Large Language Models in Bio-Ontology Research: A Review
Abstract
1. Introduction
2. Comparison to Prior Reviews
2.1. Scope and Audience
2.2. Ontology Engineering vs. Application-Centric Views
2.3. Alignment and Mapping: LLMs vs. Classical Matchers
2.4. LLMs, Knowledge Graphs, and Grounding
2.5. Risk, Evaluation, and Governance
2.6. Summary of Differentiators
- (a)
- A bio-ontology-centric synthesis of LLM use across creation, curation, mapping/ alignment, and KG integration, rather than general biomedical applications;
- (b)
- A methods-to-workflow bridge that surfaces where LLM suggestions enter curator pipelines and how ontology logic/IDs constrain outputs;
- (c)
- A precision view of alignment in biomedical settings (phenotypes, diseases, anatomy), emphasizing curation cost and governance when adopting LLM-generated mappings;
- (d)
- A grounding and reproducibility focus, consolidating practices (Retrieval Augmented Generation (RAG) to canonical IDs, reasoner checks, audit trails) that are specific to ontology/KG ecosystems.
2.7. Where This Review Adds Unique Value
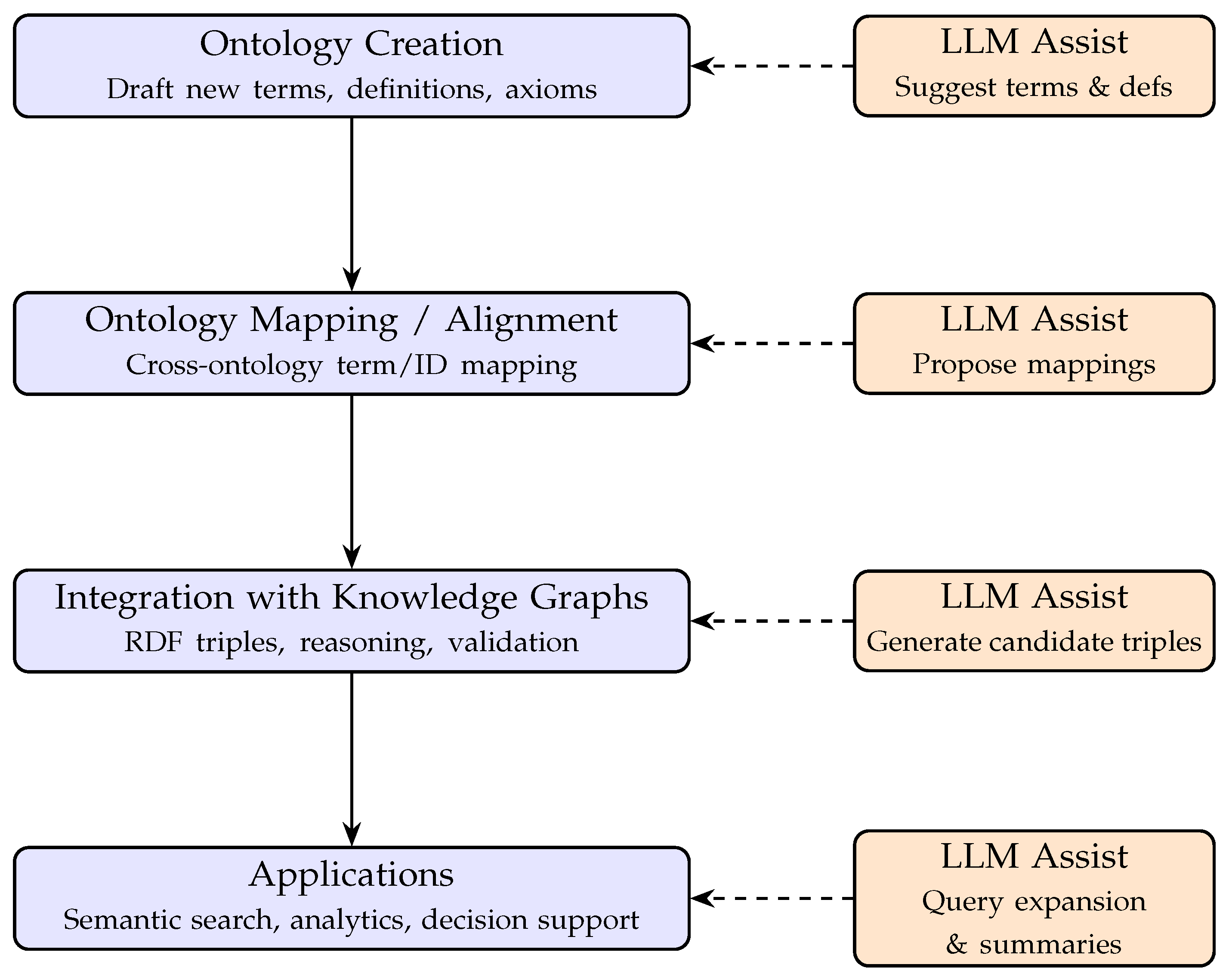
3. Background
3.1. LLM-Assisted Ontology Creation and Curation
3.2. Ontology Mapping and Integration
3.3. Text Mining and Semantic Search with Ontologies
3.4. Knowledge Graphs and Ontology Integration
3.5. Comparative Analysis: Scalability, Maturity, and Limitations
3.6. Milestones in LLM-Assisted Ontology Engineering: Strengths and Trade-Offs
4. Domain-Specific BioLLMs: Challenges, Limitations, and Future Directions
4.1. Challenges
4.1.1. Data Scarcity and Quality
4.1.2. Bias and Ethical Concerns
4.1.3. Hallucination and Factual Consistency
4.1.4. Computational Challenges in Fine-Tuning
5. Limitations
5.1. Data Issues
5.2. Algorithmic Issues
5.3. Clinical Application Issues
5.4. Future Directions
5.4.1. Domain Adaptation and Continuous Learning
5.4.2. Integration with Ontologies and Knowledge Bases
5.4.3. Evaluation Metrics and Benchmarks for BioLLMs
5.4.4. Hybrid Neural–Symbolic Reasoning Models
5.4.5. Interactive and Collaborative Curation Tools
6. Conclusions
- Hybrid neuro-symbolic approaches. One of the most promising avenues is combining the probabilistic power of LLMs with the logical rigor of ontologies and reasoning systems. Neuro-symbolic methods could allow LLMs to propose candidate terms and relations, while symbolic reasoners check consistency against established ontology axioms. Early work already shows that hybrid models outperform standalone LLMs in entity recognition and mapping tasks. Developing robust pipelines for ontology-grounded reasoning will help ensure factual reliability and logical coherence in AI-assisted ontology curation.
- Domain-specific evaluation and benchmarking. Generic NLP benchmarks are insufficient for biomedical ontology tasks, where the stakes include clinical safety and scientific validity. Future research must develop ontology-focused evaluation metrics that assess logical consistency, ontology coverage, and alignment with curated gold standards. Resources such as BLURB and BioASQ provide a foundation, but dedicated benchmarks for ontology creation, mapping accuracy, and definition quality will be needed to guide model improvement. Equally important is incorporating human expert evaluation, where domain specialists assess whether AI-generated ontology content is clinically meaningful and biologically accurate.
