Peripheral Blood TCR Clonotype Diversity as a Biomarker for Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Bioinformatics and Statistical Analyses for the TCR Repertoire
2.3. Supervised Machine Learning
2.4. Feature Importance Measures
3. Results
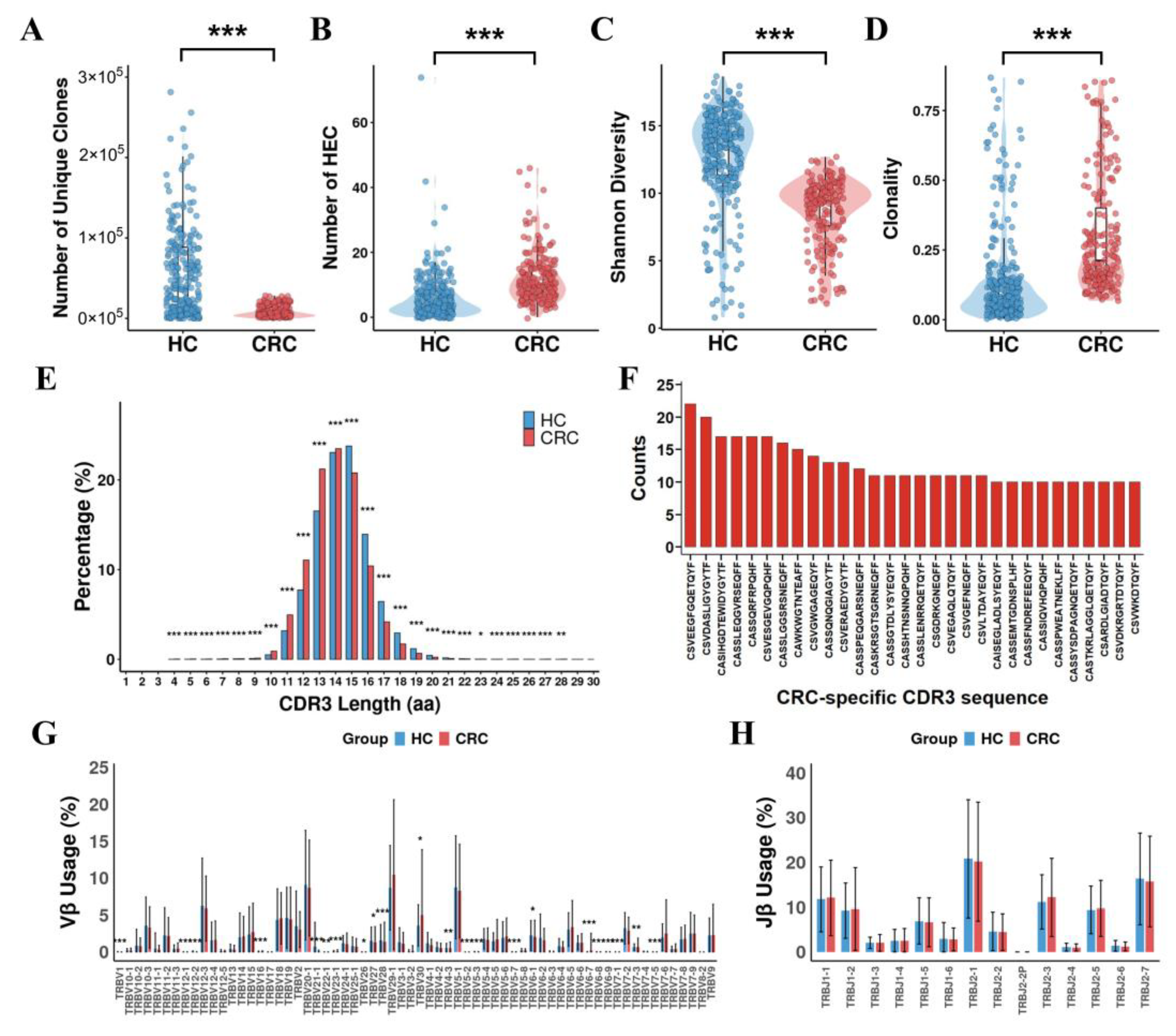
3.1. Comparison of TCR Repertoires Between CRC Patients and Healthy Controls
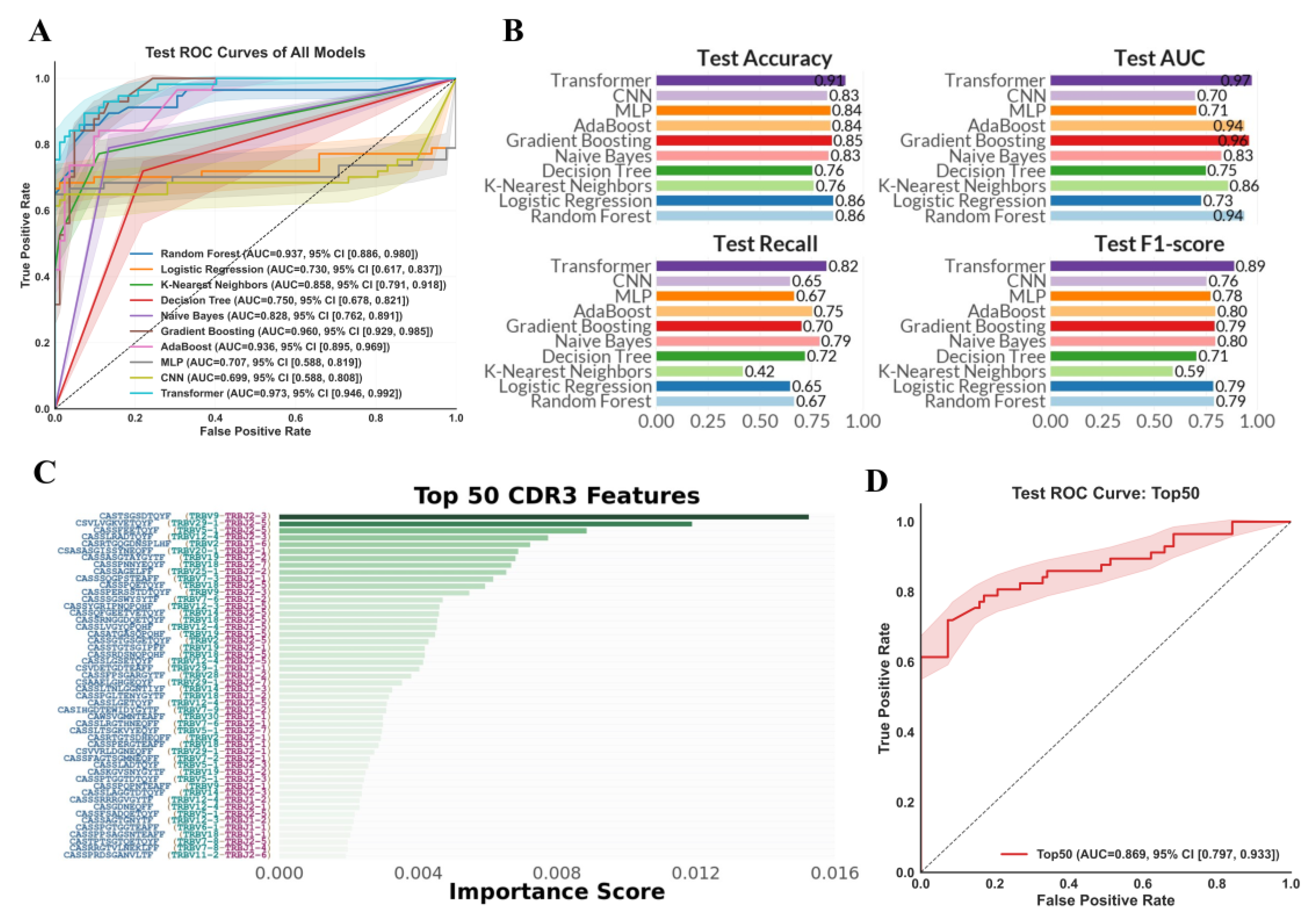
3.2. Performance of TCR Repertoire-Based ML Models for CRC Diagnosis
3.3. Identification of Key TCR Repertoire Biomarkers
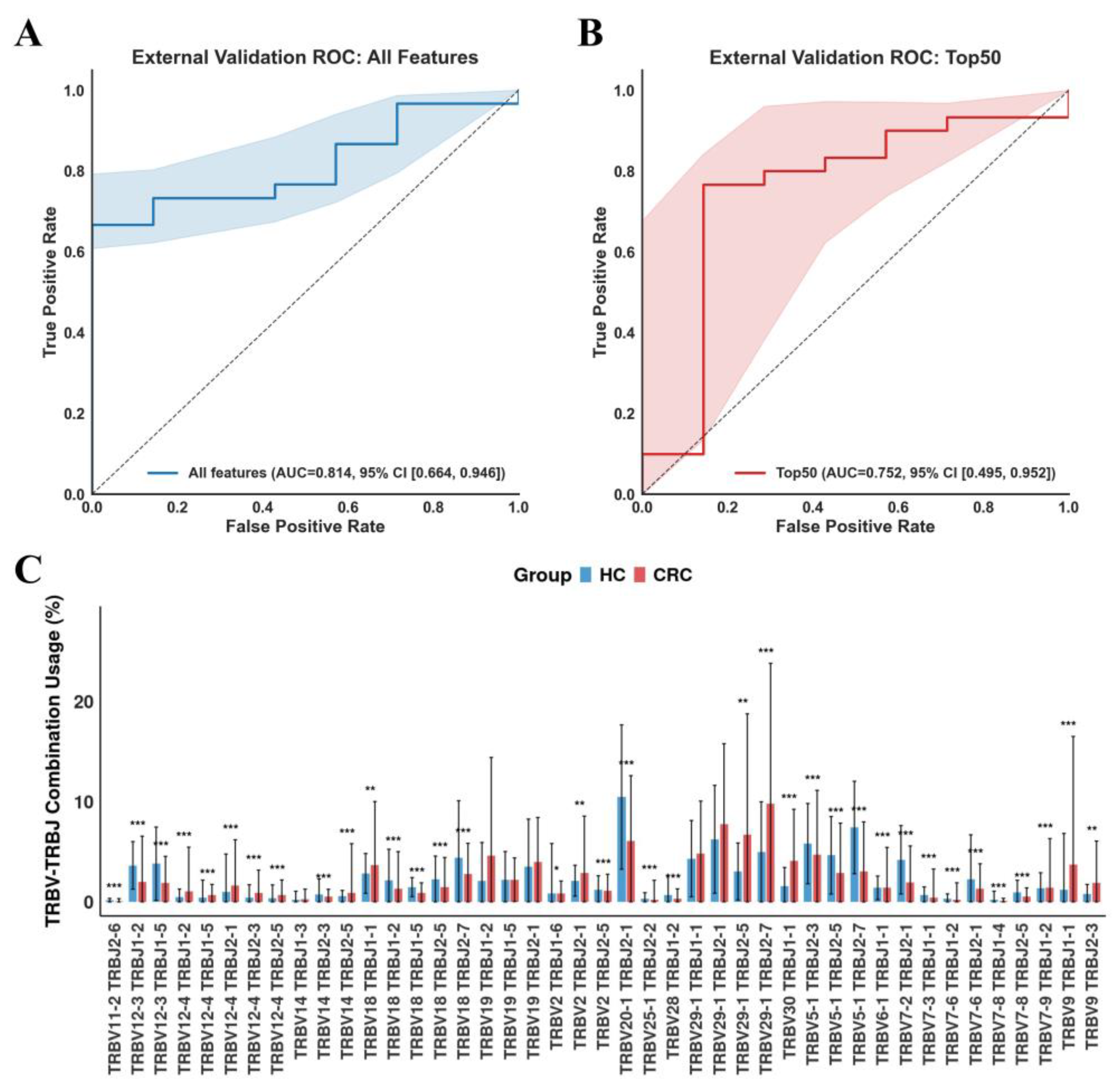
3.4. Validation of TCR Repertoire-Based Models for CRC Diagnosis in Independent Cohorts
4. Discussion
4.1. Peripheral Blood TCR Repertoire Profiling Enables Accurate, Non-Invasive CRC Diagnosis
4.2. Key TCR Repertoire Features Underpin Robust Performance
4.3. Clinical Implications, Limitations, and Future Directions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. Przegląd Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Bell, C.C.; Gilan, O. Principles and mechanisms of non-genetic resistance in cancer. Br. J. Cancer 2020, 122, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Piper, M.A.; Perdue, L.A.; Rutter, C.M.; Webber, E.M.; O’Connor, E.; Smith, N.; Whitlock, E.P. Screening for colorectal cancer: Updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 2016, 315, 2576–2594, Erratum in JAMA 2021, 325, 1978–1998. [Google Scholar] [CrossRef]
- Jia, Q.; Zhou, J.; Chen, G.; Shi, Y.; Yu, H.; Guan, P.; Lin, R.; Jiang, N.; Yu, P.; Li, Q.-J. Diversity index of mucosal resident T lymphocyte repertoire predicts clinical prognosis in gastric cancer. Oncoimmunology 2015, 4, e1001230. [Google Scholar] [CrossRef]
- Han, Y.; Liu, X.; Wang, Y.; Wu, X.; Guan, Y.; Li, H.; Chen, X.; Zhou, B.; Yuan, Q.; Ou, Y. Identification of characteristic TRB V usage in HBV-associated HCC by using differential expression profiling analysis. Oncoimmunology 2015, 4, e1021537. [Google Scholar] [CrossRef]
