A Multi-Task Ensemble Strategy for Gene Selection and Cancer Classification
Abstract
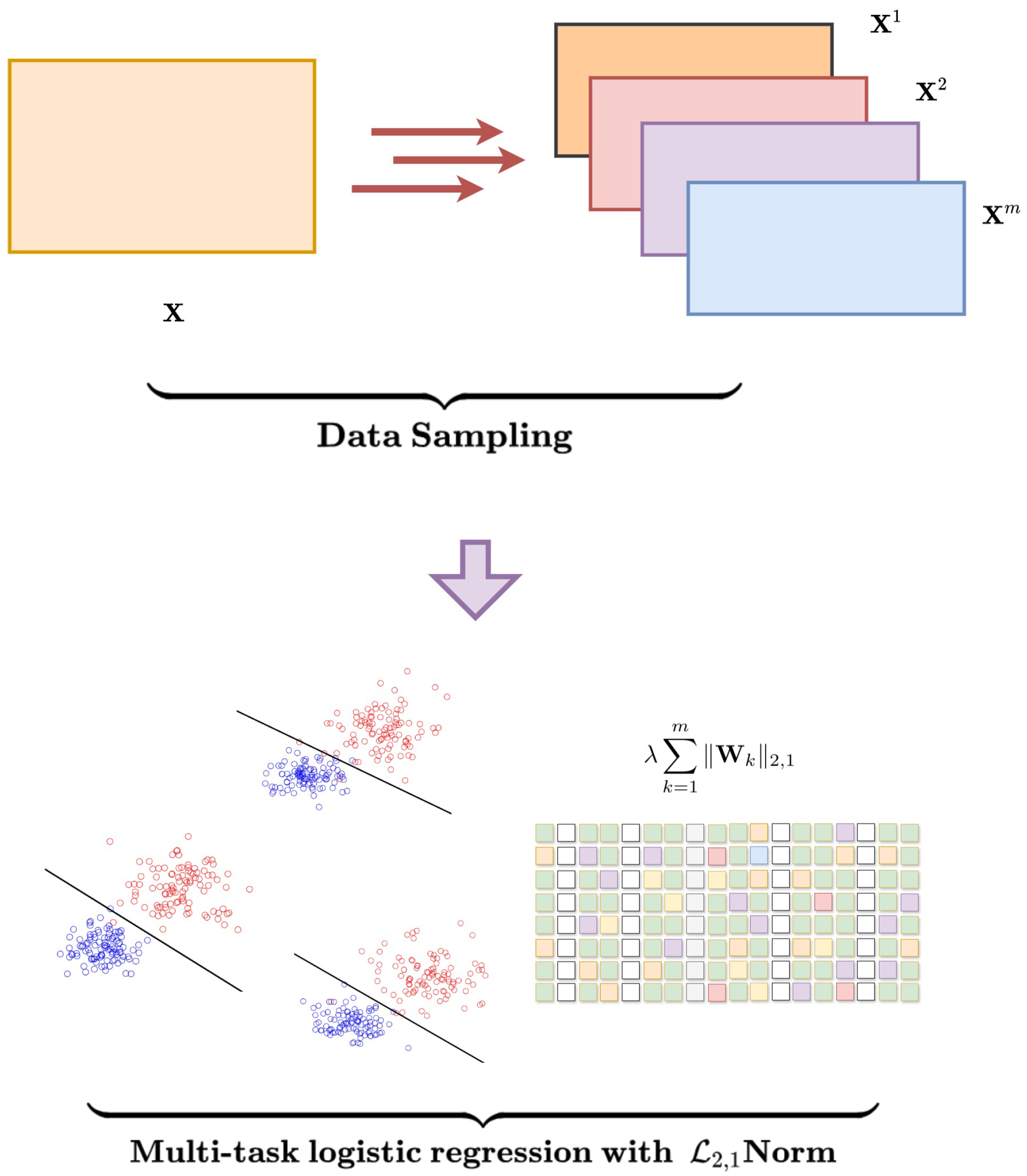
1. Introduction
- We propose a novel ensemble-based framework that systematically enhances gene selection stability by leveraging a sampling approach combined with multi-task learning.
- The use of regularization supports the selection of a compact yet informative gene set across multiple sampling subsets, reducing redundancy and improving model interpretability.
- Experimental results validate the effectiveness of our proposed ensemble approach, demonstrating that it achieves superior performance in terms of classification and gene selection consistency across datasets.
2. Methodology
3. Optimization
| Algorithm 1 Optimization |
|
Computational and Convergence Analysis
- The sequence generated by the proximal gradient descent algorithm satisfies:
- The objective value sequence is non-increasing.
- , where is the global minimum.
- Every limit point of is a minimizer of .