- Interactive and collaborative curation platforms. Ontology development is a community-driven process, and the next generation of tools should reflect this. Embedding LLMs into real-time, collaborative editing platforms could support curators by suggesting terms, drafting definitions, and surfacing candidate mappings while still allowing experts to accept, reject, or modify suggestions. Early prototypes such as conversational ontology editors demonstrate the potential of such systems. Expanding these into multi-user platforms—where groups of experts and AI collaborate simultaneously—could reduce workload, improve transparency, and accelerate consensus building.
- Bias detection and mitigation. Biomedical datasets carry known demographic biases, such as under-representation of women in cardiology studies or lighter skin tones in dermatology images. If uncorrected, these biases can propagate into ontology terms, definitions, and mappings, perpetuating inequities. Future work should focus on bias-aware LLM training and auditing, where generated ontology content is evaluated for representational fairness and coverage across diverse populations. This will require both technical solutions (balanced corpora, fairness audits) and sociotechnical frameworks that embed equity considerations into ontology development practices.
- Continuous learning and adaptability. Biomedical knowledge evolves rapidly, with new diseases, drugs, and molecular mechanisms emerging every year. Static models quickly become outdated. A key direction is developing LLMs capable of continual domain adaptation, ingesting new literature, clinical data, and curated ontologies without catastrophic forgetting. Coupled with incremental ontology updating workflows, such systems could ensure that biomedical ontologies remain current and responsive to emerging discoveries.
- Ethical and governance frameworks. Finally, as LLMs become more tightly integrated into biomedical knowledge infrastructures, questions of accountability, authorship, and governance will grow in importance. Policies are needed to define how AI-generated content is validated, attributed, and disseminated. Ontology communities may need to establish standards for documenting AI contributions, auditing decision-making, and ensuring that ethical safeguards are systematically applied.
6.1. Actionable Recommendations for Practice (Next 12–18 Months)
- Adopt hybrid pipelines: pair LLM proposals with ontology reasoners and ID validators before acceptance; require curator sign-off for any new class, definition, or mapping.
- Track curator ROI: instrument workflows to log “minutes-per-accepted-edit” and reject reasons to quantify true time savings.
- Harden grounding: enforce identifier normalization (e.g., HPO/GO/DO/MONDO) and fail closed when IDs are uncertain; no free-text outputs into production.
- Gate mappings: require two independent signals (lexical/structural + LLM judgment) for cross-ontology equivalence; demote to relatedTo when confidence is borderline.
- Bias checks: run stratified audits (sex, skin tone, ancestry, rare disease) on LLM-suggested terms/mappings; escalate gaps to curators with templated remediation.
- Reproducibility: pin model, prompt, temperature, retrieval index, and ontology snapshot in an audit trail; re-run spot checks each release.
6.2. Operational Metrics and Target Bands
- Identifier Fidelity Rate (IFR): proportion of LLM suggestions with correct canonical IDs. Target: ≥95%.
- Axiom Consistency Violations per 1000 additions (ACV@1k): reasoner-detected contradictions post-merge. Target: ≤1.