- Postow, M.A.; Manuel, M.; Wong, P.; Yuan, J.; Dong, Z.; Liu, C.; Perez, S.; Tanneau, I.; Noel, M.; Courtier, A. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J. Immunother. Cancer 2015, 3, 23. [Google Scholar] [CrossRef]
- Rahman, S.F.; Olm, M.R.; Morowitz, M.J.; Banfield, J.F. Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. MSystems 2018, 3, e00123-17. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Liu, Q.; Lau, R.I.; Zhang, J.; Xu, Z.; Yeoh, Y.K.; Leung, T.W.; Tang, W.; Zhang, L.; Liang, J.Q. Faecal microbiome-based machine learning for multi-class disease diagnosis. Nat. Commun. 2022, 13, 6818. [Google Scholar] [CrossRef]
- Manandhar, I.; Alimadadi, A.; Aryal, S.; Munroe, P.B.; Joe, B.; Cheng, X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G328–G337. [Google Scholar] [CrossRef]
- Katayama, Y.; Yokota, R.; Akiyama, T.; Kobayashi, T.J. Machine learning approaches to TCR repertoire analysis. Front. Immunol. 2022, 13, 858057. [Google Scholar] [CrossRef]
- Ostmeyer, J.; Christley, S.; Rounds, W.H.; Toby, I.; Greenberg, B.M.; Monson, N.L.; Cowell, L.G. Statistical classifiers for diagnosing disease from immune repertoires: A case study using multiple sclerosis. BMC Bioinform. 2017, 18, 401. [Google Scholar] [CrossRef]
- Beshnova, D.; Ye, J.; Onabolu, O.; Moon, B.; Zheng, W.; Fu, Y.-X.; Brugarolas, J.; Lea, J.; Li, B. De novo prediction of cancer-associated T cell receptors for noninvasive cancer detection. Sci. Transl. Med. 2020, 12, eaaz3738. [Google Scholar] [CrossRef]
- Xu, Y.; Qian, X.; Zhang, X.; Lai, X.; Liu, Y.; Wang, J. DeepLION: Deep multi-instance learning improves the prediction of cancer-associated T cell receptors for accurate cancer detection. Front. Genet. 2022, 13, 860510. [Google Scholar] [CrossRef]