- Since is convex with Lipschitz continuous gradient, we have:
- Combining with the update rule using the proximal operator, we obtain that:
4. Experiment
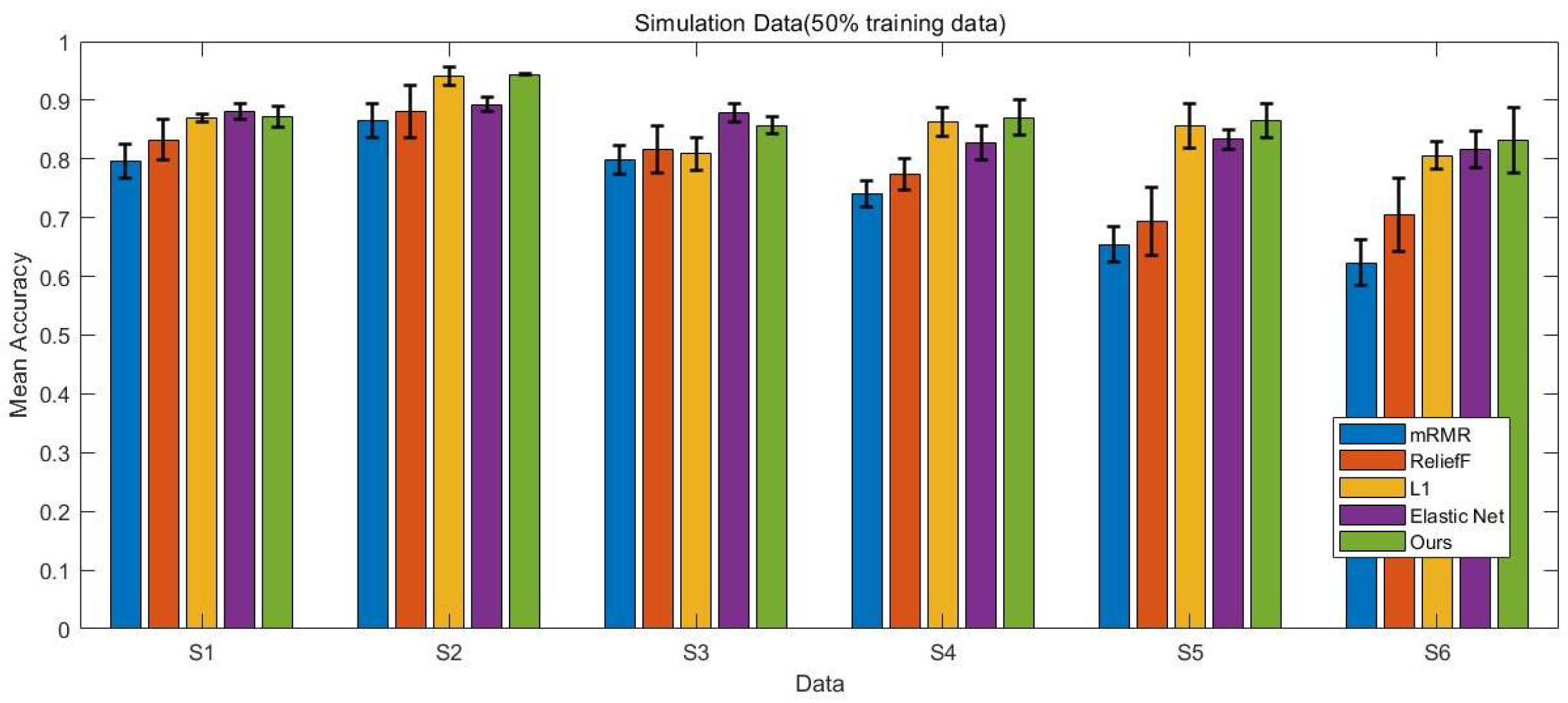
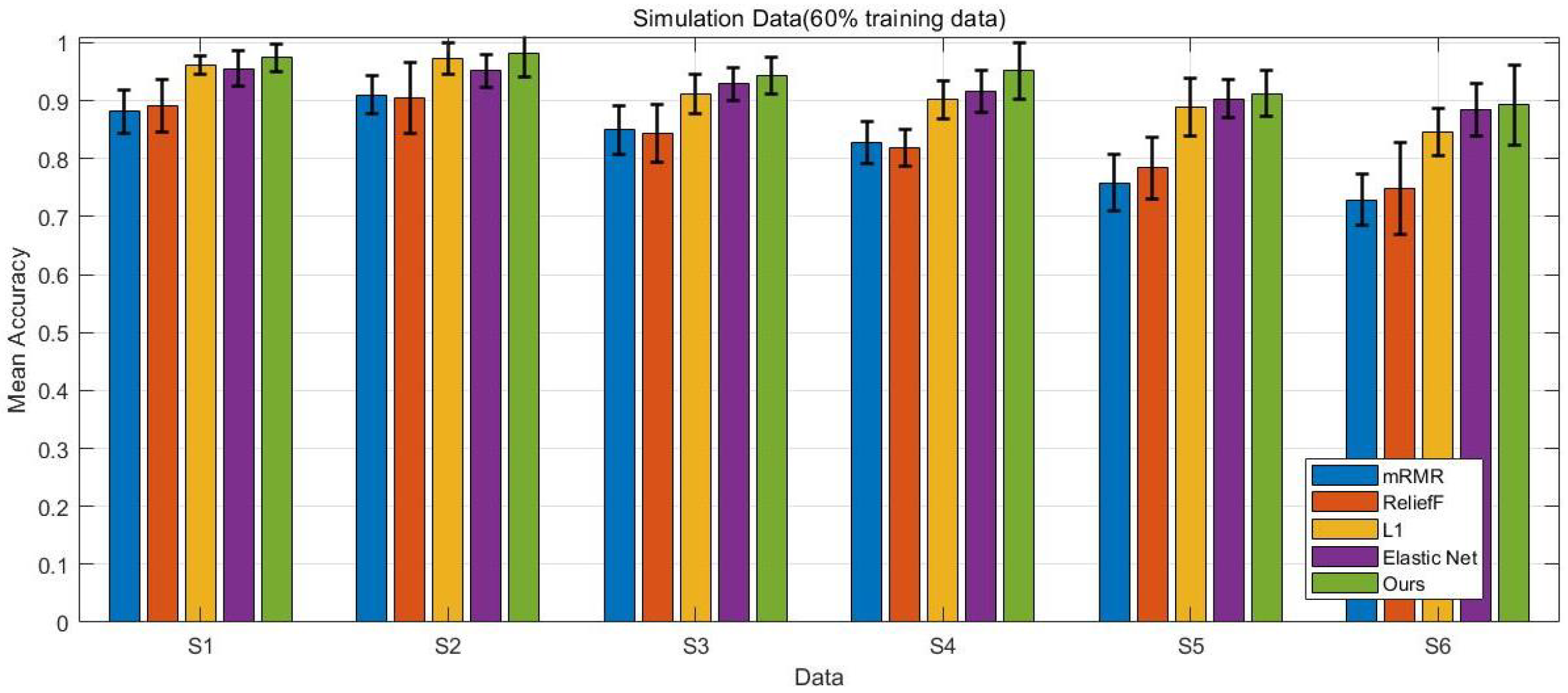
4.1. Simulation Data