- Curator Minutes per Accepted Edit (CMS@Edit): average human time to review and accept. Target: ≤5 min for definitions, ≤8 for mappings.
- Mapping Precision@K with curator-in-the-loop (P@KHITL): precision of candidate alignments at top-K. Target: ≥0.90 at K = 5 on biomedical tracks.
- Hallucinated-ID Rate (HR-ID): fraction of suggestions containing fabricated or non-resolvable IDs. Target: =0 in production.
- Run-to-Run Stability (RS@seed): agreement of accepted suggestions across re-runs with fixed seeds/snapshots. Target: ≥98%.
6.3. Minimal Reporting Checklist for LLM-Assisted Curation
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.W.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef] [PubMed]
- Eilbeck, K.; Lewis, S.E.; Mungall, C.J.; Yandell, M.; Stein, L.; Durbin, R.; Ashburner, M. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 2005, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Degtyarenko, K.; De Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2007, 36, D344–D350. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Havrilla, J.M.; Singaravelu, A.; Driscoll, D.M.; Minkovsky, L.; Helbig, I.; Medne, L.; Wang, K.; Krantz, I.; Desai, B.R. PheNominal: An EHR-integrated web application for structured deep phenotyping at the point of care. BMC Med. Inform. Decis. Mak. 2022, 22, 198. [Google Scholar] [CrossRef]
- Dahdul, W.; Dececchi, T.A.; Ibrahim, N.; Lapp, H.; Mabee, P. Moving the mountain: Analysis of the effort required to transform comparative anatomy into computable anatomy. Database 2015, 2015, bav040. [Google Scholar] [CrossRef]
- Smith, B.; Ashburner, M.; Rosse, C.; Bard, J.; Bug, W.; Ceusters, W.; Goldberg, L.J.; Eilbeck, K.; Ireland, A.; Mungall, C.J.; et al. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007, 25, 1251–1255. [Google Scholar] [CrossRef]
- Stearns, M.Q.; Price, C.; Spackman, K.A.; Wang, A.Y. SNOMED clinical terms: Overview of the development process and project status. In Proceedings of the AMIA Symposium. American Medical Informatics Association, Arlington, VA, USA, 3–7 November 2001; pp. 662–666. [Google Scholar]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef]
- Dahdul, W.; Manda, P.; Cui, H.; Balhoff, J.P.; Dececchi, T.A.; Ibrahim, N.; Lapp, H.; Vision, T.; Mabee, P.M. Annotation of phenotypes using ontologies: A gold standard for the training and evaluation of natural language processing systems. Database 2018, 2018, bay110. [Google Scholar] [CrossRef]
- Liu, J.; Yang, M.; Yu, Y.; Xu, H.; Li, K.; Zhou, X. Large language models in bioinformatics: Applications and perspectives. Bioinformatics 2024, 40, btae030. [Google Scholar] [CrossRef]
- Smith, N.; Yuan, X.; Melissinos, C.; Moghe, G. FuncFetch: An LLM-assisted workflow enables mining thousands of enzyme-substrate interactions from published manuscripts. Bioinformatics 2024, 41, btae756. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: A pretrained language model for scientific text. arXiv 2019, arXiv:1903.10676. [Google Scholar] [CrossRef]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthc. (HEALTH) 2021, 3, 1–23. [Google Scholar] [CrossRef]
- Luo, R.; Sun, L.; Xia, Y.; Qin, T.; Zhang, S.; Poon, H.; Liu, T.Y. BioGPT: Generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 2022, 23, bbac409. [Google Scholar] [CrossRef] [PubMed]
- Caufield, J.; Hegde, H.; Emonet, V.; Harris, N.; Joachimiak, M.; Matentzoglu, N.; Kim, H.; Moxon, S.; Reese, J.; Haendel, M. Structured Prompt Interrogation and Recursive Extraction of Semantics (SPIRES): A method for populating knowledge bases using zero-shot learning. Bioinformatics 2024, 40, btae104. [Google Scholar] [CrossRef] [PubMed]
- Toro, S.; Anagnostopoulos, A.V.; Bello, S.M.; Blumberg, K.; Cameron, R.; Carmody, L.; Diehl, A.D.; Dooley, D.M.; Duncan, W.D.; Fey, P.; et al. Dynamic Retrieval Augmented Generation of Ontologies using Artificial Intelligence (DRAGON-AI). J. Biomed. Semant. 2024, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Bizon, C.; Cox, S.; Balhoff, J.; Kebede, Y.; Wang, P.; Morton, K.; Fecho, K.; Tropsha, A. ROBOKOP KG and KGB: Integrated knowledge graphs from federated sources. J. Chem. Inf. Model. 2019, 59, 4968–4973. [Google Scholar] [CrossRef]
