Author Contributions
Conceptualization, T.D.H., C.G.I., B.T.N., T.V.C. and T.C.T.; Methodology, T.D.H., C.G.I., Q.T.M.N., T.M.D.B. and T.C.T.; Software, T.N.H.N., T.M.D.B., V.M.N., H.C.T. and V.H.P.; Validation, T.D.H., C.G.I., Q.T.M.N., E.J., V.H.P., L.S. and T.C.T.; Formal analysis, T.D.H., T.N.H.N., V.H.P. and T.C.T.; Investigation, T.D.H., C.G.I., Q.T.M.N., T.N.H.N., V.M.N., K.Q.T., H.C.T., V.H.P. and T.C.T.; Resources, B.T.N., T.N.H.N., T.M.D.B., K.D.C.N., E.J., V.H.P. and T.C.T.; Data curation, T.D.H., V.M.N., K.Q.T., H.C.T., K.D.C.N., L.S. and T.C.T.; Writing—original draft, T.D.H. and C.G.I.; Writing—review & editing, C.G.I.; Visualization, C.G.I., T.V.C. and K.D.C.N.; Supervision, B.T.N., E.J., L.S. and T.C.T.; Project administration, T.C.T.; Funding acquisition, T.C.T. All authors have read and agreed to the published version of the manuscript.
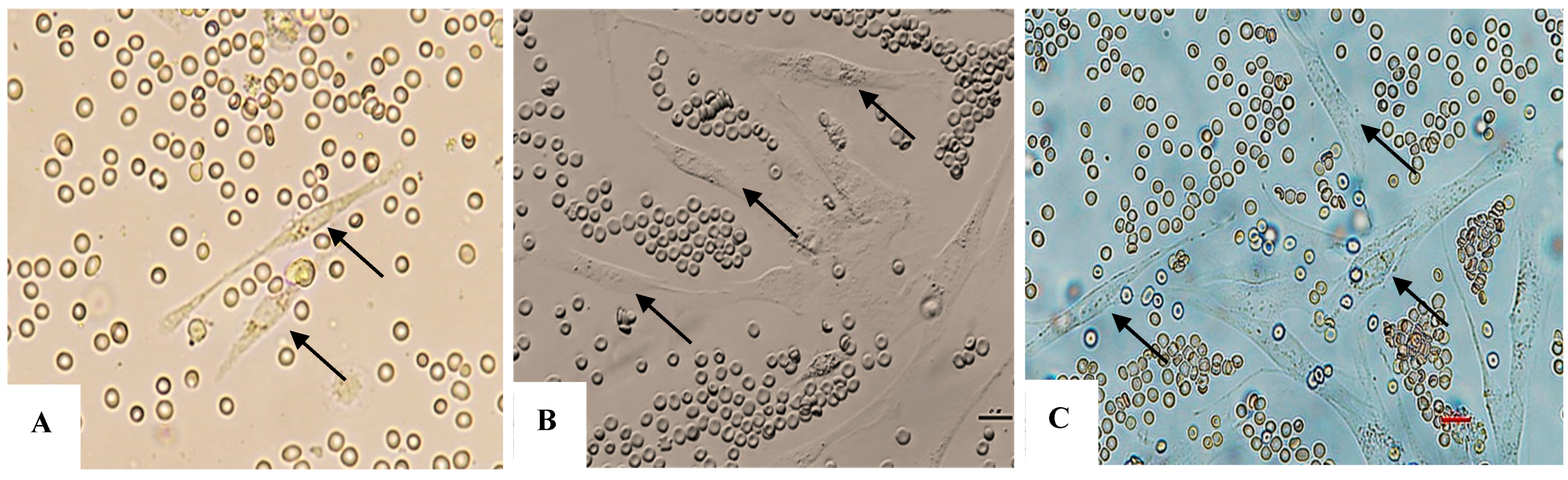
Figure 1.
(A–C): AT-MSCs at day 4, 5, and 6; there are MSCs with typical spindle-like fibroblast shapes (black arrows).
Figure 1.
(A–C): AT-MSCs at day 4, 5, and 6; there are MSCs with typical spindle-like fibroblast shapes (black arrows).
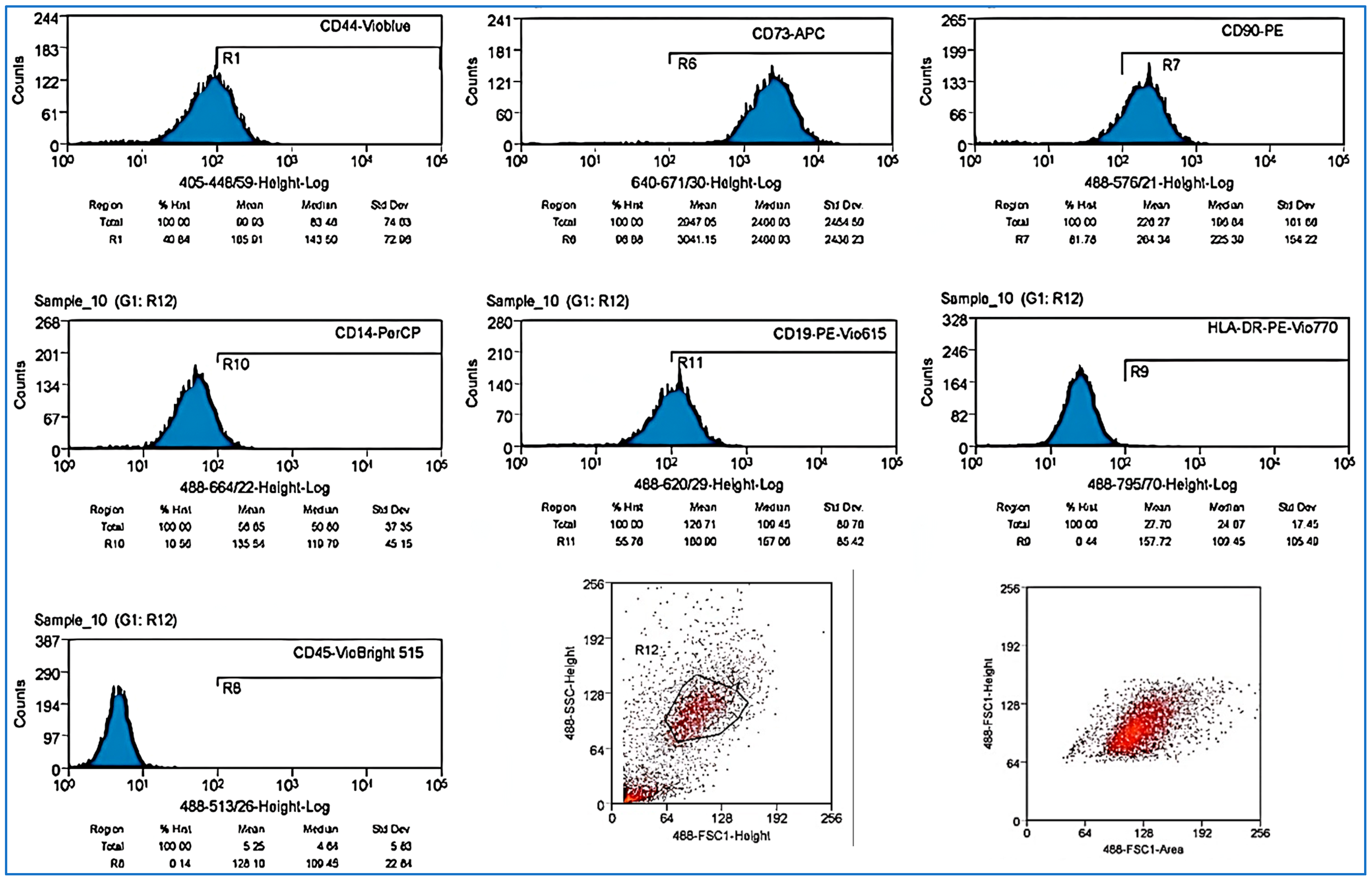
Figure 2.
Results of evaluation using flow cytometry to identify AT-MSCs CD-44, CD-73, CD-90, CD-19, and CD14low, while showing negative results for CD45 and HLA-DR.
Figure 2.
Results of evaluation using flow cytometry to identify AT-MSCs CD-44, CD-73, CD-90, CD-19, and CD14low, while showing negative results for CD45 and HLA-DR.
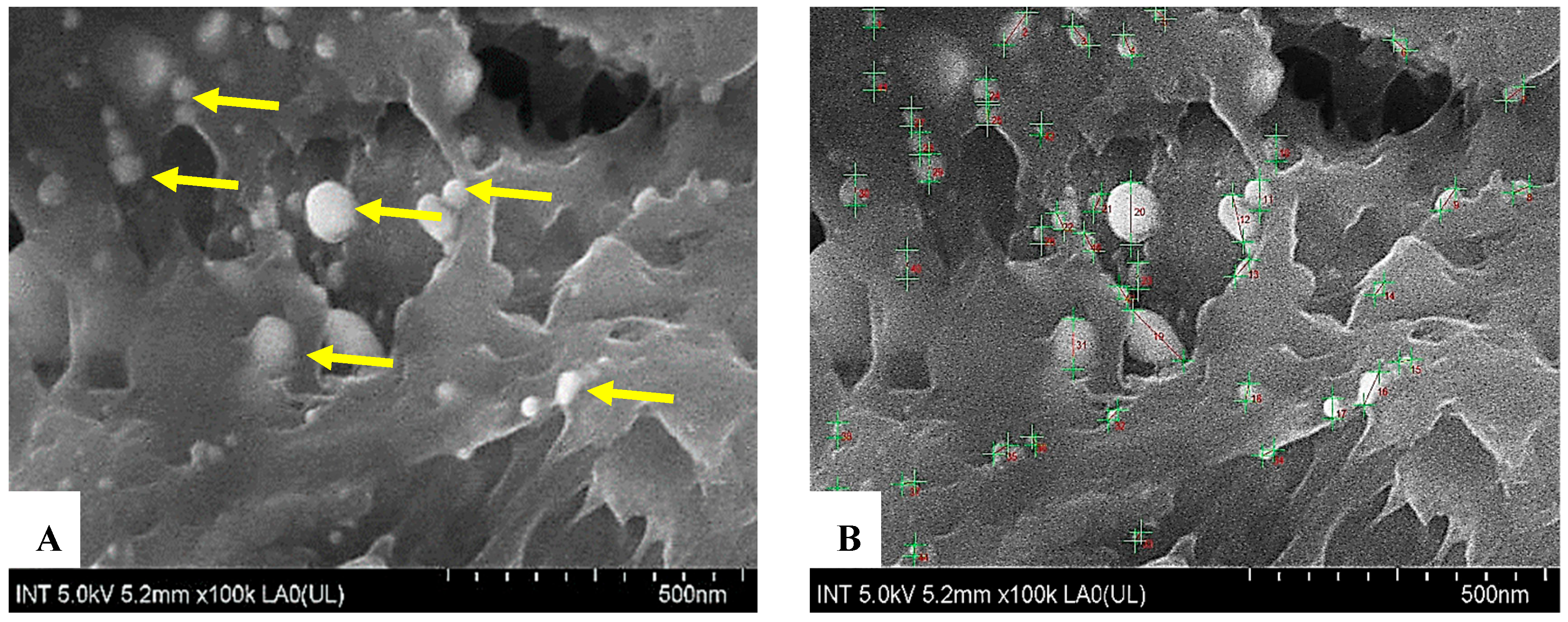
Figure 3.
FE-SEM images of exosomes. Using a scanning electron microscope to observe the morphology and size of exosome vesicles revealed that these vesicles are spherical (yellow arrows, (A)), with an average size of approximately 40 nm, falling within the 30–200 nm range specified by ISEV standards (B).
Figure 3.
FE-SEM images of exosomes. Using a scanning electron microscope to observe the morphology and size of exosome vesicles revealed that these vesicles are spherical (yellow arrows, (A)), with an average size of approximately 40 nm, falling within the 30–200 nm range specified by ISEV standards (B).
Figure 4.
(A,B) Exosome particle size measurement by Dynamic Light Scattering (DLS). The measurement results indicate that exosome particles have an average size of approximately 154.5 nm, falling within the 30–200 nm range according to ISEV standards. (B) The DLS measurements demonstrate that the exosomes have an average size of approximately 154.50 nm, which is within the typical range for exosomes (30–200 nm). This confirms that the sample contains particles with characteristics corresponding to exosomes, without contamination from larger extracellular vesicles such as microvesicles (>200 nm). The red line shows a cumulative distribution curve that matches the ‘Subsize (%)’ axis on the right side of the graph. This curve displays the total percentage of particles with diameters at or below a certain value. At each point on the red curve, the x-axis shows the particle diameter, and the right y-axis shows the percentage of the sample with diameters up to that value.
Figure 4.