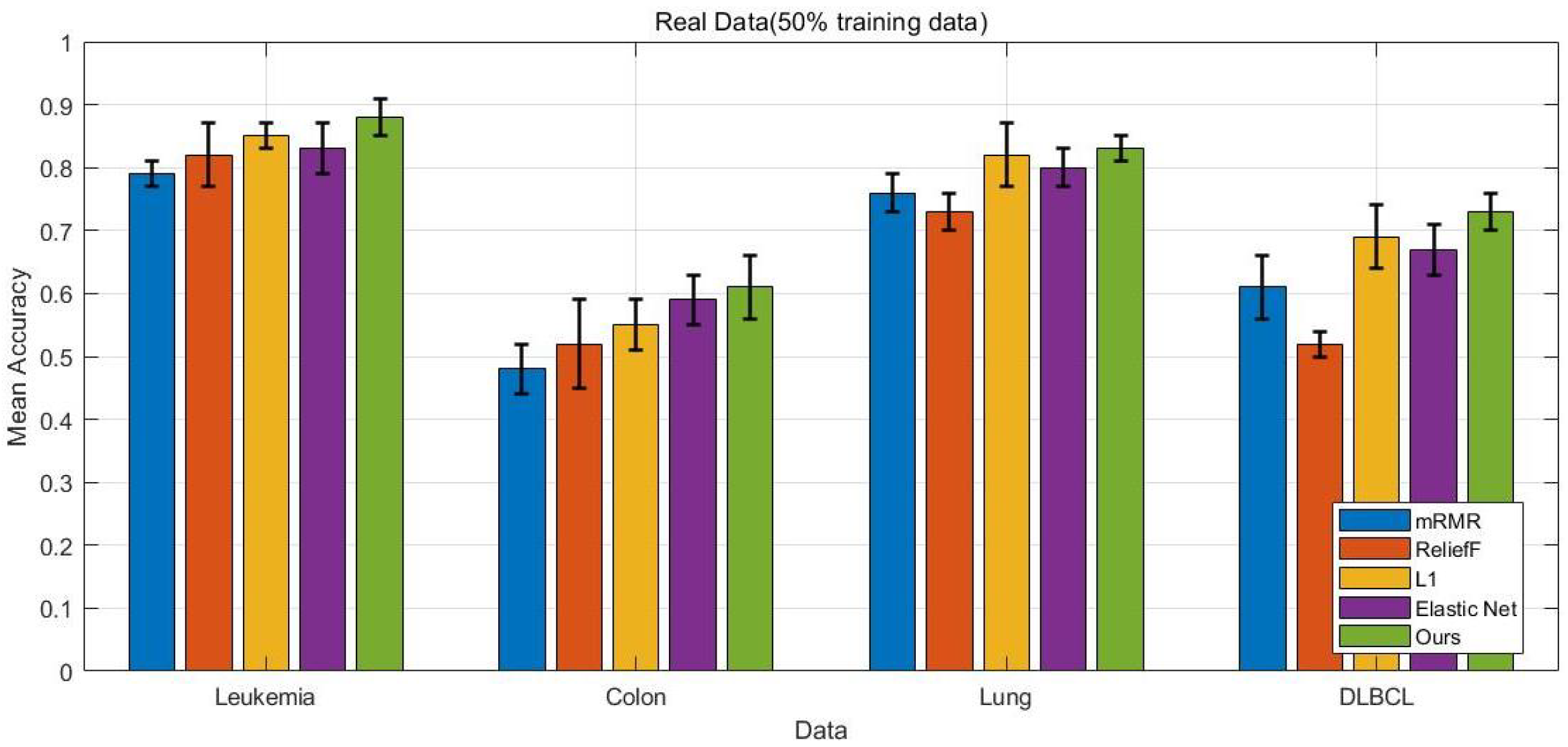
4.2. Real Gene Expression Data
- Leukemia Dataset: The preprocessed Leukemia dataset comprises 3571 genes and 72 samples, including 47 cases of acute lymphoblastic leukemia (ALL) and 25 cases of acute myeloid leukemia (AML).