- Consortium, G.O. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Schriml, L.M.; Munro, J.B.; Schor, M.; Olley, D.; McCracken, C.; Felix, V.; Baron, J.A.; Jackson, R.; Bello, S.M.; Bearer, C.; et al. The human disease ontology 2022 update. Nucleic Acids Res. 2022, 50, D1255–D1261. [Google Scholar] [CrossRef] [PubMed]
- Fecho, K.; Bizon, C.; Miller, F.; Schurman, S.; Schmitt, C.; Xue, W.; Morton, K.; Wang, P.; Tropsha, A. A biomedical knowledge graph system to propose mechanistic hypotheses for real-world environmental health observations: Cohort study and informatics application. JMIR Med. Inform. 2021, 9, e26714. [Google Scholar] [CrossRef] [PubMed]
- Unni, D.R.; Moxon, S.A.; Bada, M.; Brush, M.; Bruskiewich, R.; Caufield, J.H.; Clemons, P.A.; Dancik, V.; Dumontier, M.; Fecho, K.; et al. Biolink Model: A universal schema for knowledge graphs in clinical, biomedical, and translational science. Clin. Transl. Sci. 2022, 15, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jin, Q.; Yeganova, L.; Lai, P.T.; Zhu, Q.; Chen, X.; Yang, Y.; Chen, Q.; Kim, W.; Comeau, D.C.; et al. Opportunities and challenges for ChatGPT and large language models in biomedicine and health. Brief. Bioinform. 2024, 25, bbad493. [Google Scholar] [CrossRef]
- Reese, J.; Röttger, P.; Thielk, M.; Hellmann, S.; Weng, C.; Xu, J.; Knecht, C.; Bedrick, S.; Fosler-Lussier, E.; Brockmeier, A.; et al. On the limitations of large language models in clinical diagnosis. medRxiv 2024. [Google Scholar] [CrossRef]
- Armitage, R. Implications of Large Language Models for Clinical Practice: Ethical Analysis Through the Principlism Framework. J. Eval. Clin. Pract. 2025, 31, e14250. [Google Scholar] [CrossRef]
- Lu, Z.; Peng, Y. Large language models in biomedicine and health. J. Am. Med. Inform. Assoc. 2024, 31, 1801–1811. [Google Scholar] [CrossRef]
- Bian, H. LLM-empowered knowledge graph construction: A survey. arXiv 2025, arXiv:2510.20345. [Google Scholar] [CrossRef]
- Yu, E.; Chu, X.; Zhang, W.; Meng, X.; Yang, Y.; Ji, X.; Wu, C. Large Language Models in Medicine: Applications, Challenges, and Future Directions. Int. J. Med. Sci. 2025, 22, 2792–2801. [Google Scholar] [CrossRef]
- Li, Q.; Nunes, B.P.; Sakor, A.; Jin, Y.; d’Aquin, M. Large Language Models for Ontology Engineering: A Systematic Literature Review. Semant. Web J. 2025. under review. Available online: https://www.semantic-web-journal.net/content/large-language-models-ontology-engineering-systematic-literature-review (accessed on 12 November 2025).
- Cui, H.; Lu, J.; Xu, R.; Wang, S.; Ma, W.; Yu, Y.; Yu, S.; Kan, X.; Ling, C.; Zhao, L.; et al. A Review on Knowledge Graphs for Healthcare. arXiv 2025, arXiv:2306.04802. [Google Scholar] [CrossRef]
- Xu, R.; Jiang, P.; Luo, L.; Xiao, C.; Cross, A.; Pan, S.; Sun, J.; Yang, C. A Survey on Unifying Large Language Models and Knowledge Graphs in Biomedicine. ACM Comput. Surv. 2025, in press. [Google Scholar]
- Giglou, H.B.; D’Souza, J.; Engel, F.; Auer, S. LLMs4OM: Matching Ontologies with Large Language Models. In Proceedings of the The Semantic Web—ESWC 2024 Satellite Events, Chalkida, Greece, 26–27 May 2024. [Google Scholar] [CrossRef]
- Taboada, M.; Martinez, D.; Arideh, M.; Mosquera, R. Ontology matching with large language models and prioritized depth-first search. Inf. Fusion 2025, 123, 103254. [Google Scholar] [CrossRef]
- Wang, X.; Ye, P.; Wu, G.; Feng, J.; Qiu, H.; Li, H.; Zhou, L.; Tang, B.; Li, Y.; Sun, K.; et al. A Survey for Large Language Models in Biomedicine. Artif. Intell. Med. 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Joachimiak, M.P.; Miller, M.A.; Caufield, J.H.; Ly, R.; Harris, N.L.; Tritt, A.; Mungall, C.J.; Bouchard, K.E. The Artificial Intelligence Ontology: LLM-assisted construction of AI concept hierarchies. Appl. Ontol. 2024, 19, 408–418. [Google Scholar] [CrossRef]
- Vendetti, J.; Harris, N.L.; Dorf, M.V.; Skrenchuk, A.; Caufield, J.H.; Gonçalves, R.S.; Graybeal, J.B.; Hegde, H.; Redmond, T.; Mungall, C.J.; et al. BioPortal: An open community resource for sharing, searching, and utilizing biomedical ontologies. Nucleic Acids Res. 2025, gkaf402. [Google Scholar] [CrossRef]
- Kommineni, V.K.; König-Ries, B.; Samuel, S. From human experts to machines: An LLM supported approach to ontology and knowledge graph construction. arXiv 2024, arXiv:2403.08345. [Google Scholar] [CrossRef]