- Sidhom, J.-W.; Oliveira, G.; Ross-MacDonald, P.; Wind-Rotolo, M.; Wu, C.J.; Pardoll, D.M.; Baras, A.S. Deep learning reveals predictive sequence concepts within immune repertoires to immunotherapy. Sci. Adv. 2022, 8, eabq5089. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Hsu, H.-C.; Lee, Y.-S.; Liu, H.; Tan, B.C.-M.; Chin, C.-Y.; Chang, I.Y.-F.; Yang, C.-Y. Longitudinal high-throughput sequencing of the T-cell receptor repertoire reveals dynamic change and prognostic significance of peripheral blood TCR diversity in metastatic colorectal cancer during chemotherapy. Front. Immunol. 2022, 12, 743448. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Hou, W.; Ding, Y.; Zhu, Y.; Zheng, J.; Huang, Q.; Cao, Z.; Xie, R.; Wei, Q. Colorectal cancer–associated T cell receptor repertoire abnormalities are linked to gut microbiome shifts and somatic cell mutations. Gut Microbes 2023, 15, 2263934. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.N.; Wang, H.; Jiang, R.; Wang, F.; Chen, C.; Xu, Z.; Xiao, R. Characterization of the T-cell receptor repertoire associated with lymph node metastasis in colorectal cancer. Front. Oncol. 2024, 14, 1354533. [Google Scholar] [CrossRef]
- Malik, A.; Sayed, A.A.; Han, P.; Tan, M.M.; Watt, E.; Constantinescu-Bercu, A.; Cocker, A.T.; Khoder, A.; Saputil, R.C.; Thorley, E. The role of CD8+ T-cell clones in immune thrombocytopenia. Blood J. Am. Soc. Hematol. 2023, 141, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Martini, G.R.; Pogorelyy, M.V.; Minervina, A.A.; Degenhardt, F.; Wendorff, M.; Sari, S.; Mayr, G.; Fazio, A.; Dowds, C.M. A novel unconventional T cell population enriched in Crohn’s disease. Gut 2022, 71, 2194–2204. [Google Scholar] [CrossRef]