- Colon Dataset: The Colon microarray dataset contains 2000 genes, with 22 samples from normal tissues and 40 samples from cancerous tissues.
- Lung Dataset: This dataset consists of 12,533 genes across 181 tissue samples, including 31 mesothelioma (MPM) samples and 150 adenocarcinoma (ADCA) samples.
- DLBCL Dataset: The used DLBCL dataset contains 6285 genes and comprises 58 samples of diffuse large B-cell lymphoma (DLBCL) and 19 samples of follicular lymphoma (FL).
4.3. Competing Methods
4.4. Experimental Setup
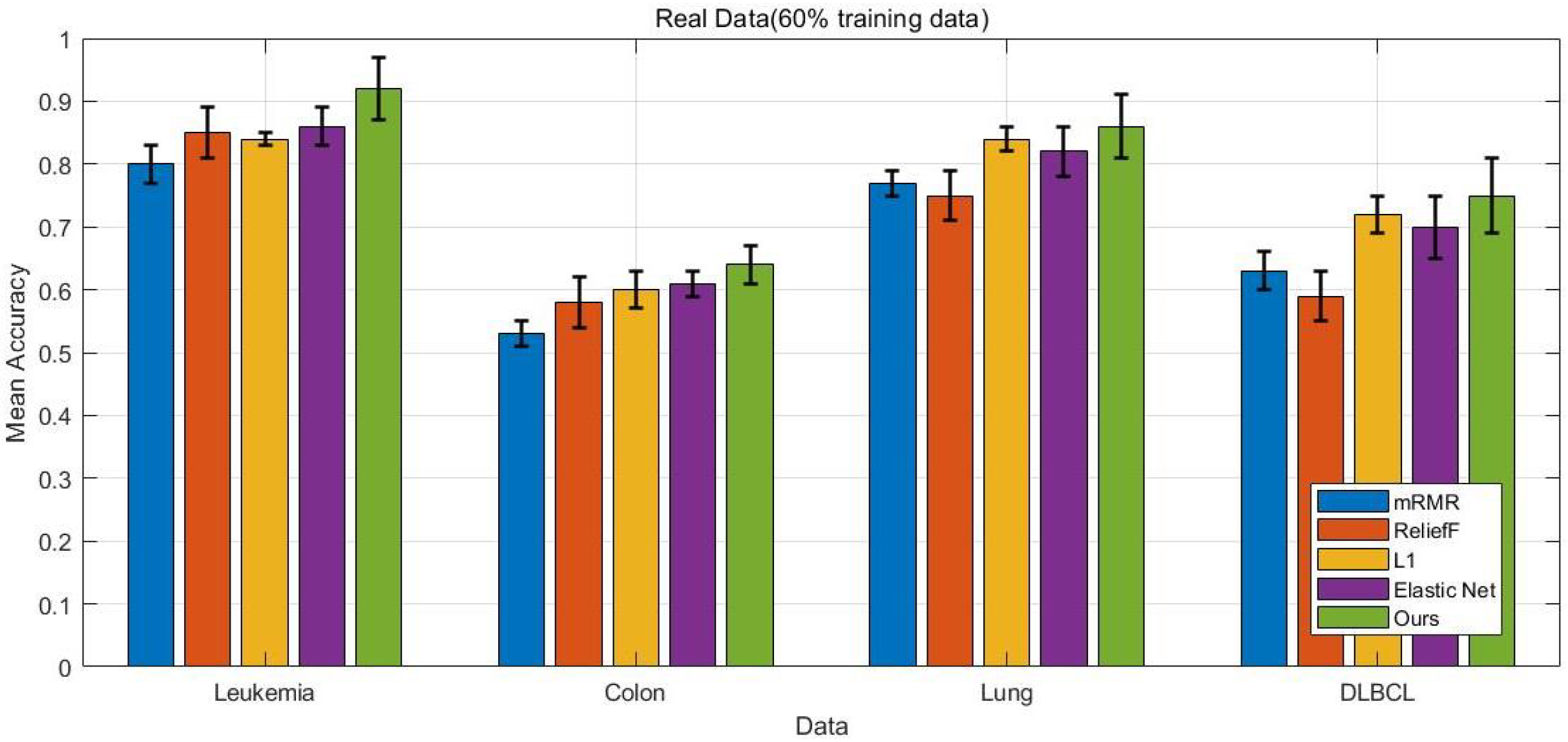
4.5. Comparison Results
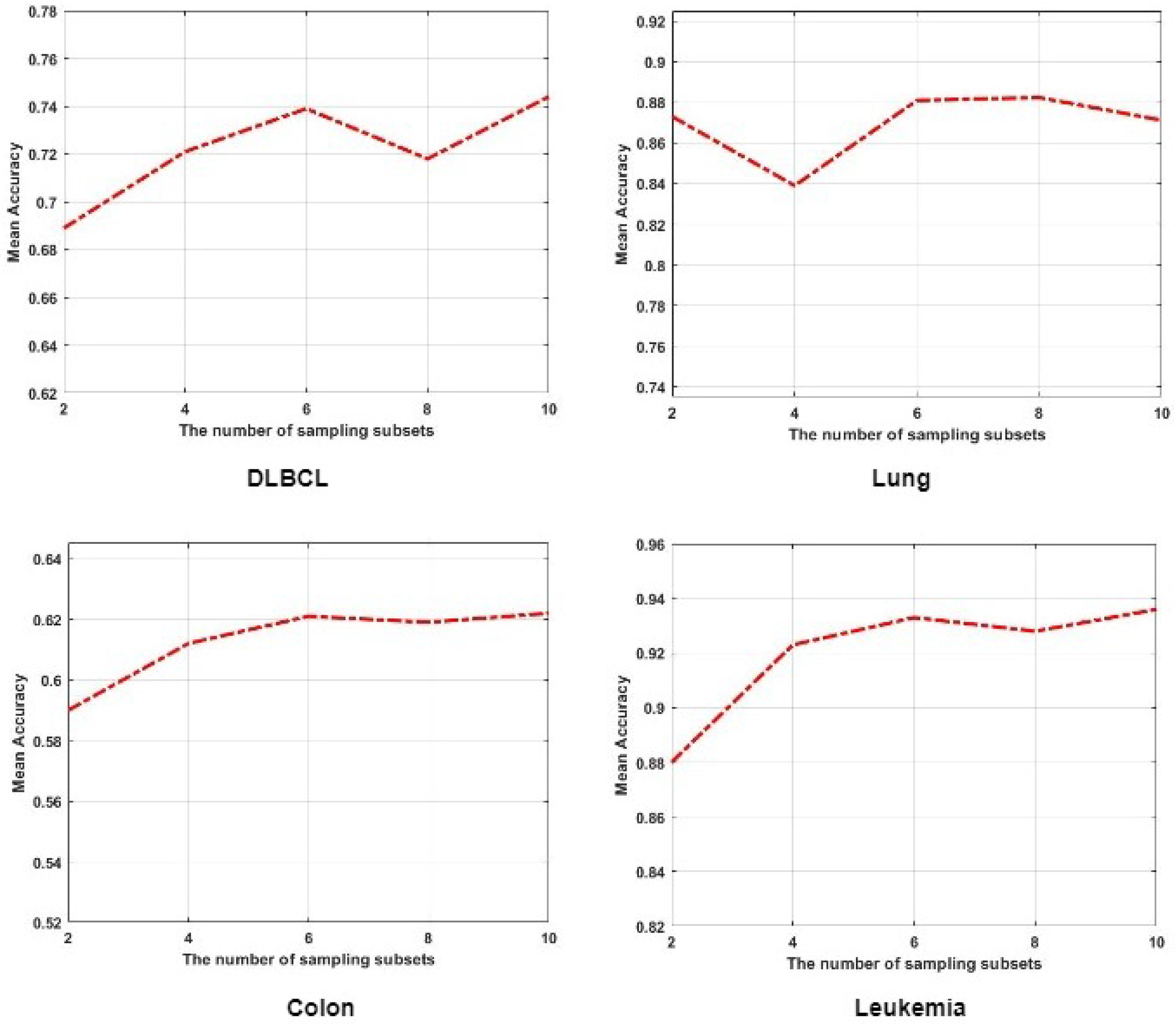
4.6. The Influence of Multi-Task Ensemble Learning Strategy
4.7. The Effectiveness of Gene Selection
4.8. Biological Analysis of the Selected Genes
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdulqader, D.M.; Abdulazeez, A.M.; Zeebaree, D.Q. Machine learning supervised algorithms of gene selection: A review. Mach. Learn. 2020, 62, 233–244. [Google Scholar]
- Pashaei, E.; Pashaei, E. Gene selection using hybrid dragonfly black hole algorithm: A case study on RNA-seq COVID-19 data. Anal. Biochem. 2021, 627, 114242. [Google Scholar] [CrossRef]
- Perscheid, C.; Grasnick, B.; Uflacker, M. Integrative gene selection on gene expression data: Providing biological context to traditional approaches. J. Integr. Bioinform. 2019, 16, 20180064. [Google Scholar] [CrossRef]
- Azadifar, S.; Rostami, M.; Berahmand, K.; Moradi, P.; Oussalah, M. Graph-based relevancy-redundancy gene selection method for cancer diagnosis. Comput. Biol. Med. 2022, 147, 105766. [Google Scholar] [CrossRef]
- Alhenawi, E.; Al-Sayyed, R.; Hudaib, A.; Mirjalili, S. Feature selection methods on gene expression microarray data for cancer classification: A systematic review. Comput. Biol. Med. 2022, 140, 105051. [Google Scholar] [CrossRef]
- Alomari, O.A.; Makhadmeh, S.N.; Al-Betar, M.A.; Alyasseri, Z.A.A.; Doush, I.A.; Abasi, A.K.; Awadallah, M.A.; Zitar, R.A. Gene selection for microarray data classification based on gray wolf optimizer enhanced with TRIZ-inspired operators. Knowl.-Based Syst. 2021, 223, 107034. [Google Scholar] [CrossRef]
- Khurma, R.; Castillo, P.; Sharieh, A.; Aljarah, I. New fitness functions in binary Harris Hawks optimization for gene selection in microarray datasets. In Proceedings of the 12th International Joint Conference on Computer Intelligence, Budapest, Hungary, 2–4 November 2020; pp. 139–146. [Google Scholar]
- Sánchez-Maroño, N.; Alonso-Betanzos, A.; Tombilla-Sanromán, M. Filter methods for feature selection: A comparative study. In Proceedings of the International Conference on Intelligent Data Engineering and Automated Learning, Birmingham, UK, 16–19 December 2007; pp. 178–187. [Google Scholar]