(A,B) Exosome particle size measurement by Dynamic Light Scattering (DLS). The measurement results indicate that exosome particles have an average size of approximately 154.5 nm, falling within the 30–200 nm range according to ISEV standards. (B) The DLS measurements demonstrate that the exosomes have an average size of approximately 154.50 nm, which is within the typical range for exosomes (30–200 nm). This confirms that the sample contains particles with characteristics corresponding to exosomes, without contamination from larger extracellular vesicles such as microvesicles (>200 nm). The red line shows a cumulative distribution curve that matches the ‘Subsize (%)’ axis on the right side of the graph. This curve displays the total percentage of particles with diameters at or below a certain value. At each point on the red curve, the x-axis shows the particle diameter, and the right y-axis shows the percentage of the sample with diameters up to that value.
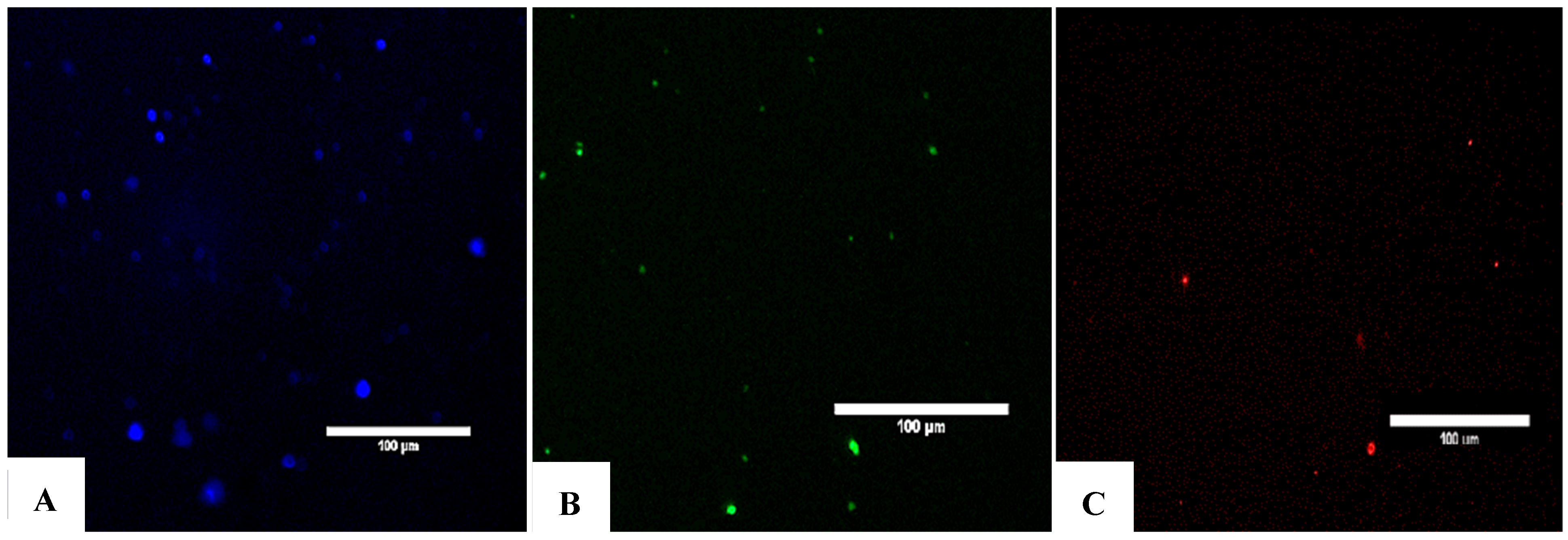
Figure 5.
Detection of exosomes as evidenced by the expression of the canonical markers CD9, CD63, and CD81. (A) Immunofluorescence staining of CD9. (B) Immunofluorescence staining of CD63. (C) Immunofluorescence staining of CD81. To evaluate the presence of exosomes in the product suspension, exosomes with fluorescently labeled antibodies were stained against CD9, CD63, and CD81 after incubating the vesicles with beads for flow cytometry preparation. The results were observed under a confocal fluorescence microscope, as shown above. (A) Fluorescent staining with anti-CD9 antibody. (B) Fluorescent staining with anti-CD63 antibody. (C) Fluorescent staining with anti-CD81 antibody. Colors indicate the density of events, such as exosomes, on the flow cytometry plot. The provided percentages—91.36% for CD9, 81.06% for CD63, and 64.73% for CD81—represent the proportion of exosomes within a specific gated region, likely R1, that are positive for each of these markers.
Figure 5.
Detection of exosomes as evidenced by the expression of the canonical markers CD9, CD63, and CD81. (A) Immunofluorescence staining of CD9. (B) Immunofluorescence staining of CD63. (C) Immunofluorescence staining of CD81. To evaluate the presence of exosomes in the product suspension, exosomes with fluorescently labeled antibodies were stained against CD9, CD63, and CD81 after incubating the vesicles with beads for flow cytometry preparation. The results were observed under a confocal fluorescence microscope, as shown above. (A) Fluorescent staining with anti-CD9 antibody. (B) Fluorescent staining with anti-CD63 antibody. (C) Fluorescent staining with anti-CD81 antibody. Colors indicate the density of events, such as exosomes, on the flow cytometry plot. The provided percentages—91.36% for CD9, 81.06% for CD63, and 64.73% for CD81—represent the proportion of exosomes within a specific gated region, likely R1, that are positive for each of these markers.
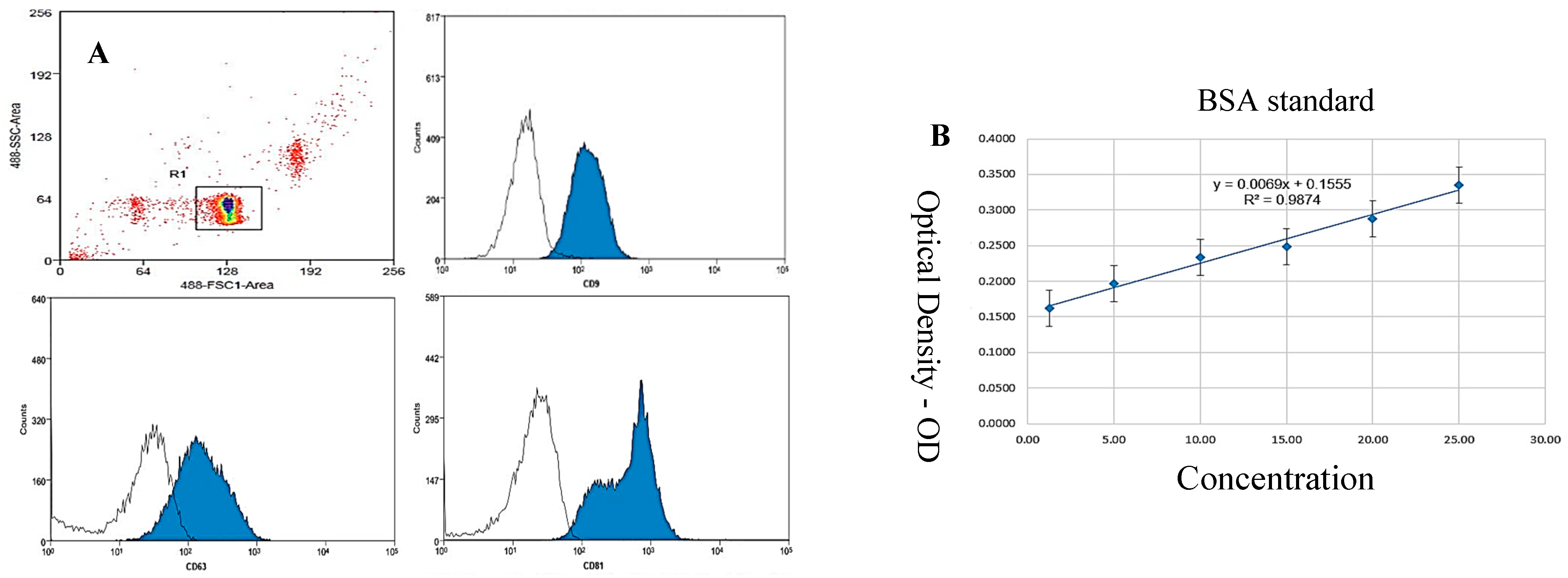
Figure 6.
(A,B): Flow cytometry results for the expression of characteristic CD9, CD63, and CD81 markers for exosome identification. The results indicate the presence of these markers on vesicles within the exosome product suspension, with CD9 expression at 91.36%, CD63 at 81.06%, and CD81 at 64.73%. The observations confirm the expression of these markers in the exosome product suspension post-isolation. To quantify the presence of these characteristic exosome proteins, flow cytometry was employed to assess the expression of CD9, CD63, and CD81 markers. (B) Illustration of results from the Bradford assay for quantifying protein concentration in the exosome suspension (x = by absorption; y = concentration). Bradford assay for quantification of protein concentration in the exosome suspension. The calibration curve is depicted with absorbance on the x-axis and protein concentration on the y-axis. The results provide a quantitative assessment of protein levels within the exosome preparation. Colors indicate the density of events, such as exosomes, on the flow cytometry plot. The provided percentages—91.36% for CD9, 81.06% for CD63, and 64.73% for CD81—represent the proportion of exosomes within a specific gated region, likely R1, that are positive for each of these markers.
Figure 6.
(A,B): Flow cytometry results for the expression of characteristic CD9, CD63, and CD81 markers for exosome identification. The results indicate the presence of these markers on vesicles within the exosome product suspension, with CD9 expression at 91.36%, CD63 at 81.06%, and CD81 at 64.73%. The observations confirm the expression of these markers in the exosome product suspension post-isolation. To quantify the presence of these characteristic exosome proteins, flow cytometry was employed to assess the expression of CD9, CD63, and CD81 markers. (B) Illustration of results from the Bradford assay for quantifying protein concentration in the exosome suspension (x = by absorption; y = concentration). Bradford assay for quantification of protein concentration in the exosome suspension. The calibration curve is depicted with absorbance on the x-axis and protein concentration on the y-axis. The results provide a quantitative assessment of protein levels within the exosome preparation. Colors indicate the density of events, such as exosomes, on the flow cytometry plot. The provided percentages—91.36% for CD9, 81.06% for CD63, and 64.73% for CD81—represent the proportion of exosomes within a specific gated region, likely R1, that are positive for each of these markers.
![Bioengineering 12 01129 g006 Bioengineering 12 01129 g006]()
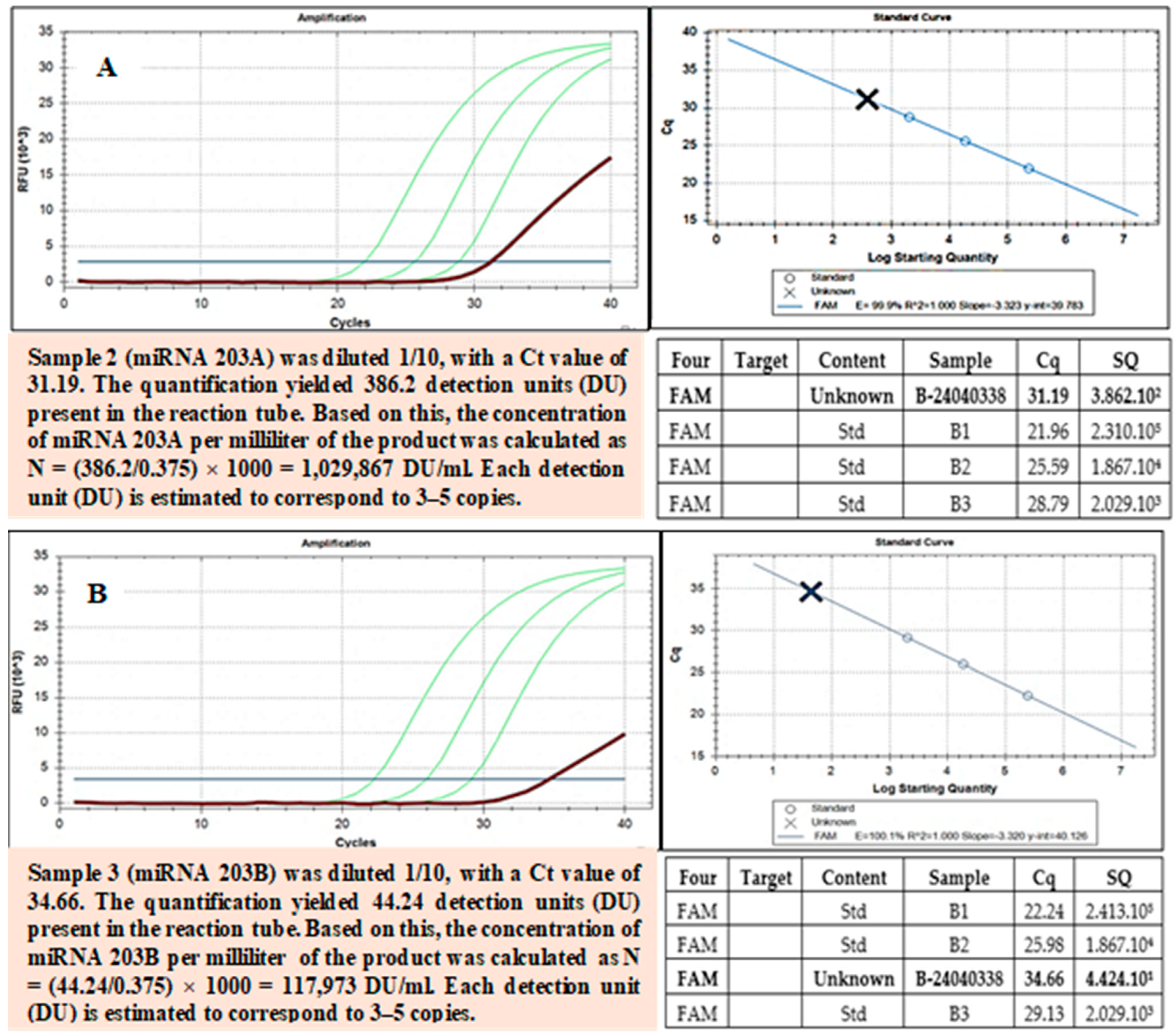
Figure 7.