- Mukanova, A.; Milosz, M.; Dauletkaliyeva, A.; Nazyrova, A.; Yelibayeva, G.; Kuzin, D.; Kussepova, L. LLM-powered natural language text processing for ontology enrichment. Appl. Sci. 2024, 14, 5860. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, E.; Cuenca Grau, B. Logmap: Logic-based and scalable ontology matching. In Proceedings of the International Semantic Web Conference, Bonn, Germany, 23–27 October 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 273–288. [Google Scholar]
- Cruz, I.F.; Stroe, C.; Caimi, F.; Fabiani, A.; Pesquita, C.; Couto, F.M.; Palmonari, M. Using AgreementMaker to align ontologies for OAEI 2011. In Proceedings of the ISWC International Workshop on Ontology Matching (OM), Bonn, Germany, 24 October 2011; Volume 814, pp. 114–121. [Google Scholar]
- Faria, D.; Santos, E.; Balasubramani, B.S.; Silva, M.C.; Couto, F.M.; Pesquita, C. Agreementmakerlight. Semant. Web 2025, 16, SW–233304. [Google Scholar] [CrossRef]
- Pour, M.A.N.; Algergawy, A.; Amini, R.; Faria, D.; Fundulaki, I.; Harrow, I.; Hertling, S.; Jiménez-Ruiz, E.; Jonquet, C.; Karam, N.; et al. Results of the ontology alignment evaluation initiative 2020. In Proceedings of the OM 2020—15th International Workshop on Ontology Matching, CEUR Proceedings, Virtual Event, 2 November 2020; Volume 2788, pp. 92–138. [Google Scholar]
- Norouzi, S.S.; Mahdavinejad, M.S.; Hitzler, P. Conversational ontology alignment with chatgpt. arXiv 2023, arXiv:2308.09217. [Google Scholar] [CrossRef]
- Matentzoglu, N.; Caufield, J.H.; Hegde, H.B.; Reese, J.T.; Moxon, S.; Kim, H.; Harris, N.L.; Haendel, M.A.; Mungall, C.J. Mappergpt: Large language models for linking and mapping entities. arXiv 2023, arXiv:2310.03666. [Google Scholar] [CrossRef]
- Ruan, W.; Lyu, Y.; Zhang, J.; Cai, J.; Shu, P.; Ge, Y.; Lu, Y.; Gao, S.; Wang, Y.; Wang, P.; et al. Large Language Models for Bioinformatics. arXiv 2025, arXiv:2501.06271. [Google Scholar] [CrossRef]
- Cavalleri, E.; Soto Gomez, M.; Pashaeibarough, A.; Malchiodi, D.; Caufield, J.; Reese, J.; Mungall, C.; Robinson, P.N.; Casiraghi, E.; Valentini, G.; et al. SPIREX: Improving LLM-based relation extraction from RNA-focused scientific literature using graph machine learning. In Proceedings of the Workshops at the 50th International Conference on Very Large Data Bases, Guangzhou, China, 25 August 2024; pp. 1–11. [Google Scholar]
- Pour, M.A.N.; Algergawy, A.; Blomqvist, E.; Buche, P.; Chen, J.; Cotovio, P.G.; Coulet, A.; Cufi, J.; Dong, H.; Faria, D.; et al. Results of the OAEI 2024 Campaign. In Proceedings of the CEUR Workshop Proceedings (OM@ISWC), Baltimore, ML, USA, 11 November 2024. [Google Scholar]
- Qiang, Z.; Taylor, K.; Wang, W.; Jiang, J. OAEI-LLM: A Benchmark Dataset for Understanding Large Language Model Hallucinations in Ontology Matching. arXiv 2024, arXiv:2409.14038. [Google Scholar] [CrossRef]
- Matentzoglu, N.; Balhoff, J.P.; Bello, S.M.; Bizon, C.; Brush, M.; Callahan, T.J.; Chute, C.G.; Duncan, W.D.; Evelo, C.T.; Gabriel, D.; et al. SSSOM: The Simple Standard for Sharing Ontology Mappings. Database 2022, 2022, baac035. [Google Scholar] [CrossRef] [PubMed]
- Hier, D.B.; Platt, S.K.; Obafemi-Ajayi, T. Predicting Failures of LLMs to Link Biomedical Ontology Terms to Identifiers. arXiv 2025, arXiv:2509.04458. [Google Scholar] [CrossRef]
- Olasunkanmi, O.; Satursky, M.; Yi, H.; Bizon, C.; Lee, H.; Ahalt, S. RELATE: Relation Extraction in Biomedical Abstracts with LLMs and Ontology Constraints. arXiv 2025, arXiv:2509.19057. [Google Scholar] [CrossRef]
- Ronzano, F.; Nanavati, J. Towards Ontology-Enhanced Representation Learning for LLMs. arXiv 2024, arXiv:2405.20527. [Google Scholar] [CrossRef]
- Mehenni, A.; Zouaq, A. Ontology-Constrained Generation for Biomedical Summarization and Extraction. arXiv 2024, arXiv:2407.03624. [Google Scholar] [CrossRef]
- Sänger, P.; Leser, U. Knowledge-Augmented Pre-trained Language Models for Biomedical Relation Extraction. Artif. Intell. Med. 2025. ahead of print. [Google Scholar] [CrossRef]
- Liu, Z.; Gan, C.; Wang, J.; Zhang, Y.; Bo, Z.; Sun, M.; Chen, H.; Zhang, W. OntoTune: Ontology-Driven Self-training for Aligning Large Language Models. arXiv 2025, arXiv:2502.05478. [Google Scholar] [CrossRef]