- Zahoor, J.; Zafar, K. Classification of microarray gene expression data using an infiltration tactics optimization (ITO) algorithm. Genes 2020, 11, 819. [Google Scholar] [CrossRef]
- Kononenko, I. Estimating attributes: Analysis and extensions of RELIEF. In Proceedings of the European Conference on Machine Learning, Catania, Italy, 6–8 April 1994; pp. 171–182. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, C.; Li, T. Gene selection algorithm by combining reliefF and mRMR. BMC Genom. 2008, 9, S27. [Google Scholar] [CrossRef] [PubMed]
- Omuya, E.O.; Okeyo, G.O.; Kimwele, M.W. Feature selection for classification using principal component analysis and information gain. Expert Syst. Appl. 2021, 174, 114765. [Google Scholar] [CrossRef]
- Gong, H.; Li, Y.; Zhang, J.; Zhang, B.; Wang, X. A new filter feature selection algorithm for classification task by ensembling pearson correlation coefficient and mutual information. Eng. Appl. Artif. Intell. 2024, 131, 107865. [Google Scholar] [CrossRef]
- Ali, L.; Leung, M.-F.; Khan, M.A.; Nour, R.; Imrana, Y.; Vasilakos, A.V. Chiga-Net: A genetically optimized neural network with refined deeply extracted features using χ2 statistical score for trustworthy Parkinson’s disease detection. Neurocomputing 2025, 624, 129450. [Google Scholar] [CrossRef]
- El Aboudi, N.; Benhlima, L. Review on wrapper feature selection approaches. In Proceedings of the 2016 International Conference on Engineering & MIS (ICEMIS), Agadir, Morocco, 22–24 September 2016; pp. 1–5. [Google Scholar]
- Ghosh, M.; Begum, S.; Sarkar, R.; Chakraborty, D.; Maulik, U. Recursive memetic algorithm for gene selection in microarray data. Expert Syst. Appl. 2019, 116, 172–185. [Google Scholar] [CrossRef]
- Sahebi, G.; Movahedi, P.; Ebrahimi, M.; Pahikkala, T.; Plosila, J.; Tenhunen, H. GeFeS: A generalized wrapper feature selection approach for optimizing classification performance. Comput. Biol. Med. 2020, 125, 103974. [Google Scholar] [CrossRef]
- Hassan, A.; Paik, J.H.; Khare, S.R.; Hassan, S.A. A wrapper feature selection approach using Markov blankets. Pattern Recognit. 2025, 158, 111069. [Google Scholar] [CrossRef]
- Sağbaş, E.A. A novel two-stage wrapper feature selection approach based on greedy search for text sentiment classification. Neurocomputing 2024, 590, 127729. [Google Scholar] [CrossRef]
- Sun, L.; Kong, X.; Xu, J.; Zhai, Z.; Xue, R.; Zhang, S. A hybrid gene selection method based on ReliefF and ant colony optimization algorithm for tumor classification. Sci. Rep. 2019, 9, 8978. [Google Scholar] [CrossRef]