The RT-PCR confirmed the presence of miRNA-3196 in exosome samples. miRNA-3196 is recognized for its role in regulating biological pathways associated with inflammation, metabolism, and cancer. miRNA-3196 is implicated in the regulation of biological pathways related to inflammation, metabolism, and cancer. RT-PCR analysis confirmed the presence of miRNA-3196 in the exosome samples. Green curves: These are the amplification curves of the target miRNA from different replicates. Red or dark curve: This represents the negative control or a baseline/reference curve.
Figure 7.
The RT-PCR confirmed the presence of miRNA-3196 in exosome samples. miRNA-3196 is recognized for its role in regulating biological pathways associated with inflammation, metabolism, and cancer. miRNA-3196 is implicated in the regulation of biological pathways related to inflammation, metabolism, and cancer. RT-PCR analysis confirmed the presence of miRNA-3196 in the exosome samples. Green curves: These are the amplification curves of the target miRNA from different replicates. Red or dark curve: This represents the negative control or a baseline/reference curve.
Figure 8.
(A,B) miRNA-203 is a key regulator of cellular differentiation and is implicated in conditions such as cancer, chronic inflammation, and tissue regeneration. RT-PCR confirmed the presence of miRNA-203 (A,B). Green curves: These are the amplification curves of the target miRNA from different replicates. Red or dark curve: This represents the negative control or a baseline/reference curve.
Figure 8.
(A,B) miRNA-203 is a key regulator of cellular differentiation and is implicated in conditions such as cancer, chronic inflammation, and tissue regeneration. RT-PCR confirmed the presence of miRNA-203 (A,B). Green curves: These are the amplification curves of the target miRNA from different replicates. Red or dark curve: This represents the negative control or a baseline/reference curve.
Figure 9.
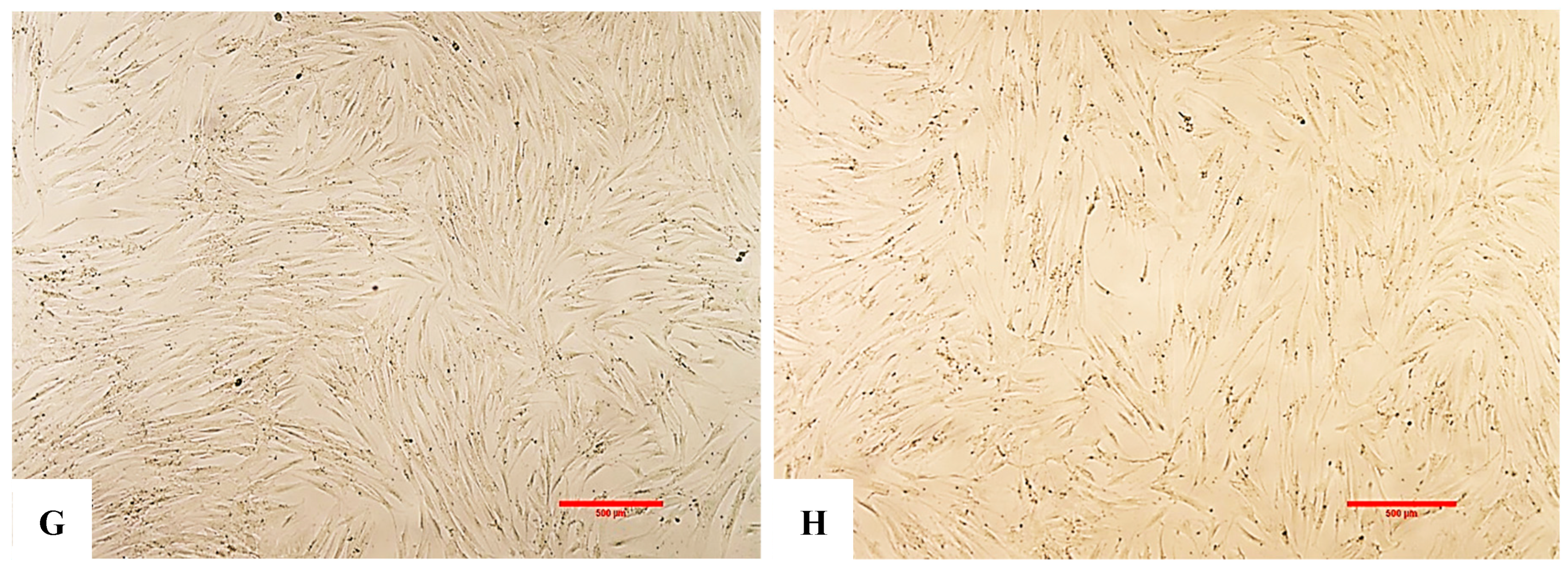
(A,B) Representative images of fibroblasts captured using an inverted microscope at 4× magnification. (A) shows the negative control (no exosomes and no H2O2), while (B) represents the positive control or EXO-0 condition (no exosomes + H2O2). A marked difference in cell density was observed between the two groups, with the negative control displaying significantly higher cell populations compared to the positive control treated with H2O2 (p < 0.001, F > 2.5). (C) (EXO-0.1) corresponds to cells treated with 0.1 µg/mL exosomes + H2O2, while (D) (EXO-0.5) represents cells treated with 0.5 µg/mL exosomes + H2O2. A clear difference in cell density was observed between them, with (C) exhibiting significantly lesser cell populations compared to (D) (p < 0.001, F > 2.5). (E) (EXO-1) corresponds to cells treated with 1 µg/mL exosomes + H2O2, while (F) (EXO-5) represents cells treated with 5 µg/mL exosomes + H2O2. A pronounced difference in cell density was observed between them, with (F) exhibiting significantly higher cell populations (p < 0.001, F > 2.5). (G) shows the negative control (no exosomes and no H2O2), while (H) represents the positive control or EXO-0 condition (no exosomes + H2O2). A significant difference in cell density was observed, with the negative control (G) displaying markedly higher cell populations compared to the positive control (H) (p < 0.001, F > 2.5).
Figure 9.
(A,B) Representative images of fibroblasts captured using an inverted microscope at 4× magnification. (A) shows the negative control (no exosomes and no H2O2), while (B) represents the positive control or EXO-0 condition (no exosomes + H2O2). A marked difference in cell density was observed between the two groups, with the negative control displaying significantly higher cell populations compared to the positive control treated with H2O2 (p < 0.001, F > 2.5). (C) (EXO-0.1) corresponds to cells treated with 0.1 µg/mL exosomes + H2O2, while (D) (EXO-0.5) represents cells treated with 0.5 µg/mL exosomes + H2O2. A clear difference in cell density was observed between them, with (C) exhibiting significantly lesser cell populations compared to (D) (p < 0.001, F > 2.5). (E) (EXO-1) corresponds to cells treated with 1 µg/mL exosomes + H2O2, while (F) (EXO-5) represents cells treated with 5 µg/mL exosomes + H2O2. A pronounced difference in cell density was observed between them, with (F) exhibiting significantly higher cell populations (p < 0.001, F > 2.5). (G) shows the negative control (no exosomes and no H2O2), while (H) represents the positive control or EXO-0 condition (no exosomes + H2O2). A significant difference in cell density was observed, with the negative control (G) displaying markedly higher cell populations compared to the positive control (H) (p < 0.001, F > 2.5).
![Bioengineering 12 01129 g009a Bioengineering 12 01129 g009a]()
![Bioengineering 12 01129 g009b Bioengineering 12 01129 g009b]()
Figure 10.
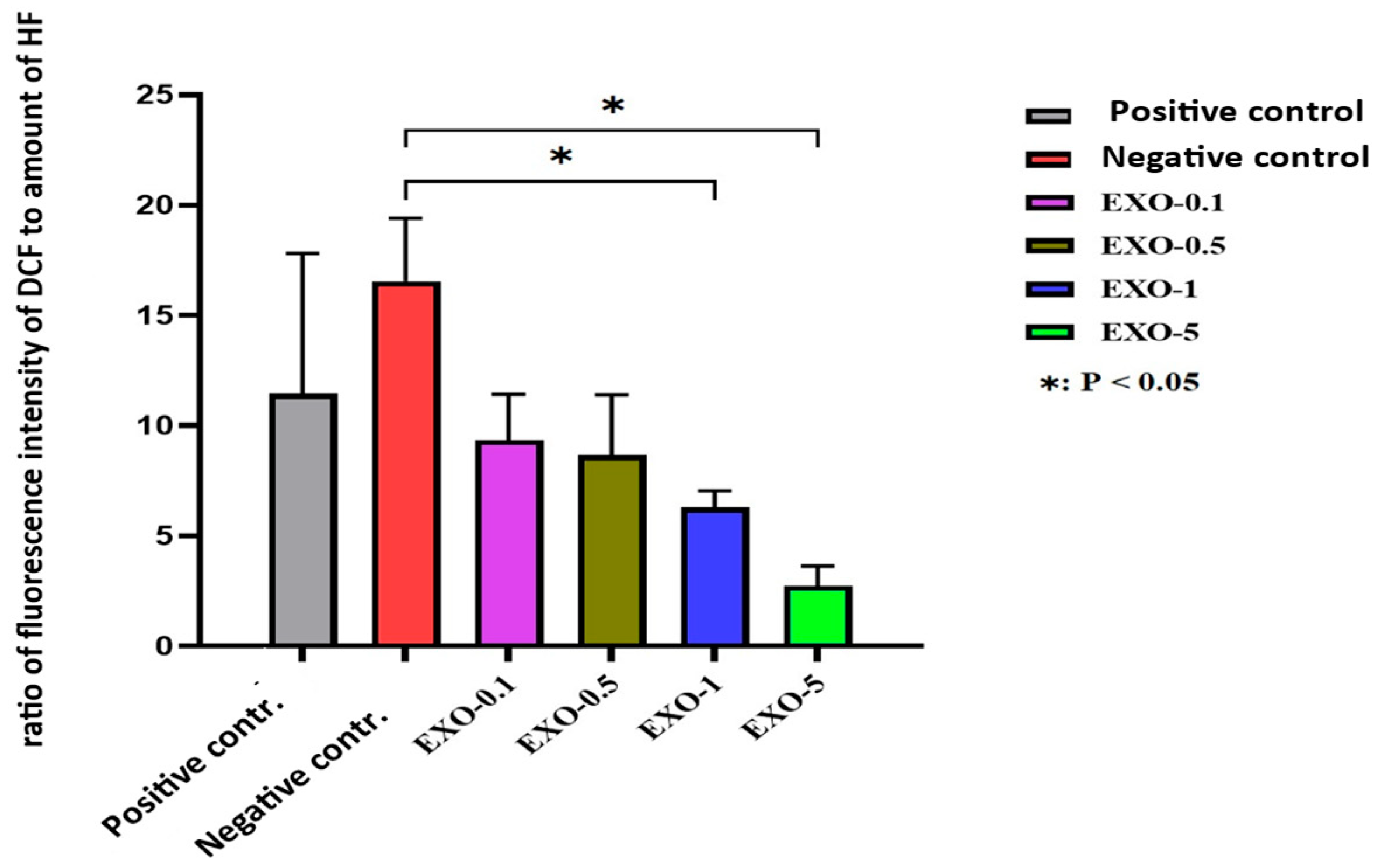
Graph depicting fibroblast proliferation over time across different experimental conditions. Fibroblast proliferation increased following treatment with exosomes at varying concentrations, indicating that exosomes confer protective effects against H2O2-induced cellular oxidative stress. The data suggested that exosome concentration supports fibroblast proliferation over time, with proliferation increasing progressively from lower (0.1 µg/mL) to higher (5.0 µg/mL) concentrations. Notably, the 5.0 µg/mL concentration yielded the highest proliferation rate among the exosome-treated groups by day 9.