- Von Niederhäusern, V.; Ghraichy, M.; Trück, J. Applicability of T cell receptor repertoire sequencing analysis to unbalanced clinical samples–comparing the T cell receptor repertoire of GATA2 deficient patients and healthy controls. Swiss Med. Wkly. 2023, 153, 40046. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2, lqaa078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Highly effective batch effect correction method for RNA-seq count data. Comput. Struct. Biotechnol. J. 2025, 27, 58–64. [Google Scholar] [CrossRef]
- Zhang, K.; Erkan, E.P.; Jamalzadeh, S.; Dai, J.; Andersson, N.; Kaipio, K.; Lamminen, T.; Mansuri, N.; Huhtinen, K.; Carpén, O. Longitudinal single-cell RNA-seq analysis reveals stress-promoted chemoresistance in metastatic ovarian cancer. Sci. Adv. 2022, 8, eabm1831. [Google Scholar] [CrossRef] [PubMed]
- Gurun, B.; Horton, W.; Murugan, D.; Zhu, B.; Leyshock, P.; Kumar, S.; Byrne, K.T.; Vonderheide, R.H.; Margolin, A.A.; Mori, M. An open protocol for modeling T Cell Clonotype repertoires using TCRβ CDR3 sequences. BMC Genom. 2023, 24, 349. [Google Scholar] [CrossRef] [PubMed]
- Lun, A.T.; Calero-Nieto, F.J.; Haim-Vilmovsky, L.; Göttgens, B.; Marioni, J.C. Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Res. 2017, 27, 1795–1806. [Google Scholar] [CrossRef]
- Qi, Y. Random forest for bioinformatics. In Ensemble Machine Learning: Methods and Applications; Springer: Berlin/Heidelberg, Germany, 2012; pp. 307–323. [Google Scholar]
- Uddin, S.; Khan, A.; Hossain, M.E.; Moni, M.A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 281. [Google Scholar] [CrossRef]
- Bansal, M.; Goyal, A.; Choudhary, A. A comparative analysis of K-nearest neighbor, genetic, support vector machine, decision tree, and long short term memory algorithms in machine learning. Decis. Anal. J. 2022, 3, 100071. [Google Scholar] [CrossRef]