- Song, Y.; Chen, J.; Schmidt, R.A. GenOM: Ontology Matching with Description Generation and Large Language Model. arXiv 2025, arXiv:2508.10703. [Google Scholar] [CrossRef]
- Groza, T.; Caufield, H.; Gration, D.; Baynam, G.; Haendel, M.A.; Robinson, P.N.; Mungall, C.J.; Reese, J.T. An evaluation of GPT models for phenotype concept recognition. BMC Med. Inform. Decis. Mak. 2024, 24, 30. [Google Scholar] [CrossRef]
- O’Neil, S.T.; Schaper, K.; Elsarboukh, G.; Reese, J.T.; Moxon, S.A.; Harris, N.L.; Munoz-Torres, M.C.; Robinson, P.N.; Haendel, M.A.; Mungall, C.J. Phenomics Assistant: An Interface for LLM-based Biomedical Knowledge Graph Exploration. bioRxiv 2024, 2024-01. [Google Scholar] [CrossRef]
- Hamed, A.A.; Lee, B.S. From Knowledge Generation to Knowledge Verification: Examining the BioMedical Generative Capabilities of ChatGPT. arXiv 2025, arXiv:2502.14714. [Google Scholar] [CrossRef] [PubMed]
- Shlyk, D.; Groza, T.; Mesiti, M.; Montanelli, S.; Cavalleri, E. REAL: A retrieval-augmented entity linking approach for biomedical concept recognition. In Proceedings of the 23rd Workshop on Biomedical Natural Language Processing, Association for Computational Linguistics, Bangkok, Thailand, 15 August 2024; pp. 380–389. [Google Scholar]
- Sung, M.; Jeon, H.; Lee, J.; Kang, J. Biomedical entity representations with synonym marginalization. arXiv 2020, arXiv:2005.00239. [Google Scholar] [CrossRef]
- Sakor, A.; Singh, K.; Vidal, M.E. BioLinkerAI: Capturing Knowledge Using LLMs to Enhance Biomedical Entity Linking. In Proceedings of the International Conference on Web Information Systems Engineering, Lisbon, Portugal, 13–15 November 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 262–272. [Google Scholar]
- Shefchek, K.A.; Harris, N.L.; Gargano, M.; Matentzoglu, N.; Unni, D.; Brush, M.; Keith, D.; Conlin, T.; Vasilevsky, N.; Zhang, X.A.; et al. The Monarch Initiative in 2019: An integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020, 48, D704–D715. [Google Scholar] [CrossRef]
- Wang, A.; Liu, C.; Yang, J.; Weng, C. Fine-tuning Large Language Models for Rare Disease Concept Normalization. bioRxiv 2023. [Google Scholar] [CrossRef]
- Isik, E.; Lei, A. The growth of Tate-Shafarevich groups of p-supersingular elliptic curves over anticyclotomic Z_p-extensions at inert primes. arXiv 2024, arXiv:2409.02202. [Google Scholar] [CrossRef]
- Iglesias-Navarro, P.; Huertas-Company, M.; Pérez-González, P.; Knapen, J.H.; Hahn, C.; Koekemoer, A.M.; Finkelstein, S.L.; Villanueva, N.; Ramos, A.A. Simulation-based inference of galaxy properties from JWST pixels. arXiv 2025, arXiv:2506.04336. [Google Scholar] [CrossRef]
- Hier, D.B.; Do, T.S.; Obafemi-Ajayi, T. A simplified retriever to improve accuracy of phenotype normalizations by large language models. Front. Digit. Health 2025, 1495040. [Google Scholar] [CrossRef]
- Soman, K.; Rose, P.W.; Morris, J.H.; Akbas, R.E.; Smith, B.; Peetoom, B.; Villouta-Reyes, C.; Cerono, G.; Shi, Y.; Rizk-Jackson, A.; et al. Biomedical knowledge graph-optimized prompt generation for large language models. Bioinformatics 2024, 40, btae560. [Google Scholar] [CrossRef]
- Callahan, T.J.; Tripodi, I.J.; Stefanski, A.L.; Cappelletti, L.; Taneja, S.B.; Wyrwa, J.M.; Casiraghi, E.; Matentzoglu, N.A.; Reese, J.; Silverstein, J.C.; et al. An open source knowledge graph ecosystem for the life sciences. Sci. Data 2024, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Mavridis, A.; Tegos, S.; Anastasiou, C.; Papoutsoglou, M.; Meditskos, G. Large language models for intelligent RDF knowledge. Front. Artif. Intell. 2025, 8, 1546179. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Tsai, P.W.; Zhuang, Y. Matching biomedical ontologies through adaptive multi-modal multi-objective evolutionary algorithm. Biology 2021, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.; Glen, A.K.; Kvarfordt, L.G.; Womack, F.; Acevedo, L.; Yoon, T.S.; Ma, C.; Flores, V.; Sinha, M.; Chodpathumwan, Y.; et al. RTX-KG2: A system for building a semantically standardized knowledge graph for translational biomedicine. BMC Bioinform. 2022, 23, 400. [Google Scholar] [CrossRef]