- Hu, P.; Zhu, J. A filter-wrapper model for high-dimensional feature selection based on evolutionary computation. Appl. Intell. 2025, 55, 581. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, C.; Luan, X.Z.; Leung, K.S.; Chan, T.M.; Xu, Z.; Zhang, H. Sparse logistic regression with a L1/2 penalty for gene selection in cancer classification. BMC Bioinform. 2013, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 2008, 24, 1175–1182. [Google Scholar] [CrossRef]
- Algamal, Z.Y.; Lee, M.H. Regularized logistic regression with adjusted adaptive elastic net for gene selection in high dimensional cancer classification. Comput. Biol. Med. 2015, 67, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liang, Y.; Luan, X.Z.; Leung, K.S.; Chan, T.M.; Xu, Z.; Zhang, H. The L1/2 regularization method for variable selection in the Cox model. Appl. Soft Comput. 2014, 14, 498–503. [Google Scholar] [CrossRef]
- Saberi-Movahed, F.; Rostami, M.; Berahmand, K.; Karami, S.; Tiwari, P.; Oussalah, M.; Band, S.S. Dual regularized unsupervised feature selection based on matrix factorization and minimum redundancy with application in gene selection. Knowl.-Based Syst. 2022, 256, 109884. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.-G.; Lu, Y. Informative gene selection for microarray classification via adaptive elastic net with conditional mutual information. Appl. Math. Model. 2019, 71, 286–297. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L.C. Gene selection in cancer classification using sparse logistic regression with Bayesian regularization. Bioinformatics 2006, 22, 2348–2355. [Google Scholar] [CrossRef]
- Xing, Z.; Zhu, W.; Tang, C.; Wang, M. Gene selection for microarray data classification via adaptive hypergraph embedded dictionary learning. Gene 2019, 706, 188–200. [Google Scholar] [CrossRef]
- Liu, C.; Wong, H.S. Structured penalized logistic regression for gene selection in gene expression data analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 16, 312–321. [Google Scholar] [CrossRef]
- Sharma, A.; Lysenko, A.; Boroevich, K.A.; Vans, E.; Tsunoda, T. DeepFeature: Feature selection in nonimage data using convolutional neural network. Brief. Bioinform. 2021, 22, bbab297. [Google Scholar] [CrossRef]
- Zeebaree, D.Q.; Haron, H.; Abdulazeez, A.M.; Zebari, D.A. Trainable model based on new uniform LBP feature to identify the risk of the breast cancer. In Proceedings of the International Conference on Advanced Science and Engineering (ICOASE), Zakho, Iraq, 2–4 April 2019; pp. 106–111. [Google Scholar]