- Charbuty, B.; Abdulazeez, A. Classification based on decision tree algorithm for machine learning. J. Appl. Sci. Technol. Trends 2021, 2, 20–28. [Google Scholar] [CrossRef]
- Boyko, N.; Boksho, K. Application of the naive bayesian classifier in work on sentimental analysis of medical data. In Proceedings of the IDDM’2020: 3rd International Conference on Informatics & Data-Driven Medicine, Växjö, Sweden, 19–21 November 2020. [Google Scholar]
- Ning, Y.; Zhang, S.; Nie, X.; Li, G.; Zhao, G. Fall detection algorithm based on gradient boosting decision tree. In Proceedings of the 2019 IEEE International Conference on Signal Processing, Communications and Computing (ICSPCC), Dalian, China, 20–22 September 2019. [Google Scholar]
- Ding, Y.; Zhu, H.; Chen, R.; Li, R. An efficient AdaBoost algorithm with the multiple thresholds classification. Appl. Sci. 2022, 12, 5872. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Xuan, J.; Dong, Y.; Bakay, M.; Feng, Y.; Clarke, R.; Hoffman, E.P. Optimized multilayer perceptrons for molecular classification and diagnosis using genomic data. Bioinformatics 2006, 22, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Ghosh, N.; Santoni, D.; Saha, I.; Felici, G. A review on the applications of Transformer-based language models for nucleotide sequence analysis. Comput. Struct. Biotechnol. J. 2025, 27, 1244–1254. [Google Scholar] [CrossRef]
- Choi, S.R.; Lee, M. Transformer architecture and attention mechanisms in genome data analysis: A comprehensive review. Biology 2023, 12, 1033. [Google Scholar] [CrossRef]
- Kha, Q.-H.; Tran, T.-O.; Nguyen, V.-N.; Than, K.; Le, N.Q.K. An interpretable deep learning model for classifying adaptor protein complexes from sequence information. Methods 2022, 207, 90–96. [Google Scholar] [CrossRef]