- Putman, T.E.; Schaper, K.; Matentzoglu, N.; Rubinetti, V.P.; Alquaddoomi, F.S.; Cox, C.; Caufield, J.H.; Elsarboukh, G.; Gehrke, S.; Hegde, H.; et al. The Monarch Initiative in 2024: An analytic platform integrating phenotypes, genes and diseases across species. Nucleic Acids Res. 2024, 52, D938–D949. [Google Scholar] [CrossRef]
- Kollapally, N.M.; Geller, J.; Keloth, V.K.; He, Z.; Xu, J. Ontology enrichment using a large language model. J. Biomed. Inform. 2025, 19, 104865. [Google Scholar]
- Chen, Q.; Hu, Y.; Peng, X.; Xie, Q.; Jin, Q.; Gilson, A.; Singer, M.B.; Ai, X.; Lai, P.; Wang, Z.; et al. Benchmarking large language models for biomedical natural language processing applications and recommendations. Nat. Commun. 2025, 16, 56989. [Google Scholar] [CrossRef]
- Mainwood, S.; Bhandari, A.; Tyagi, S. Semantic Encoding in Medical LLMs for Vocabulary Standardisation. medRxiv 2025, 2025–2006. [Google Scholar] [CrossRef]
- Arsenyan, J.; Albayrak, S. A Comprehensive Knowledge Graph Creation Approach from EMR Notes. In Proceedings of the IEEE BigData 2024, Miami, FL, USA, 15–18 December 2024. [Google Scholar]
- Yang, H.; Li, J.; Zhang, C.; Sierra, A.P.; Shen, B. SepsisKG: Constructing a Sepsis Knowledge Graph using GPT-4. J. Biomed. Inform. 2025, in press. [Google Scholar]
- Riquelme-García, A.; Mulero-Hernández, J.; Fernández-Breis, J.T. Annotation of biological samples data to standard ontologies with support from large language models. Comput. Struct. Biotechnol. J. 2025, 27, 2155–2167. [Google Scholar] [CrossRef]
- Cavalleri, E.; Cabri, A.; Soto-Gomez, M.; Bonfitto, S.; Perlasca, P.; Gliozzo, J.; Callahan, T.J.; Reese, J.; Robinson, P.N.; Casiraghi, E.; et al. An ontology-based knowledge graph for representing interactions involving RNA molecules. Sci. Data 2024, 11, 906. [Google Scholar] [CrossRef]
- Hegde, H.; Vendetti, J.; Goutte-Gattat, D.; Caufield, J.H.; Graybeal, J.B.; Harris, N.L.; Karam, N.; Kindermann, C.; Matentzoglu, N.; Overton, J.A.; et al. A change language for ontologies and knowledge graphs. Database 2025, 2025, baae133. [Google Scholar] [CrossRef]
- Naseem, U.; Dunn, A.G.; Khushi, M.; Kim, J. Benchmarking for biomedical natural language processing tasks with a domain specific ALBERT. BMC Bioinform. 2022, 23, 144. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, S.; Lu, Z. Transfer learning in biomedical natural language processing: An evaluation of BERT and ELMo on ten benchmarking datasets. arXiv 2019, arXiv:1906.05474. [Google Scholar] [CrossRef]
- Yakimovich, A.; Beaugnon, A.; Huang, Y.; Ozkirimli, E. Labels in a haystack: Approaches beyond supervised learning in biomedical applications. Patterns 2021, 2, 100383. [Google Scholar] [CrossRef] [PubMed]
- Holdcroft, A. Gender bias in research: How does it affect evidence based medicine? J. R. Soc. Med. 2007, 100, 2–3. [Google Scholar] [CrossRef]
- Adamson, A.S.; Smith, A. Machine learning and health care disparities in dermatology. JAMA Dermatol. 2018, 154, 1247–1248. [Google Scholar] [CrossRef]
- Sjoding, M.W.; Dickson, R.P.; Iwashyna, T.J.; Gay, S.E.; Valley, T.S. Racial bias in pulse oximetry measurement. N. Engl. J. Med. 2020, 383, 2477–2478. [Google Scholar] [CrossRef]
- Bender, E.M.; Gebru, T.; McMillan-Major, A.; Shmitchell, S. On the Dangers of Stochastic Parrots: Can Language Models Be Too Big? In Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, Virtual Event, 3–10 March 2021; pp. 610–623. [Google Scholar] [CrossRef]
- Di Palo, F.; Singhi, P.; Fadlallah, B.H. Performance-Guided LLM Knowledge Distillation for Efficient Text Classification at Scale. In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing (EMNLP), Panama City, Panama, 12–16 November 2024; pp. 3675–3687. [Google Scholar] [CrossRef]
- Cross, J.L.; Choma, M.A.; Onofrey, J.A. Bias in medical AI: Implications for clinical decision-making. PLoS Digit. Health 2024, 3, e0000651. [Google Scholar] [CrossRef]
- Jha, K.; Zhang, A. Continual knowledge infusion into pre-trained biomedical language models. Bioinformatics 2022, 38, 494–502. [Google Scholar] [CrossRef]
- Garcia-Barragan, A.; Sakor, A.; Vidal, M.E.; Menasalvas, E.; Gonzalez, J.C.S.; Provencio, M.; Robles, V. NSSC: A neuro-symbolic AI system for enhancing accuracy of named entity recognition and linking from oncologic clinical notes. Med. Biol. Eng. Comput. 2024, 63, 749–772. [Google Scholar] [CrossRef]