Figure 10.
Graph depicting fibroblast proliferation over time across different experimental conditions. Fibroblast proliferation increased following treatment with exosomes at varying concentrations, indicating that exosomes confer protective effects against H2O2-induced cellular oxidative stress. The data suggested that exosome concentration supports fibroblast proliferation over time, with proliferation increasing progressively from lower (0.1 µg/mL) to higher (5.0 µg/mL) concentrations. Notably, the 5.0 µg/mL concentration yielded the highest proliferation rate among the exosome-treated groups by day 9.
Figure 18.
(
A,
B) Fibroblasts without any exposure; this served as a control to assess the baseline effect of exosomes on fibroblast cells in the absence of oxidative stress; telomere length was 10.9 kb. Fibroblasts were exposed to exosomes for 24 h, followed by 200 µM H
2O
2 treatment for 90 min; telomere length was 15.7 kb. (
C,
D) Fibroblasts were exposed to treatment with 200 µM H
2O
2 for 90 min; subsequently, exosomes were added for 24 h; the telomere measured 14.9 kb.
Figure 11: Fibroblasts were exposed to exosomes only for 24 h; telomere measured 14.8 kb. (
E) Fibroblasts were exposed to 200 µM H
2O
2 for 90 min; the resulting telomere length was 11.1 kb.
Figure 18.
(
A,
B) Fibroblasts without any exposure; this served as a control to assess the baseline effect of exosomes on fibroblast cells in the absence of oxidative stress; telomere length was 10.9 kb. Fibroblasts were exposed to exosomes for 24 h, followed by 200 µM H
2O
2 treatment for 90 min; telomere length was 15.7 kb. (
C,
D) Fibroblasts were exposed to treatment with 200 µM H
2O
2 for 90 min; subsequently, exosomes were added for 24 h; the telomere measured 14.9 kb.
Figure 11: Fibroblasts were exposed to exosomes only for 24 h; telomere measured 14.8 kb. (
E) Fibroblasts were exposed to 200 µM H
2O
2 for 90 min; the resulting telomere length was 11.1 kb.
Figure 19.
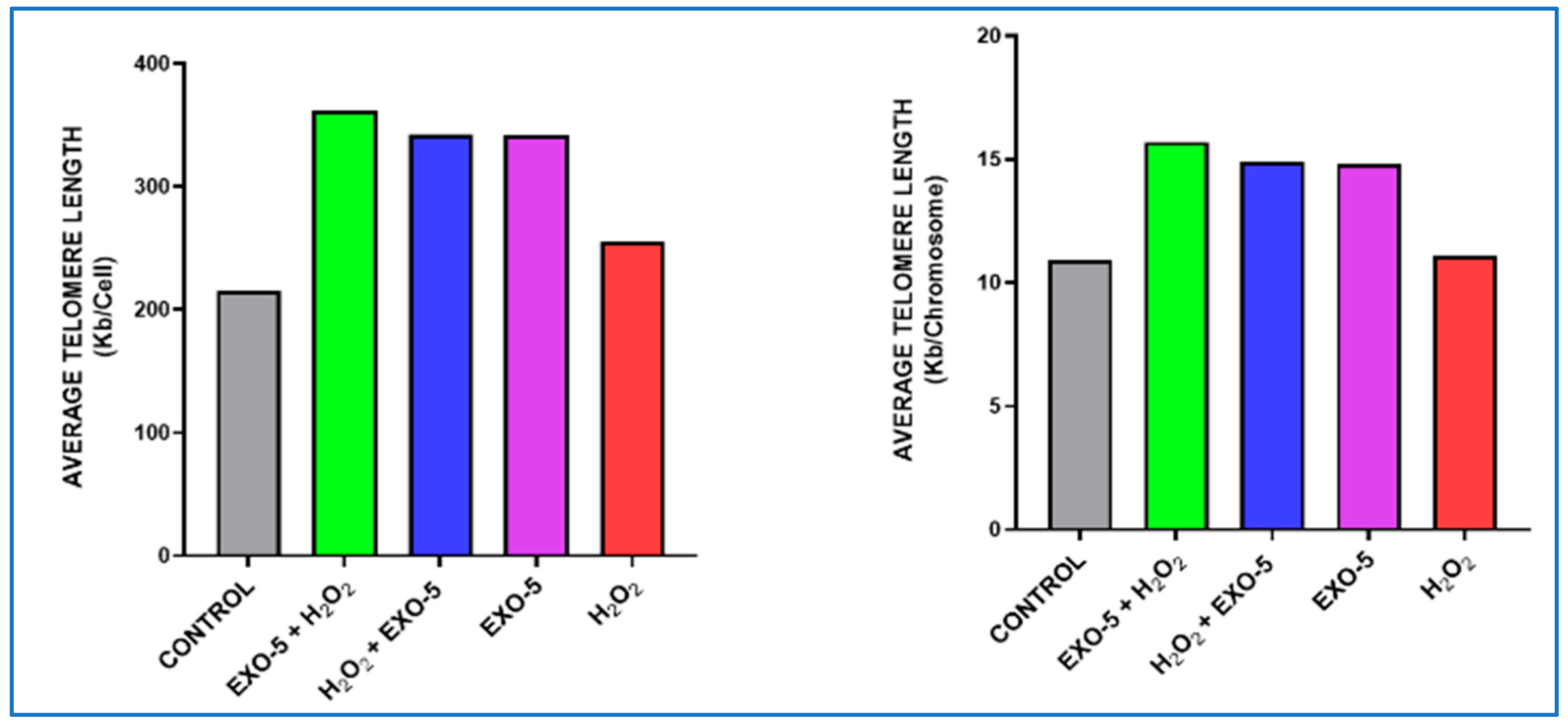
Graph evaluating telomere length per chromosome and per cell across the five experimental conditions. Condition 1: Fibroblast control group. Condition 2: hFs were incubated with exosomes at a concentration of 5.0 µg/mL and subsequently exposed to H2O2 (200 nM). Condition 3: hFs were incubated with H2O2 (200 nM) and then were exposed to exosomes at a concentration of 5.0 µg/mL. Condition 4: hFs were exposed to exosomes only at a concentration of 5.0 µg/mL. Condition 5: hFs were exposed to H2O2 (200 nM). Exosomes generally exert a clear protective and enhancement activity toward cells exposed to H2O2; eventually, the best gradient was 5.0 µg/mL (green) (p-value < 0.001).
Figure 19.
Graph evaluating telomere length per chromosome and per cell across the five experimental conditions. Condition 1: Fibroblast control group. Condition 2: hFs were incubated with exosomes at a concentration of 5.0 µg/mL and subsequently exposed to H2O2 (200 nM). Condition 3: hFs were incubated with H2O2 (200 nM) and then were exposed to exosomes at a concentration of 5.0 µg/mL. Condition 4: hFs were exposed to exosomes only at a concentration of 5.0 µg/mL. Condition 5: hFs were exposed to H2O2 (200 nM). Exosomes generally exert a clear protective and enhancement activity toward cells exposed to H2O2; eventually, the best gradient was 5.0 µg/mL (green) (p-value < 0.001).
Figure 20.
Exosomes caused an increase in telomere content, observed primarily under the influence of the highest concentration tested on each cell sample: control group 1 (dark blue), group 2 (orange), group 3 (gray), group 4 (dark yellow), and exosome dose group 5 (light blue) included in the experiment. Exosomes had a stronger effect on the tested parameter both before and after H2O2, even at the lowest concentration (p value < 0.001).
Figure 20.
Exosomes caused an increase in telomere content, observed primarily under the influence of the highest concentration tested on each cell sample: control group 1 (dark blue), group 2 (orange), group 3 (gray), group 4 (dark yellow), and exosome dose group 5 (light blue) included in the experiment. Exosomes had a stronger effect on the tested parameter both before and after H2O2, even at the lowest concentration (p value < 0.001).
Figure 21.
The Q-Q plots indicate that the full sample did not deviate from linearity due to the influence of one sample (Experiment 5); data are normally distributed (p-value is < 0.001).
Figure 21.
The Q-Q plots indicate that the full sample did not deviate from linearity due to the influence of one sample (Experiment 5); data are normally distributed (p-value is < 0.001).
Figure 22.
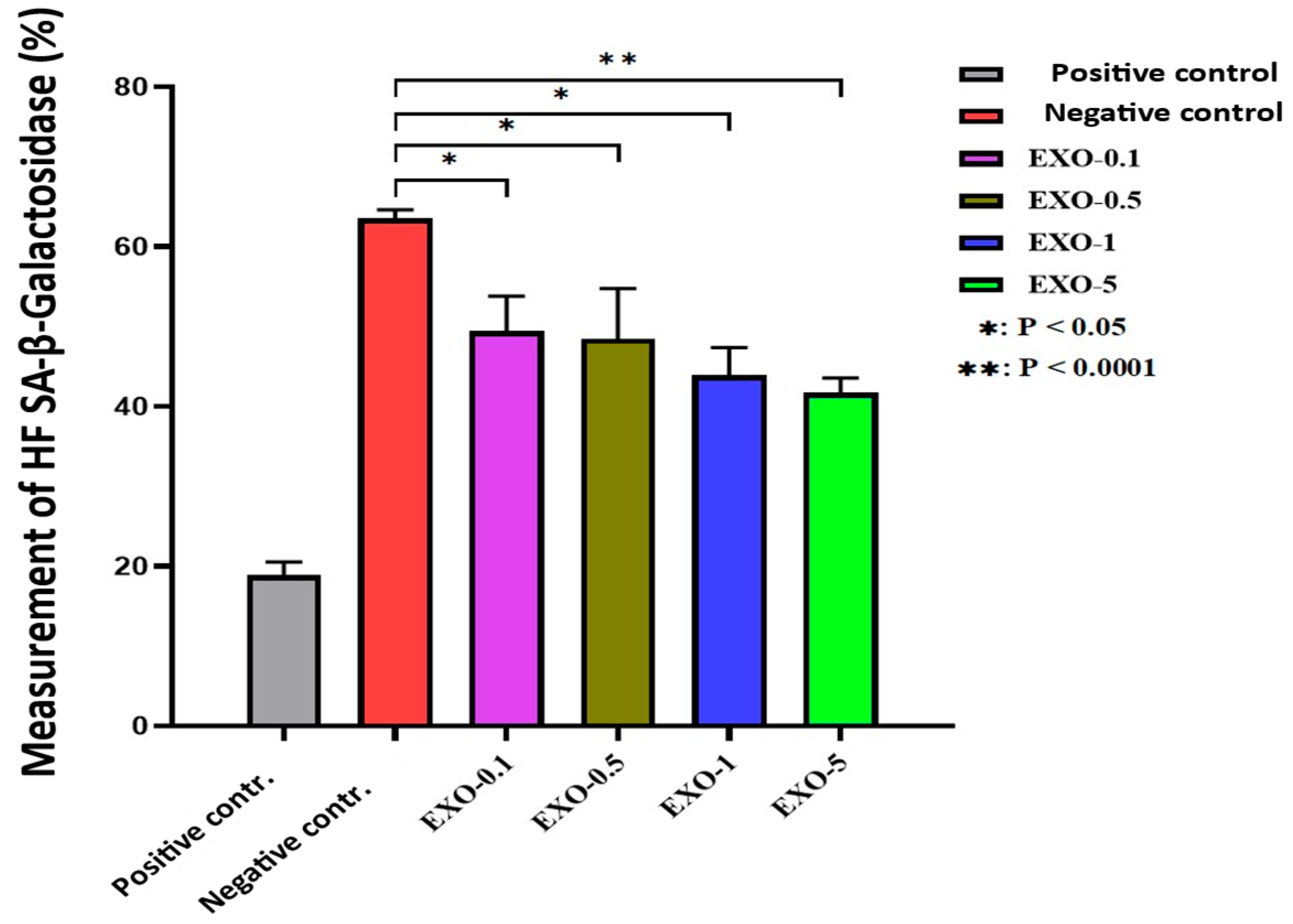
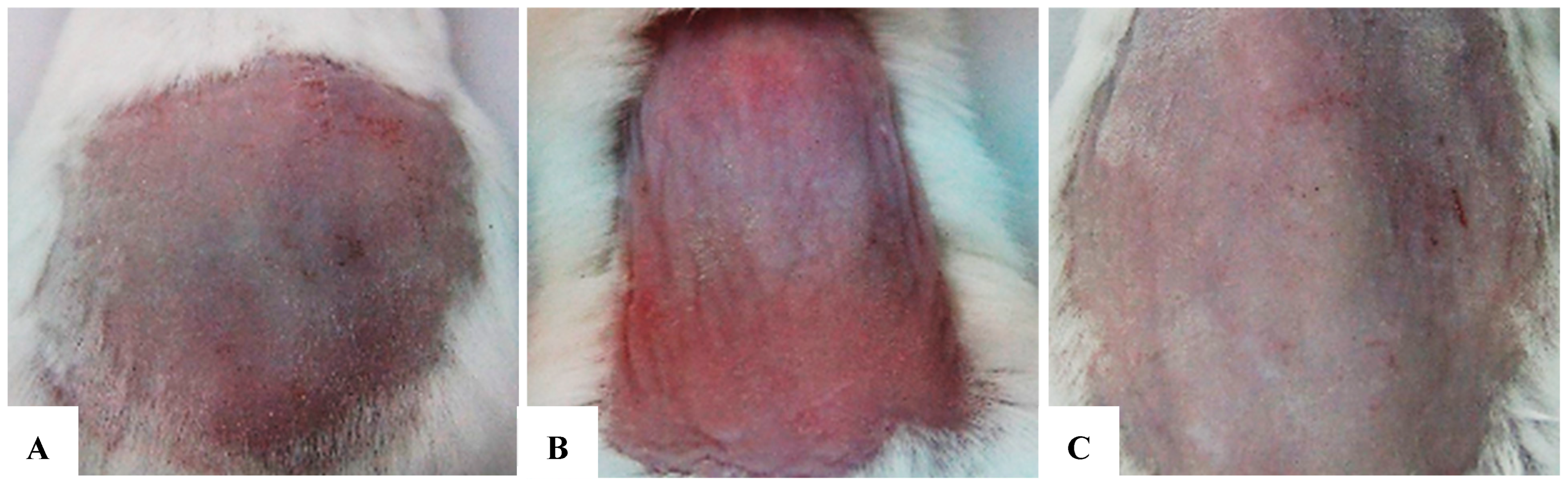
(A–C) Comparison of morphological assessments at 2 weeks. (A) Negative control group. (B) Positive control group. (C) Research group (treated with exosomes prior to each UVB exposure). After 2 weeks, distinct differences in the test area’s morphology were clear across the three groups, particularly in terms of color, elasticity, and inflammation reduction. The exosome-treated group exhibited substantial improvement compared to the untreated group, confirmed by reduced inflammatory patterns, scaling, and enhanced elasticity. The untreated positive control group showed persistent severe damage, with dry, rough skin and slow recovery. The negative control group kept healthy skin with no signs of damage or inflammation.
