Programmed cell death pathways in hearing loss: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2020, 53, e12915. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, J.; Luan, F.; Gu, G.; Zhao, R.; Wang, Q.; Dong, Z.; Tang, J.; Wang, W.; Sun, J.; et al. Sensorineural Hearing Loss and Mitochondrial Apoptosis of Cochlear Spiral Ganglion Neurons in Fibroblast Growth Factor 13 Knockout Mice. Front. Cell. Neurosci. 2021, 15, 658586. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Chao, J.R.; Kim, C.; Kim, B.; Thi-Thanh Nguyen, P.; Jung, H.; Chang, J.; Lee, J.H.; Suh, J.G. Hearing loss through apoptosis of the spiral ganglion neurons in apolipoprotein E knockout mice fed with a western diet. Biochem. Biophys. Res. Commun. 2020, 523, 692–698. [Google Scholar] [CrossRef]
- Li, T.; Guo, R.; Zong, Q.; Ling, G. Application of molecular docking in elaborating molecular mechanisms and interactions of supramolecular cyclodextrin. Carbohydr. Polym. 2022, 276, 118644. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wu, M.B.; Chen, Z.J.; Chen, H.; Lin, J.P.; Yang, L.R. Fragment-based drug discovery and molecular docking in drug design. Curr. Pharm. Biotechnol. 2015, 16, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Halim, A.; Tian, B.; Luo, Q.; Song, G. MT1-MMP downregulation via the PI3K/Akt signaling pathway is required for the mechanical stretching-inhibited invasion of bone-marrow-derived mesenchymal stem cells. J. Cell. Physiol. 2019, 234, 14133–14144. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lan, Y.; Zhang, L.; Ye, X.; Shen, Q.; Mo, G.; Chen, X. Genistein exerts anti-colorectal cancer actions: Clinical reports, computational and validated findings. Aging 2023, 15, 3678–3689. [Google Scholar] [CrossRef]
- Thakkar, D.; Singh, S.; Wairkar, S. Advanced Delivery Strategies of Nintedanib for Lung Disorders and Beyond: A Comprehensive Review. AAPS PharmSciTech 2024, 25, 150. [Google Scholar] [CrossRef]
- Schmidt, A.F.; Finan, C.; Gordillo-Marañón, M.; Asselbergs, F.W.; Freitag, D.F.; Patel, R.S.; Tyl, B.; Chopade, S.; Faraway, R.; Zwierzyna, M.; et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 2020, 11, 3255. [Google Scholar] [CrossRef]
| Characteristics | Total (N) | HR (95% CI) | p Value |
|---|---|---|---|
| LATS1 | 1 | 1.81 (1.39–2.38) | 0.006 |
| TEF | 1 | 0.60 (0.47–0.78) | 0.006 |
| LMNB2 | 1 | 2.33 (1.55–3.51) | 0.011 |
| OGFR | 1 | 0.65 (0.53–0.82) | 0.027 |
| EIF2AK3 | 1 | 0.77 (0.67–0.89) | 0.048 |
| Drug | p-Value | Odds Ratio | Combined Score | Genes |
|---|---|---|---|---|
| Ampyrone HL60 DOWN | 0.009 | 24.669 | 117.1276 | OGFR; LMNB2 |
| Atrazine CTD 00005450 | 0.012 | 17.2329 | 76.7860 | OGFR; TEF; LMNB2 |
| PARAOXON CTD 00006470 | 0.014 | 93.5449 | 397.178 | LMNB2 |
| A443654 LINCS | 0.021 | 64.379 | 249.824 | LATS1 |
| Genistein CTD 00007324 | 0.021 | 15.270 | 59.0690 | OGFR; LMNB2 |
| Arsenenous acid CTD 00000922 | 0.023 | 14.610 | 55.357 | OGFR; LMNB2 |
| SU-14813 Kinome Scan | 0.029 | 45.634 | 161.7507 | LATS1 |
| AST-487 Kinome Scan | 0.029 | 45.009 | 158.928 | LATS1 |
| Nintedanib FDA | 0.031 | 42.121 | 146.008 | LATS1 |
| BIBF-1120 (derivative) Kinome Scan | 0.031 | 42.121 | 146.0084 | LATS1 |
| Target | PDB ID | Drug | Binding Energy (kcal/mol) |
|---|---|---|---|
| LMNB2 | 2LLL | Atrazine | −63.766 |
| LMNB2 | 2LLL | Ampyrone | −67.623 |
| LMNB2 | 2LLL | Genistein | −86.926 |
| LMNB2 | 2LLL | PARAOXON | −67.057 |
| LATS1 | 7LWH | A443654 | −7.949 |
| TEF | 8CAA | Ampyrone | −7.836 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.; Tong, Q.; Liu, Y.; Qin, M.; Sun, S. Identification of Potential Therapeutic Targets for Sensorineural Hearing Loss and Evaluation of Drug Development Potential Using Mendelian Randomization Analysis. Bioengineering 2025, 12, 126. https://doi.org/10.3390/bioengineering12020126
Ding S, Tong Q, Liu Y, Qin M, Sun S. Identification of Potential Therapeutic Targets for Sensorineural Hearing Loss and Evaluation of Drug Development Potential Using Mendelian Randomization Analysis. Bioengineering. 2025; 12(2):126. https://doi.org/10.3390/bioengineering12020126
Chicago/Turabian StyleDing, Shun, Qiling Tong, Yixuan Liu, Mengyao Qin, and Shan Sun. 2025. "Identification of Potential Therapeutic Targets for Sensorineural Hearing Loss and Evaluation of Drug Development Potential Using Mendelian Randomization Analysis" Bioengineering 12, no. 2: 126. https://doi.org/10.3390/bioengineering12020126
APA StyleDing, S., Tong, Q., Liu, Y., Qin, M., & Sun, S. (2025). Identification of Potential Therapeutic Targets for Sensorineural Hearing Loss and Evaluation of Drug Development Potential Using Mendelian Randomization Analysis. Bioengineering, 12(2), 126. https://doi.org/10.3390/bioengineering12020126