- Zhang, B.; Carriero, V.A.; Schreiberhuber, K.; Tsaneva, S.; González, L.S.; Kim, J.; de Berardinis, J. OntoChat: A framework for conversational ontology engineering using language models. In Proceedings of the European Semantic Web Conference, Chalkida, Greece, 26–30 May 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 102–121. [Google Scholar]
- Matentzoglu, N.; Malone, J.; Mungall, C.; Stevens, R. A community-driven roadmap for ontology development in biology. BMC Bioinform. 2018, 19, 155. [Google Scholar]
- Samek, W.; Montavon, G.; Lapuschkin, S.; Anders, C.J.; Müller, K.R. Explaining deep neural networks and beyond: A review of methods and applications. Proc. IEEE 2021, 109, 247–278. [Google Scholar] [CrossRef]
- Pesquita, C.; Ferreira, J.; Couto, F.M. Interactive tools for ontology curation: Challenges and opportunities. J. Biomed. Semant. 2023, 14, 5. [Google Scholar]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]
| Task | Representative LLM Tools/Methods | Strengths | Weaknesses/Limitations |
|---|---|---|---|
| Ontology creation and enrichment | GPT-4, ChatGPT; DRAGON-AI [19]; ontology enrichment pipelines [76] | Draft new terms and definitions; accelerate ontology expansion; capture synonyms from literature | Risk of hallucinated terms or fabricated IDs; needs human validation; limited logical constraint handling |
| Ontology mapping and alignment | MILA [35]; GenOM (2025) [60]; embedding-based alignment methods | Leverage LLM embeddings for cross-ontology matching; capture semantic similarity beyond string overlap | Sensitive to prompt design; error-prone with rare terms; struggles with semantic disambiguation |
| Text mining and semantic search | GPT-3.5/GPT-4 for NER; SPIRES [18]; FuncFetch [13] | Handle biomedical synonyms; flexible extraction from unstructured text; improve recall for retrieval | Struggle with ambiguity (e.g., hypertension vs. hypotension); variable precision; ontology ID grounding remains challenging |
| Ontology alignment with knowledge graphs | RELATE [53]; LLM-assisted RDF generation [72] | Bridge unstructured and structured data; generate candidate triples; support semantic integration | Require symbolic reasoning validation; scalability issues; spurious relations without ontology constraints |
| Curation and interactive editing | Conversational editing prototypes; collaborative LLM–ontology platforms | Reduce curator workload; provide interactive drafts; support human-in-the- loop workflows | Dependence on expert oversight; reproducibility issues; lack of standardized evaluation metrics |
| Method Class | Demonstrated at Scale? | Key Strengths | Key Limitations |
|---|---|---|---|
| Rule-based/Heuristic Matchers (e.g., AML, LogMap) | Yes (production deployments) | High precision, deterministic behavior, easy reproducibility | Low recall for semantically distant terms; limited adaptability to new ontologies; minimal contextual understanding. |
| Hybrid LLM-Assisted Matchers (e.g., MapperGPT, MILA, SPIREX) | Emerging (pilot-scale validation) | Improved recall and contextual sensitivity; curator-in-the-loop validation; scalable through selective prompting | Requires manual oversight; lacks standardized runtime benchmarks; higher compute cost than rule-based systems. |
| Pure LLM-Based Systems (e.g., OntoTune, GenOM prototypes) | Limited (proof-of-concept) | Deep semantic reasoning, cross-domain generalization, ontology-grounded representation learning | Prone to hallucinations; poor reproducibility; logical constraint violations; unsuitable for regulated environments without validation layers. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manda, P. Large Language Models in Bio-Ontology Research: A Review. Bioengineering 2025, 12, 1260. https://doi.org/10.3390/bioengineering12111260
Manda P. Large Language Models in Bio-Ontology Research: A Review. Bioengineering. 2025; 12(11):1260. https://doi.org/10.3390/bioengineering12111260
Chicago/Turabian StyleManda, Prashanti. 2025. "Large Language Models in Bio-Ontology Research: A Review" Bioengineering 12, no. 11: 1260. https://doi.org/10.3390/bioengineering12111260
APA StyleManda, P. (2025). Large Language Models in Bio-Ontology Research: A Review. Bioengineering, 12(11), 1260. https://doi.org/10.3390/bioengineering12111260