Figure 22.
(A–C) Comparison of morphological assessments at 2 weeks. (A) Negative control group. (B) Positive control group. (C) Research group (treated with exosomes prior to each UVB exposure). After 2 weeks, distinct differences in the test area’s morphology were clear across the three groups, particularly in terms of color, elasticity, and inflammation reduction. The exosome-treated group exhibited substantial improvement compared to the untreated group, confirmed by reduced inflammatory patterns, scaling, and enhanced elasticity. The untreated positive control group showed persistent severe damage, with dry, rough skin and slow recovery. The negative control group kept healthy skin with no signs of damage or inflammation.
Figure 25.
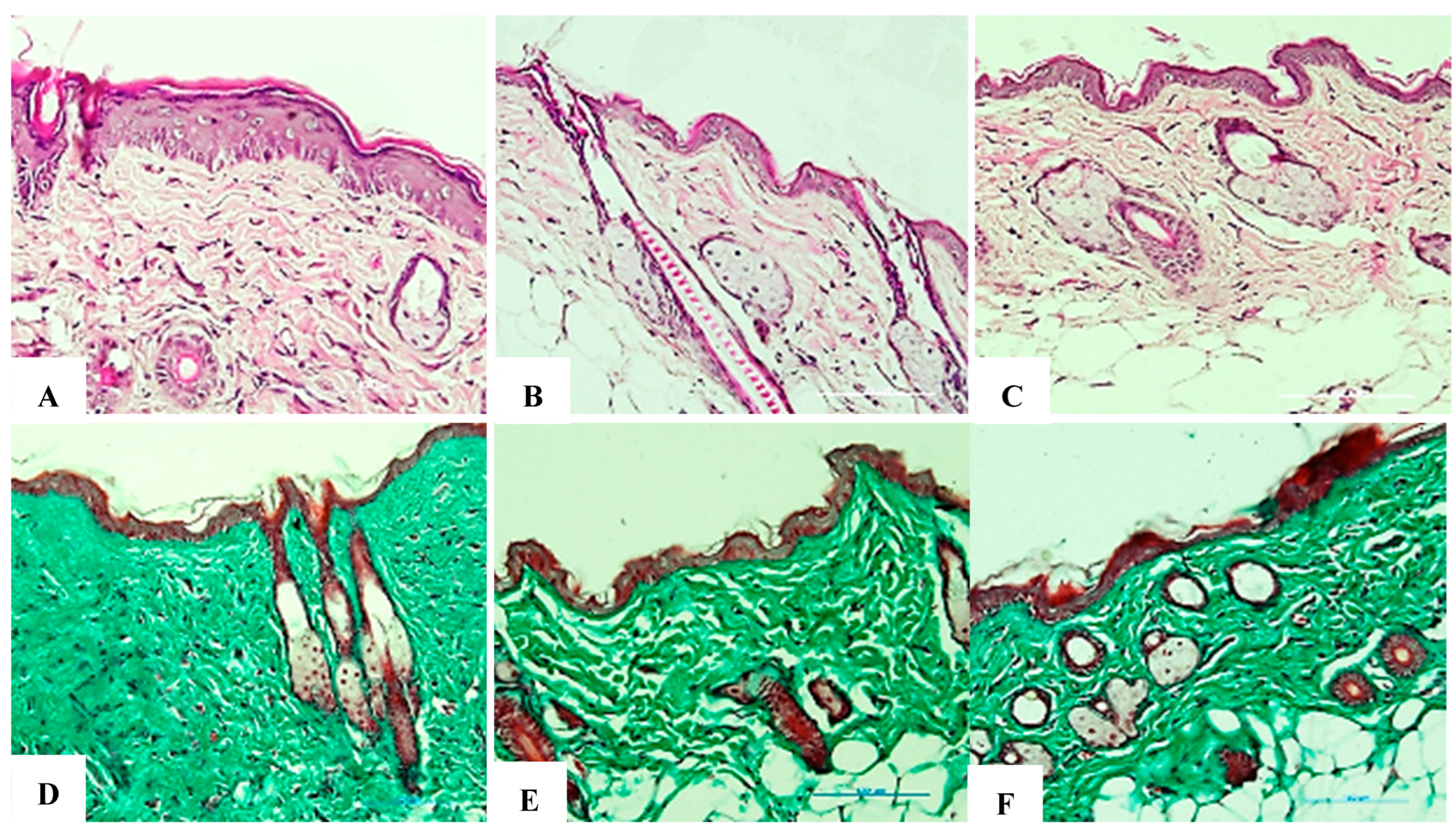
(A–F). Comparison of histological results for collagen at 2 weeks. (A,D) Negative control group stained with H&E and Trichrome, respectively (20× objective). (B,E) Positive control group stained with H&E and Trichrome, respectively (20× objective). (C,F) Research group (treated with exosomes before each UVB exposure) stained with H&E and Trichrome, respectively (20× objective). The negative control group showed an intact collagen structure reflecting healthy skin; the positive control group showed that collagen density was significantly reduced, with marked structural breakdown due to UVB exposure, which caused inflammation and early signs of aging. The research team demonstrated that collagen density was higher than that of the positive control group, indicating early recovery. Collagenous fibers showed a phase of reorganization but did not reach optimal levels. This suggests that exosomes stimulated collagen regeneration after 2 weeks, although further time is needed for full recovery.
Figure 25.
(A–F). Comparison of histological results for collagen at 2 weeks. (A,D) Negative control group stained with H&E and Trichrome, respectively (20× objective). (B,E) Positive control group stained with H&E and Trichrome, respectively (20× objective). (C,F) Research group (treated with exosomes before each UVB exposure) stained with H&E and Trichrome, respectively (20× objective). The negative control group showed an intact collagen structure reflecting healthy skin; the positive control group showed that collagen density was significantly reduced, with marked structural breakdown due to UVB exposure, which caused inflammation and early signs of aging. The research team demonstrated that collagen density was higher than that of the positive control group, indicating early recovery. Collagenous fibers showed a phase of reorganization but did not reach optimal levels. This suggests that exosomes stimulated collagen regeneration after 2 weeks, although further time is needed for full recovery.
![Bioengineering 12 01129 g025 Bioengineering 12 01129 g025]()
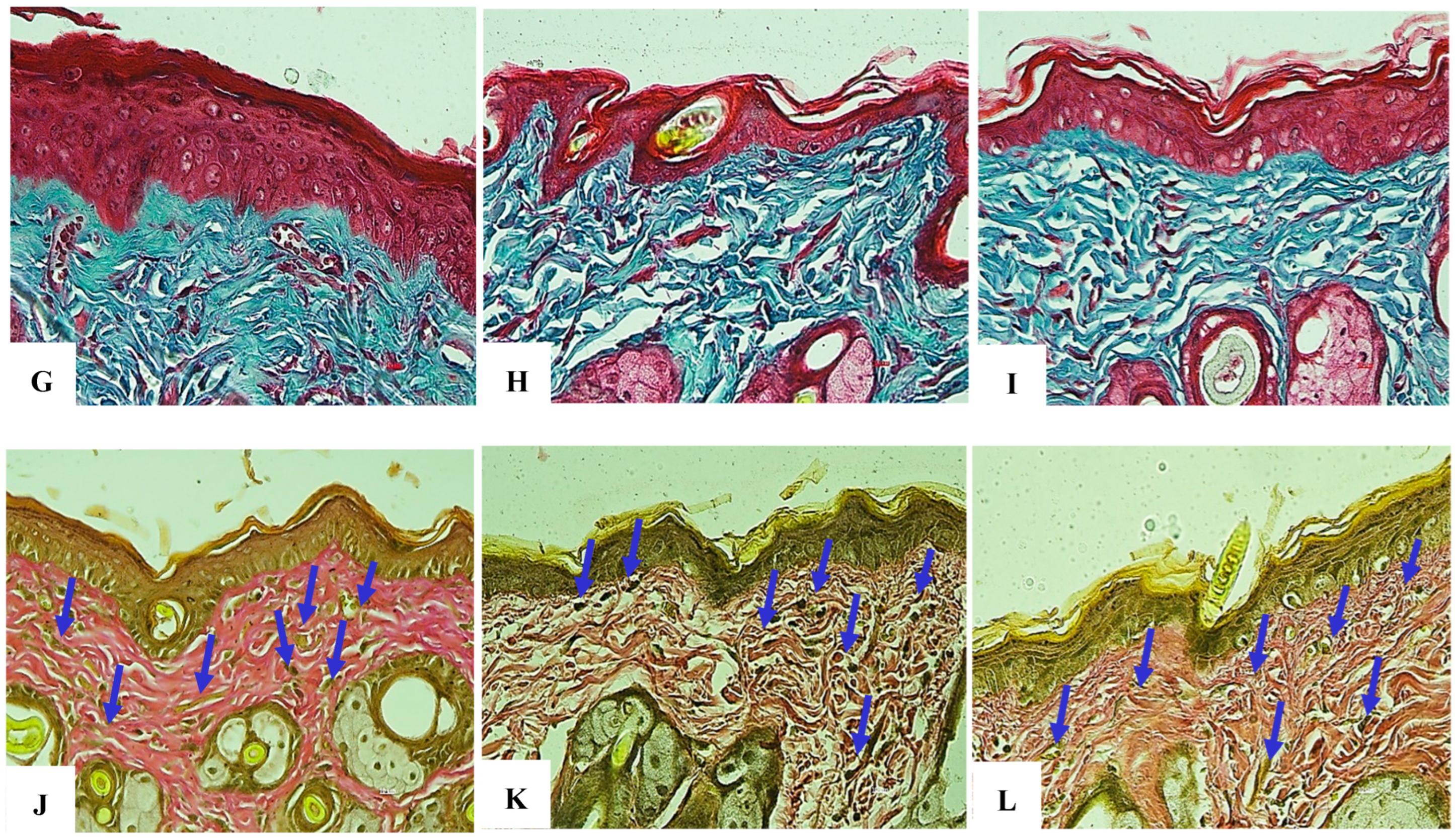
Figure 26.
(A–F). Comparison of histological collagen structure assessments at the 4-week time point. (A,D) Negative control group stained with H&E and Trichrome, respectively (20× objective). (B,E) Positive control group stained with H&E and Trichrome, respectively (20× objective). (C,F) Research group (treated with exosomes prior to each UVB exposure) stained with H&E and Trichrome, respectively (20× objective).
Figure 26.
(A–F). Comparison of histological collagen structure assessments at the 4-week time point. (A,D) Negative control group stained with H&E and Trichrome, respectively (20× objective). (B,E) Positive control group stained with H&E and Trichrome, respectively (20× objective). (C,F) Research group (treated with exosomes prior to each UVB exposure) stained with H&E and Trichrome, respectively (20× objective).
Figure 27.