- Eluri, N.R.; Kancharla, G.R.; Dara, S.; Dondeti, V. Cancer data classification by quantum-inspired immune clone optimization-based optimal feature selection using gene expression data: Deep learning approach. Data Technol. Appl. 2022, 56, 247–282. [Google Scholar] [CrossRef]
- Rukhsar, L.; Bangyal, W.H.; Khan, M.S.A.; Ibrahim, A.A.A.; Nisar, K.; Rawat, D.B. Analyzing RNA-seq gene expression data using deep learning approaches for cancer classification. Appl. Sci. 2022, 12, 1850. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, W.; Li, W.; Wang, L.; Feng, C. GAMB-GNN: Graph neural networks learning from gene structure relations and Markov blanket ranking for cancer classification in microarray data. Chemom. Intell. Lab. Syst. 2023, 232, 104713. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shorfuzzaman, M. Early Diabetic Retinopathy Cyber-Physical Detection System Using Attention-Guided Deep CNN Fusion. IEEE Trans. Netw. Sci. Eng. 2025, 12, 1898–1910. [Google Scholar] [CrossRef]
- Song, Y.; Liu, Y.; Lin, Z.; Zhou, J.; Li, D.; Zhou, T.; Leung, M.-F. Learning from AI-generated annotations for medical image segmentation. IEEE Trans. Consum. Electron. 2025, 71, 1473–1481. [Google Scholar] [CrossRef]
- Li, Y.; Hao, W.; Zeng, H.; Wang, L.; Xu, J.; Routray, S.; Jhaveri, R.H.; Gadekallu, T.R. Cross-Scale Texture Supplementation for Reference-based Medical Image Super-Resolution. IEEE J. Biomed. Health Inform. (Early Access) 2021, 1–15. [Google Scholar] [CrossRef]
- Wang, L.; Cai, J.; Wang, T.; Zhao, J.; Gadekallu, T.R.; Fang, K. Pine wilt disease detection based on uav remote sensing with an improved yolo model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 19230–19242. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, H.; Hossain, M.S.; Chen, Y.; Wang, H.; Yin, Y. Scaled Background Swap: Video Augmentation for Action Quality Assessment with Background Debiasing. ACM Trans. Multimed. Comput. Commun. Appl. 2025, 21, 1–18. [Google Scholar] [CrossRef]
- Li, G.; Wang, Z.; Sun, J.; Zhang, Q. Objective Extraction for Simplifying Many-Objective Solution Sets. IEEE Trans. Emerg. Top. Comput. Intell. 2024, 8, 337–349. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Q. Multitask Feature Selection for Objective Reduction. In Proceedings of the International Conference on Evolutionary Multi-Criterion Optimization, Shenzhen, China, 28–31 March 2021; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2021; Volume 12654, pp. 77–88. [Google Scholar]
- Nagra, A.A.; Khan, A.H.; Abubakar, M.; Faheem, M.; Rasool, A.; Masood, K.; Hussain, M. A gene selection algorithm for microarray cancer classification using an improved particle swarm optimization. Sci. Rep. 2024, 14, 19613. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, T.M.; Kecman, V. Gene extraction for cancer diagnosis by support vector machines. Int. Conf. Artif. Neural Netw. 2005, 35, 185–194. [Google Scholar]