- Kha, Q.-H.; Le, V.-H.; Hung, T.N.K.; Nguyen, N.T.K.; Le, N.Q.K. Development and validation of an explainable machine learning-based prediction model for drug–food interactions from chemical structures. Sensors 2023, 23, 3962. [Google Scholar] [CrossRef]
- Soares, F.; Becker, K.; Anzanello, M.J. A hierarchical classifier based on human blood plasma fluorescence for non-invasive colorectal cancer screening. Artif. Intell. Med. 2017, 82, 1–10. [Google Scholar] [CrossRef]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; López-Fernández, H.; Iglesias, Á.; Cubiella, J.; Fdez-Riverola, F.; Reboiro-Jato, M.; Glez-Pena, D. Deep neural networks approaches for detecting and classifying colorectal polyps. Neurocomputing 2021, 423, 721–734. [Google Scholar] [CrossRef]
- Cui, J.-H.; Lin, K.-R.; Yuan, S.-H.; Jin, Y.-B.; Chen, X.-P.; Su, X.-K.; Jiang, J.; Pan, Y.-M.; Mao, S.-L.; Mao, X.-F. TCR repertoire as a novel indicator for immune monitoring and prognosis assessment of patients with cervical cancer. Front. Immunol. 2018, 9, 2729. [Google Scholar] [CrossRef]
- Sanz-Pamplona, R.; Melas, M.; Maoz, A.; Schmit, S.L.; Rennert, H.; Lejbkowicz, F.; Greenson, J.K.; Sanjuan, X.; Lopez-Zambrano, M.; Alonso, M.H. Lymphocytic infiltration in stage II microsatellite stable colorectal tumors: A retrospective prognosis biomarker analysis. PLoS Med. 2020, 17, e1003292. [Google Scholar] [CrossRef] [PubMed]
- Simnica, D.; Akyüz, N.; Schliffke, S.; Mohme, M.; Wenserski, L.V.; Mährle, T.; Fanchi, L.F.; Lamszus, K.; Binder, M. T cell receptor next-generation sequencing reveals cancer-associated repertoire metrics and reconstitution after chemotherapy in patients with hematological and solid tumors. Oncoimmunology 2019, 8, e1644110. [Google Scholar] [CrossRef]
- Borràs, D.M.; Verbandt, S.; Ausserhofer, M.; Sturm, G.; Lim, J.; Verge, G.A.; Vanmeerbeek, I.; Laureano, R.S.; Govaerts, J.; Sprooten, J. Single cell dynamics of tumor specificity vs bystander activity in CD8+ T cells define the diverse immune landscapes in colorectal cancer. Cell Discov. 2023, 9, 114. [Google Scholar] [CrossRef]