(A–F) Comparison of elastin histological structure assessments at 2 weeks. (A,D) Negative control group stained for elastin at 20× and 40× objectives, respectively. (B,E) Positive control group stained for elastin at 20× and 40× objectives, respectively. (C,F) Research group (treated with exosomes prior to each UVB exposure) stained for elastin at 20× and 40× objectives, respectively. Elastin fiber structure is indicated by blue arrowheads.
Figure 27.
(A–F) Comparison of elastin histological structure assessments at 2 weeks. (A,D) Negative control group stained for elastin at 20× and 40× objectives, respectively. (B,E) Positive control group stained for elastin at 20× and 40× objectives, respectively. (C,F) Research group (treated with exosomes prior to each UVB exposure) stained for elastin at 20× and 40× objectives, respectively. Elastin fiber structure is indicated by blue arrowheads.
Figure 28.
(A–F) Comparison of elastin histological structure assessments at the 4-week time point. (A,D) Negative control group stained for elastin at 20× and 40× objectives, respectively. (B,E) Positive control group stained for elastin at 20× and 40× objectives, respectively. (C,F) Research group (treated with exosomes prior to each UVB exposure) stained for elastin at 20× and 40× objectives, respectively. Elastin fiber structure is indicated by blue arrowheads. (G–I) Comparative histological assessment of collagen structure at the 6-week time point. (G) Negative control group stained with H&E and Trichrome (40× objective). (H) Positive control group stained with H&E and Trichrome (40× objective). (I) Exosome-treated group (administered prior to each UVB exposure) stained with H&E and Trichrome, respectively (40× objective). (J–L) Comparative histological assessment of elastin structure at the 6-week time point. (J) Negative control group stained for elastin at 40× magnification. (K) Positive control group stained for elastin at 40× magnification. (L) Exosome-treated group (administered prior to each UVB exposure) stained for elastin at 40× magnification. Elastin fibers are indicated by blue arrowheads.
Figure 28.
(A–F) Comparison of elastin histological structure assessments at the 4-week time point. (A,D) Negative control group stained for elastin at 20× and 40× objectives, respectively. (B,E) Positive control group stained for elastin at 20× and 40× objectives, respectively. (C,F) Research group (treated with exosomes prior to each UVB exposure) stained for elastin at 20× and 40× objectives, respectively. Elastin fiber structure is indicated by blue arrowheads. (G–I) Comparative histological assessment of collagen structure at the 6-week time point. (G) Negative control group stained with H&E and Trichrome (40× objective). (H) Positive control group stained with H&E and Trichrome (40× objective). (I) Exosome-treated group (administered prior to each UVB exposure) stained with H&E and Trichrome, respectively (40× objective). (J–L) Comparative histological assessment of elastin structure at the 6-week time point. (J) Negative control group stained for elastin at 40× magnification. (K) Positive control group stained for elastin at 40× magnification. (L) Exosome-treated group (administered prior to each UVB exposure) stained for elastin at 40× magnification. Elastin fibers are indicated by blue arrowheads.
![Bioengineering 12 01129 g028a Bioengineering 12 01129 g028a]()
![Bioengineering 12 01129 g028b Bioengineering 12 01129 g028b]()
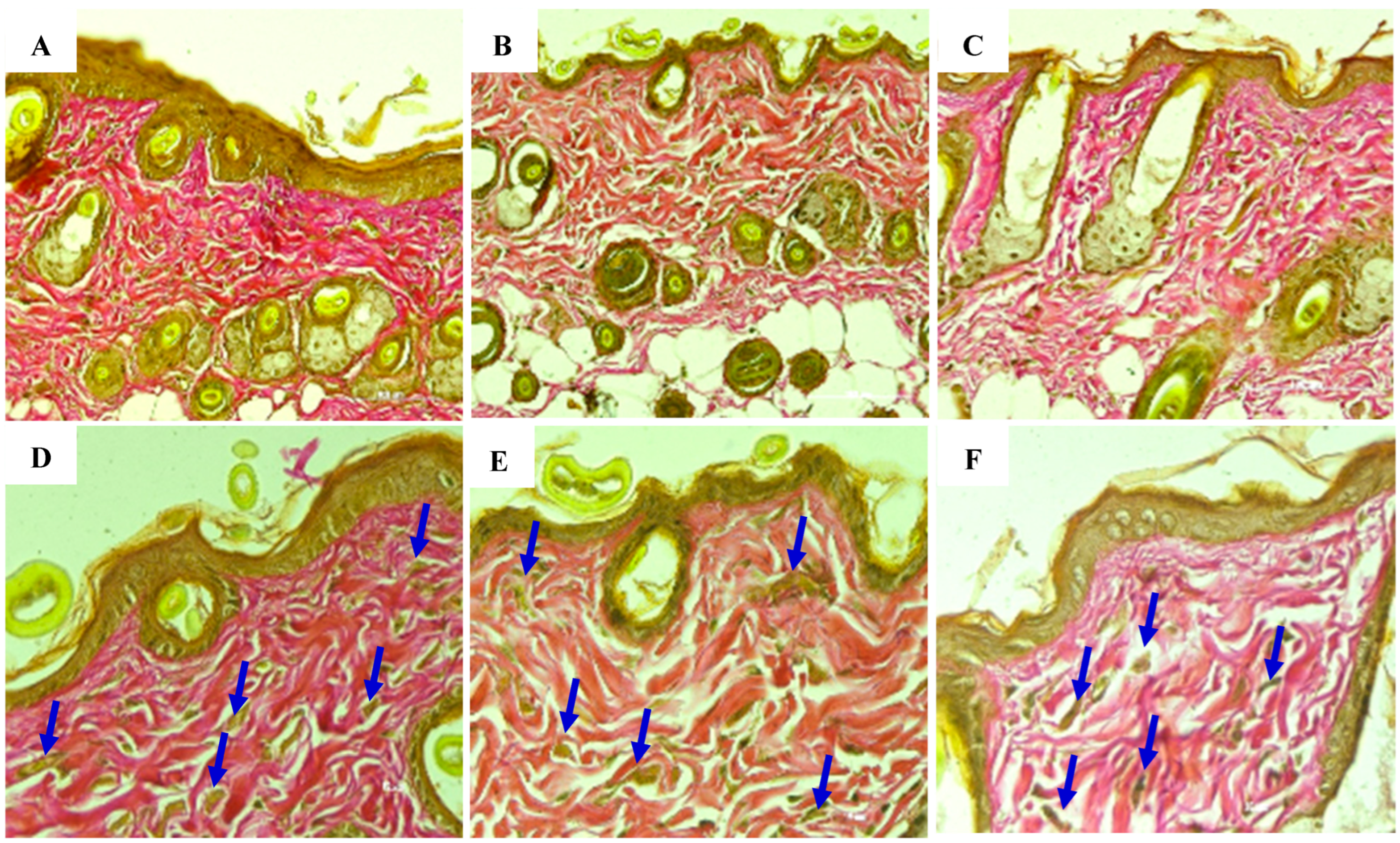
Figure 29.
(A,A1) The negative control group showed no changes in damage characteristics, meaning no damage occurred. In the positive control group, mild damage occurred in the second week, but over time the damage became more severe, resulting in extensive tissue damage by the sixth week. In the “EXO-50” group, moderate damage occurred in the second week. Over time, the damage improved, from moderate to mild by the fourth week, maintaining the status quo. This resulted in an improvement from the second to the sixth week. (A1) Histological evaluation based on the histologic wound-healing scoring system. (B,B1) Negative control group: no visible changes in the skin from week 2 to week 6. Positive control group: at week 2, there were mild but visible wrinkle lines seen even at rest. At week 4, wrinkles were well defined, with greater depth and length. At week 6, skin resulted in deep and static wrinkles with visible furrows and fixed lines. The “EXO-50” group: at week 2, models showed mild wrinkles; improvements were seen at week 4; visible at rest, fine lines were visible only during movements. At week 6, the skin improvements were persistent. (B1) Visual evaluation based on the Agrawal criteria, with a gradient ranging from 0 to 4. (C,C1). The negative control group showed no changes or disruptions in collagen arrangement. In the positive control group, we observed (from week 0 to week 2) a deep deterioration of the collagen fiber arrangement. Further worsening effects were seen at week 2 onward, resulting in disorganization and fragmentation of skin structure fibers. The “EXO-50” group showed significant improvements of the collagen fibers from week 2 onward. At week 4, loosely arranged and irregularly aligned collagen become moderately aligned and structured. Over time, the condition remined stable. (C1) Histological evaluation based on the histologic wound-healing scoring system for collagen arrangement evaluated using trichrome staining (score from 0 to 3).
Figure 29.
(A,A1) The negative control group showed no changes in damage characteristics, meaning no damage occurred. In the positive control group, mild damage occurred in the second week, but over time the damage became more severe, resulting in extensive tissue damage by the sixth week. In the “EXO-50” group, moderate damage occurred in the second week. Over time, the damage improved, from moderate to mild by the fourth week, maintaining the status quo. This resulted in an improvement from the second to the sixth week. (A1) Histological evaluation based on the histologic wound-healing scoring system. (B,B1) Negative control group: no visible changes in the skin from week 2 to week 6. Positive control group: at week 2, there were mild but visible wrinkle lines seen even at rest. At week 4, wrinkles were well defined, with greater depth and length. At week 6, skin resulted in deep and static wrinkles with visible furrows and fixed lines. The “EXO-50” group: at week 2, models showed mild wrinkles; improvements were seen at week 4; visible at rest, fine lines were visible only during movements. At week 6, the skin improvements were persistent. (B1) Visual evaluation based on the Agrawal criteria, with a gradient ranging from 0 to 4. (C,C1). The negative control group showed no changes or disruptions in collagen arrangement. In the positive control group, we observed (from week 0 to week 2) a deep deterioration of the collagen fiber arrangement. Further worsening effects were seen at week 2 onward, resulting in disorganization and fragmentation of skin structure fibers. The “EXO-50” group showed significant improvements of the collagen fibers from week 2 onward. At week 4, loosely arranged and irregularly aligned collagen become moderately aligned and structured. Over time, the condition remined stable. (C1) Histological evaluation based on the histologic wound-healing scoring system for collagen arrangement evaluated using trichrome staining (score from 0 to 3).