- Han, F.; Tang, D.; Sun, Y.W.T.; Cheng, Z.; Jiang, J.; Li, Q.W. A hybrid gene selection method based on gene scoring strategy and improved particle swarm optimization. BMC Bioinform. 2019, 20 (Suppl. S8), 289. [Google Scholar] [CrossRef] [PubMed]
| Dataset | Sample Size | Feature Dimension |
|---|---|---|
| S1 | 100 | 500 |
| S2 | 200 | 500 |
| S3 | 100 | 1000 |
| S4 | 200 | 1000 |
| S5 | 100 | 2000 |
| S6 | 200 | 2000 |
| Dataset | Sample Size | Feature Dimension |
|---|---|---|
| Leukemia | 72 | 7129 |
| Colon | 62 | 2000 |
| Lung | 181 | 12,533 |
| DLBCL | 77 | 6258 |
| Dataset | Elastic Net | mRMR | ReliefF | Ours | |
|---|---|---|---|---|---|
| S1 | 12.3/21.6 | 16.6/28.9 | 14.2/50 | 11.8/50 | 18.1/27.2 |
| S2 | 17.6/22.9 | 18.3/36.1 | 13.1/50 | 13.9/50 | 19.1/30.2 |
| S3 | 11.4/27.6 | 15.3/32.4 | 14.9/50 | 12.4/50 | 17.8/26.4 |
| S4 | 15.9/28.2 | 16.8/34.6 | 14.4/50 | 14.1/50 | 18.6/31.2 |
| S5 | 12.1/35.1 | 14.8/52.7 | 12.1/50 | 10.9/50 | 17.8/30.4 |
| S6 | 14.6/34.4 | 14.9/59.9 | 13.7/50 | 11.8/50 | 18.3/28.9 |
| Dataset | Gene ID | Description |
|---|---|---|
| Colon | H06524 | Gelsolin precursor, plasma. |
| T92451 | Tropomyosin, fibroblast and epithelial muscle-type. | |
| H20709 | Myosin light chain alkali, smooth-muscle isoform. | |
| T94579 | Human chitotriosidase precursor. | |
| R88740 | ATP synthase coupling factor 6, mitochondrial precursor (Human). | |
| Leukemia | X62654 | ME491 gene extracted from Homo sapiens antigen. |
| M23197 | CD33 antigen. | |
| M63138 | Cathepsin D, lysosomal aspartyl protease. | |
| Y00787 | Glutathione peroxidase 1. | |
| U05259 | MB-1 gene. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Lin, Z.; Zhang, J.; Leung, M.-F. A Multi-Task Ensemble Strategy for Gene Selection and Cancer Classification. Bioengineering 2025, 12, 1245. https://doi.org/10.3390/bioengineering12111245
Lin S, Lin Z, Zhang J, Leung M-F. A Multi-Task Ensemble Strategy for Gene Selection and Cancer Classification. Bioengineering. 2025; 12(11):1245. https://doi.org/10.3390/bioengineering12111245
Chicago/Turabian StyleLin, Suli, Zhizhe Lin, Jin Zhang, and Man-Fai Leung. 2025. "A Multi-Task Ensemble Strategy for Gene Selection and Cancer Classification" Bioengineering 12, no. 11: 1245. https://doi.org/10.3390/bioengineering12111245
APA StyleLin, S., Lin, Z., Zhang, J., & Leung, M.-F. (2025). A Multi-Task Ensemble Strategy for Gene Selection and Cancer Classification. Bioengineering, 12(11), 1245. https://doi.org/10.3390/bioengineering12111245