- Valpione, S.; Mundra, P.A.; Galvani, E.; Campana, L.G.; Lorigan, P.; De Rosa, F.; Gupta, A.; Weightman, J.; Mills, S.; Dhomen, N. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat. Commun. 2021, 12, 4098. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Chen, L.; Chen, Z.; Wang, X.; Jiang, R.; Zhao, K.; He, X. T cell receptor Beta-chain profiling of tumor tissue, peripheral blood and regional lymph nodes from patients with papillary thyroid carcinoma. Front. Immunol. 2021, 12, 595355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, Q.; Zhu, C.; Shi, Z.; Shao, C.; Chen, Y.; Wang, N.; Jiang, Y.; Liang, Q.; Wang, K. The intrarenal landscape of T cell receptor repertoire in clear cell renal cell cancer. J. Transl. Med. 2022, 20, 558. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, G.; Chen, T.; Ma, C.; Liu, K.; Huang, B.; Yang, G. Peripheral Blood TCR Clonotype Diversity as a Biomarker for Colorectal Cancer. Bioengineering 2025, 12, 1215. https://doi.org/10.3390/bioengineering12111215
Zhu G, Chen T, Ma C, Liu K, Huang B, Yang G. Peripheral Blood TCR Clonotype Diversity as a Biomarker for Colorectal Cancer. Bioengineering. 2025; 12(11):1215. https://doi.org/10.3390/bioengineering12111215
Chicago/Turabian StyleZhu, Gaochen, Tao Chen, Chen Ma, Kai Liu, Bihui Huang, and Guan Yang. 2025. "Peripheral Blood TCR Clonotype Diversity as a Biomarker for Colorectal Cancer" Bioengineering 12, no. 11: 1215. https://doi.org/10.3390/bioengineering12111215
APA StyleZhu, G., Chen, T., Ma, C., Liu, K., Huang, B., & Yang, G. (2025). Peripheral Blood TCR Clonotype Diversity as a Biomarker for Colorectal Cancer. Bioengineering, 12(11), 1215. https://doi.org/10.3390/bioengineering12111215