![Bioengineering 12 01129 g029a Bioengineering 12 01129 g029a]()
![Bioengineering 12 01129 g029b Bioengineering 12 01129 g029b]()
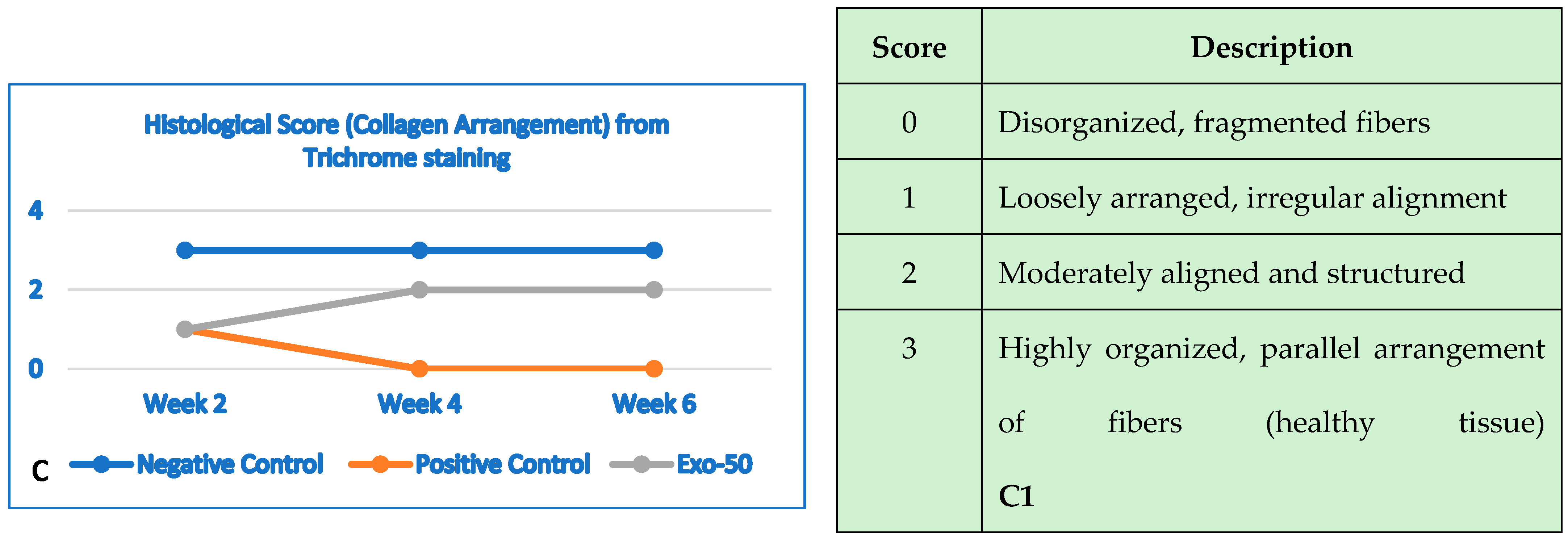
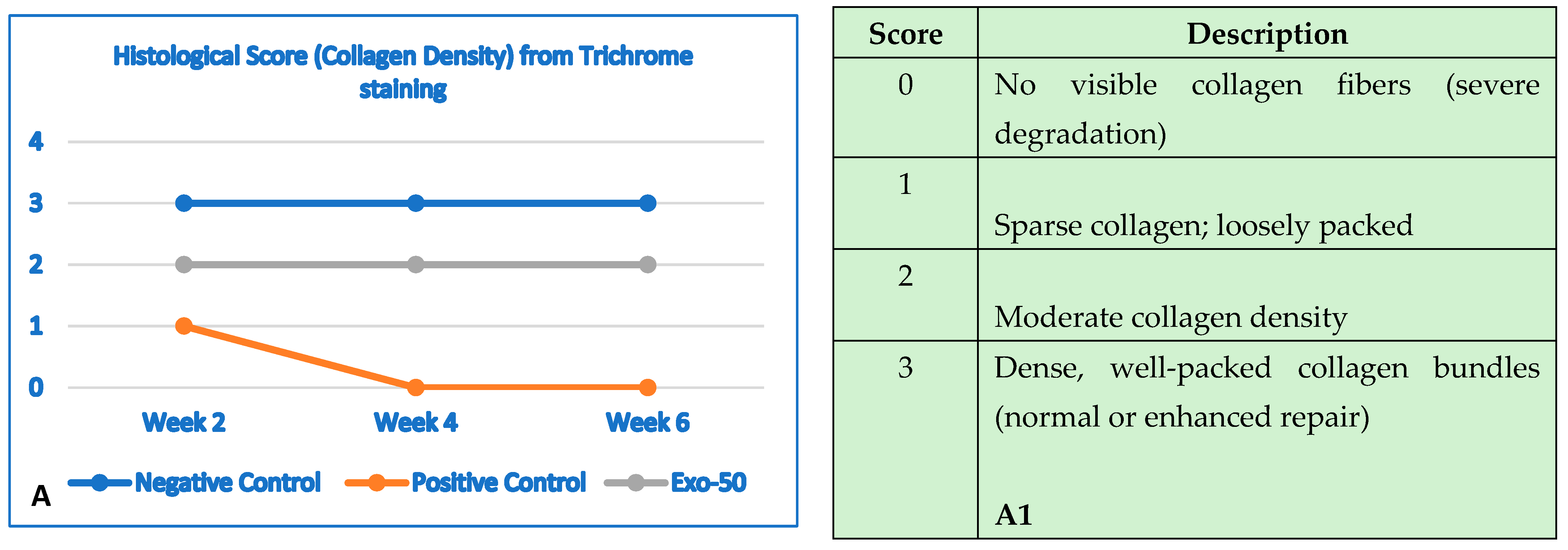
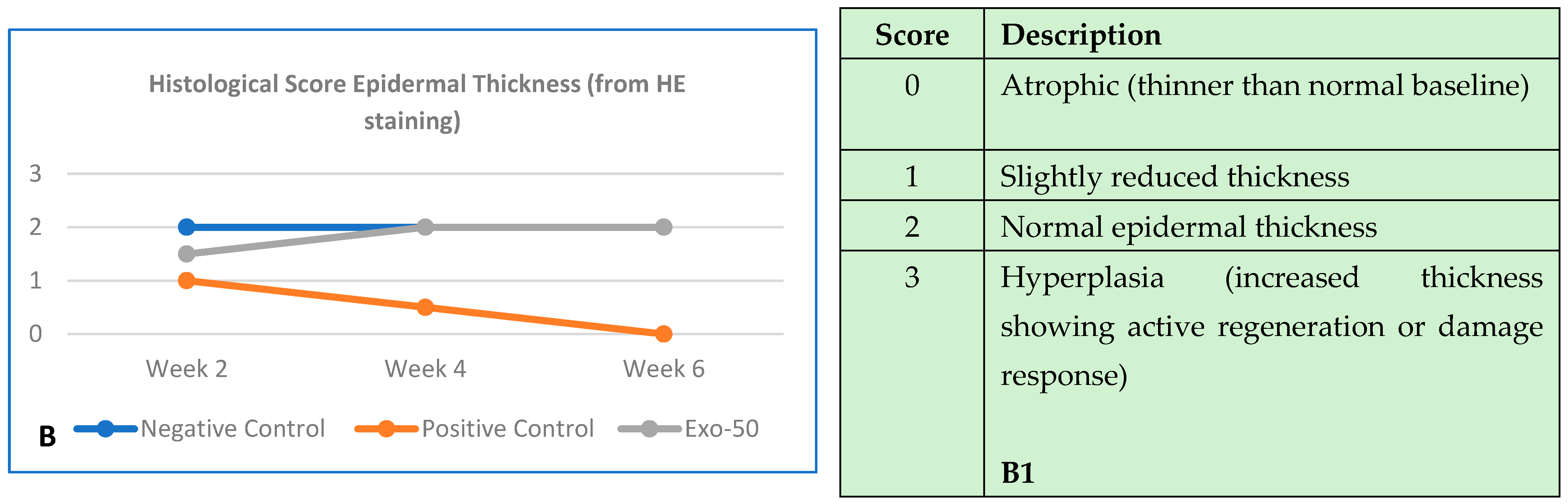
Figure 30.
(A,A1) The negative control group (blue line) showed no changes in collagen structure and organization up to 6 weeks. The positive control group (orange line) showed a gradual and significant reduction in collagen density by the end of 6 weeks. The “exo-50” group showed moderate collagen density at week 2, despite the damage, but did not experience deterioration like the positive control group (gray line). From week 2 onward, density remained moderate with no further degradation. (B) Histological evaluation based on the histologic wound-healing scoring system; here, it highlighted collagen density, evaluated by using trichrome staining (score from 0 to 3). (B,B1) The negative control group (blue line) histology outcomes showed no changes in epidermal thickness. The positive control group (orange line) showed a reduction in thickness starting by the second week throughout week 6. In the “exo-50” group, skin thickness improved in week 4, reaching normal thickness until the end of week 6. (B) Histological evaluation based the histologic wound-healing scoring system, based on HE staining (score from 0 to 3).
Figure 30.
(A,A1) The negative control group (blue line) showed no changes in collagen structure and organization up to 6 weeks. The positive control group (orange line) showed a gradual and significant reduction in collagen density by the end of 6 weeks. The “exo-50” group showed moderate collagen density at week 2, despite the damage, but did not experience deterioration like the positive control group (gray line). From week 2 onward, density remained moderate with no further degradation. (B) Histological evaluation based on the histologic wound-healing scoring system; here, it highlighted collagen density, evaluated by using trichrome staining (score from 0 to 3). (B,B1) The negative control group (blue line) histology outcomes showed no changes in epidermal thickness. The positive control group (orange line) showed a reduction in thickness starting by the second week throughout week 6. In the “exo-50” group, skin thickness improved in week 4, reaching normal thickness until the end of week 6. (B) Histological evaluation based the histologic wound-healing scoring system, based on HE staining (score from 0 to 3).
![Bioengineering 12 01129 g030a Bioengineering 12 01129 g030a]()
![Bioengineering 12 01129 g030b Bioengineering 12 01129 g030b]()
Table 1.
Data and outcomes of the experimental procedure in vivo.
Table 1.
Data and outcomes of the experimental procedure in vivo.
| Experiment Performance |
|---|
| Group | Number of Mice | Method | Visual Assessment | Histological Assessment |
|---|
| NEGATIVE CONTROL | Seven mice (from the beginning):- -
Sacrifice two mice for each 2-week period. - -
One mouse for backup, used in week 6.
| - -
No exosome exposure. - -
No UVB irradiation.
| Capture skin of two mice with digital camera at the time points of week 2, week 4, and week 6. | - -
At the time points of week 2, week 4, and week 6, conduct histological assessment (after visual assessment). - -
Histological assessment: HE staining and Trichrome staining. - -
Two mice used at each time point.
|
| POSITIVE CONTROL | Seven mice (from the beginning):- -
Sacrifice two mice for each 2-week period. - -
One mouse for backup, used in week 6.
| - -
No exosome exposure. - -
UVB irradiation.
| Capture skin of two mice with digital camera at the time points of week 2, week 4, and week 6. | - -
At the time points of week 2, week 4, and week 6, conduct histological assessment (after visual assessment). - -
Histological assessment: HE staining and Trichrome staining. - -
Two mice used at each time point.
|
| EXO-50 | Seven mice (from the beginning):- -
Sacrifice two mice for each 2-week period. - -
One mouse for backup, used in week 6.
| - -
Apply 20 μL exosomes at 50 μg/mL on shaved dorsal skin of each mouse (2 × 2 cm). - -
After 3 h from exosomes exposure, perform UVB irradiation on skin.
| Capture skin of two mice with digital camera at the time points of week 2, week 4, and week 6. | - -
At the time points of week 2, week 4, and week 6, conduct histological assessment (after visual assessment). - -
Histological assessment: HE staining and Trichrome staining. - -
Two mice used at each time point.
|
Table 2.
Skin wrinkle severity according to Agrawal criteria.
Table 2.
Skin wrinkle severity according to Agrawal criteria.
| Score | Description |
|---|
| 0 | No wrinkles or sagging; small cracks aligned with the spine. |
| 1 | Appearance of small wrinkles perpendicular to the body axis. |
| 2 | Disappearance of small wrinkles. |
| 3 | Appearance of shallow wrinkles on the back perpendicular to the body axis. |
| 4 | Presence of a few shallow wrinkles with initial signs of sagging. |
| 5 | Numerous deep, permanent wrinkles. |
| 6 | Multiple deep wrinkles with tumor/skin lesion development. |
Table 6.
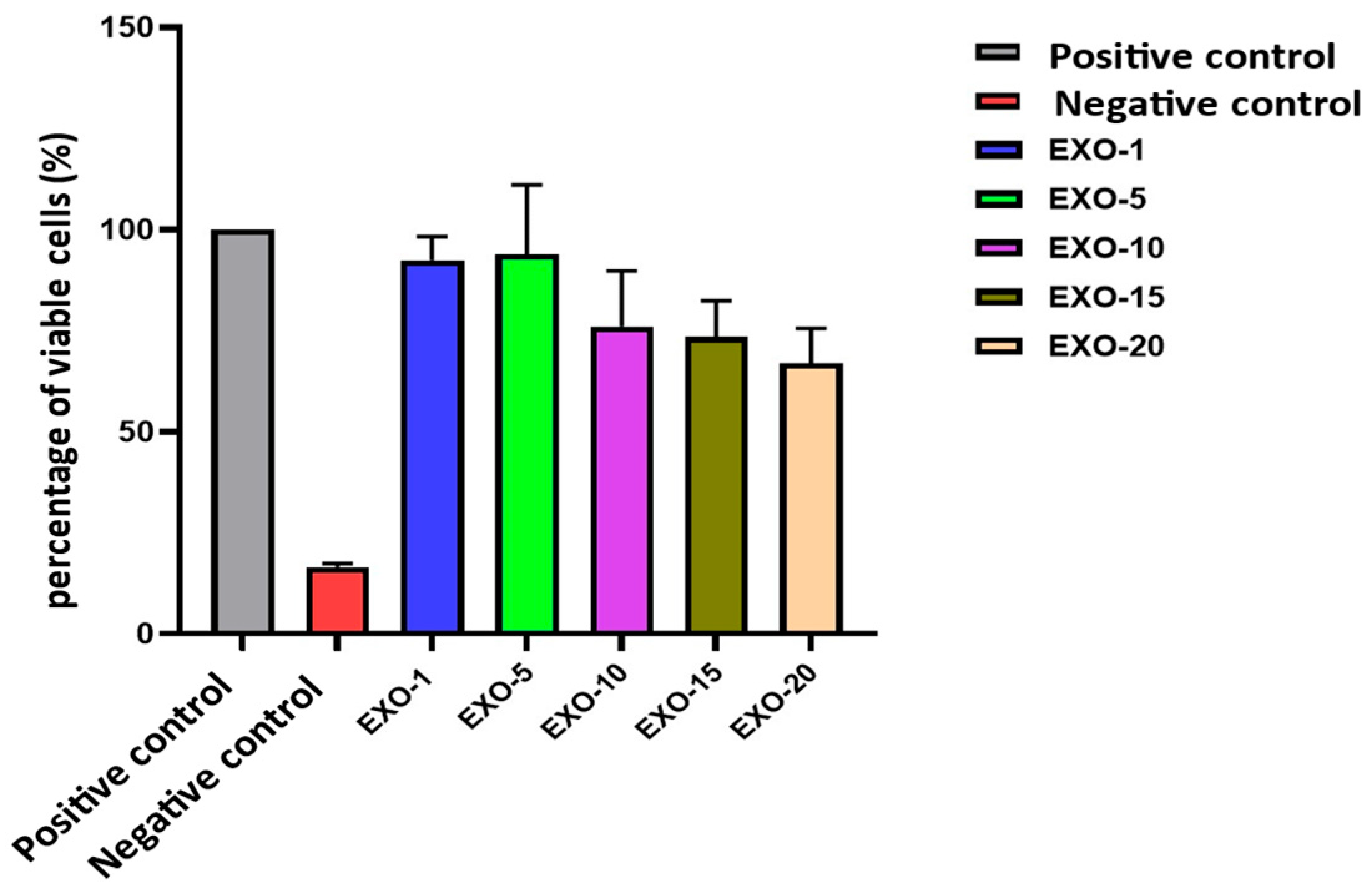
Viability assessment of exosomes on fibroblasts.
Table 6.
Viability assessment of exosomes on fibroblasts.
| No. | Experimental Group | Cell Viability (%) | Mean ± SD |
|---|
| 1 | Negative Control | 100.00 | 0.00 |
| 2 | Positive Control | 16.46 | 0.90 |
| 3 | Exo-1 | 92.42 | 5.84 |
| 4 | Exo-5 | 93.90 | 17.12 |
| 5 | Exo-10 | 76.06 | 13.72 |
| 6 | Exo-15 | 73.51 | 8.87 |
Table 7.
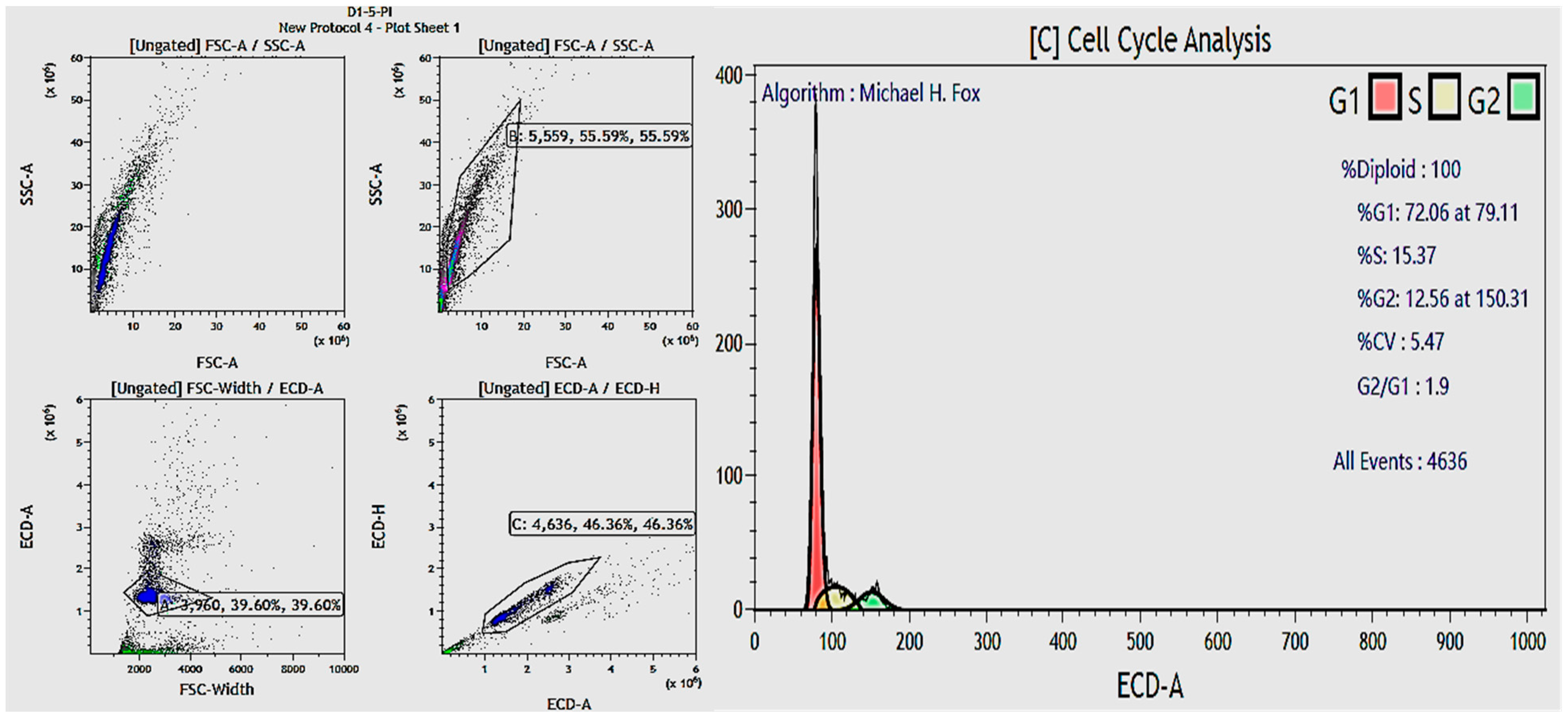
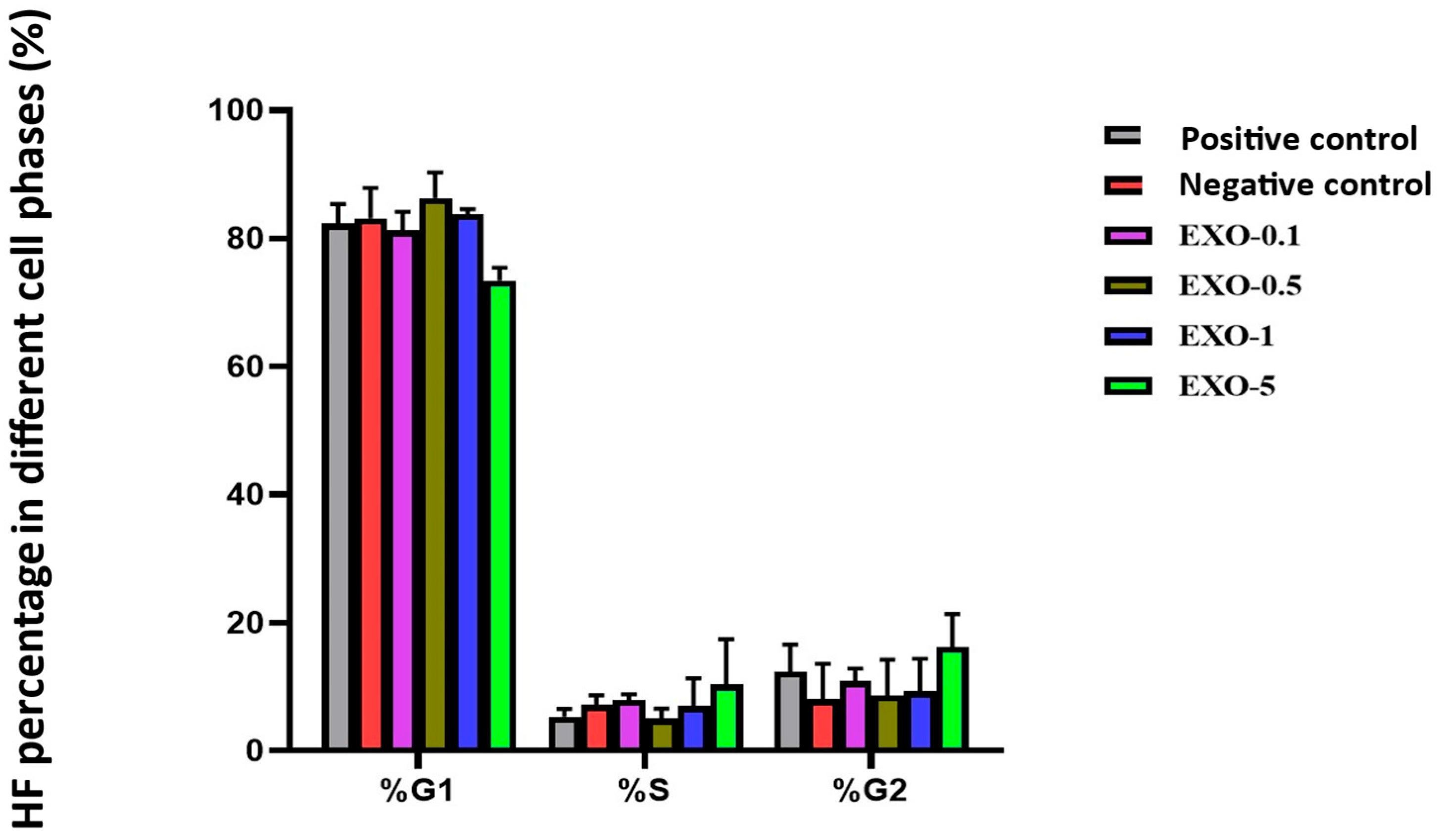
Fibroblast cell cycle evaluation.
Table 7.
Fibroblast cell cycle evaluation.
| No. | Experimental Group | % Fibroblasts in G1 Phase | % Fibroblasts in S Phase | % Fibroblasts in G2 Phase |
|---|
| 1 | Negative Control | 82.33 ± 3.05 | 5.34 ± 1.21 | 12.35 ± 4.26 |
| 2 | Positive Control | 83.16 ± 4.72 | 7.18 ± 1.51 | 8.01 ± 5.55 |
| 3 | Exo-0.1 | 81.24 ± 2.91 | 7.93 ± 0.89 | 10.84 ± 2.01 |
| 4 | Exo-0.5 | 86.34 ± 3.98 | 5.03 ± 1.58 | 8.64 ± 5.56 |
| 5 | Exo-1.0 | 83.80 ± 0.75 | 6.97 ± 4.36 | 9.24 ± 5.11 |
| 6 | Exo-5.0 | 73.47 ± 1.99 | 10.32 ± 7.14 | 16.21 ± 5.15 |
Table 11.
Comparison of collagen fiber structure in the test area at the 2-week time point.
Table 11.
Comparison of collagen fiber structure in the test area at the 2-week time point.
| Group | Collagen Density | Collagen Arrangement | Epidermal Thickness | Damage
Characteristics |
|---|
| Negative Control | Dense, intact | Well preserved and parallel to dermis | Thick dermis, no damages | Intact skin,
no inflammation |
| Positive Control | Reduced in density, numerous breaks | Disorganized,
short, fragmented | Thinned dermis due to UVB damage | Initial recovery,
mild inflammation |
| Research (exosome-treated) | Improved density compared to positive control | Reorganized,
less disordered | Thicker dermis than positive control, but below negative control | Inflammation, swelling, collagen degeneration present |
Table 12.
Comparison of collagen fiber structure of the tested area at 4 weeks.
Table 12.
Comparison of collagen fiber structure of the tested area at 4 weeks.
| Group | Collagen Density | Collagen Arrangement | Epidermal Thickness | Damage Characteristics |
|---|
| Negative Control | High, long, continuous fibers | Orderly, parallel to epidermis | Stable thickness, firm, elastic skin | Intact skin, no inflammation or damage |
| Positive Control | Further reduced, loosened structure | Short, severely disordered | Thin dermis, sagging skin, mild wrinkles | Persistent inflammation, reduced elasticity, small scars |
| Research (exosome-treated) | Recovered density, like negative control | Well organized, fewer breaks | Dermis thickness similar to negative control | No scars, good dermal recovery, lower inflammation,
less wrinkling/sagging than positive control |
Table 13.
Comparison of elastin fiber structure in the test area at 2 weeks.
Table 13.
Comparison of elastin fiber structure in the test area at 2 weeks.
| Group | Elastin Density | Elastin Fiber Structure | Skin Elasticity | Damage Characteristics |
|---|
| Negative Control | Intact, evenly distributed throughout the epithelium | Parallel, well organized | Excellent elasticity | No signs of damage, aging, or inflammation |
| Positive Control | Sharply reduced, numerous disruptions | Fragmented, shortened, heavily disrupted, forming abnormal clusters | Markedly reduced elasticity | Small wrinkles, loose connective tissue, mild sagging |
| Research (exosome-treated) | Mild recovery, higher than positive control | Longer fibers, fewer disruptions than positive control | Improved elasticity | Reduced wrinkles, firmer skin compared to positive control |
Table 14.
Comparison of elastin fiber structure in the test area at 4 weeks.
Table 14.
Comparison of elastin fiber structure in the test area at 4 weeks.
| Group | Elastin Density | Elastin Fiber Structure | Skin Elasticity | Damage Characteristics |
|---|
| Negative Control | High, long, continuous fibers | Well organized arrangement | Good elasticity | No damage, firm skin |
| Positive Control | Sharply reduced rispetto negative control, slight increase from 2 weeks | Fragmented, heavily disrupted, numerous abnormal clusters | Poor elasticity, persistent sagging | More pronounced wrinkles, reduced firmness, mild sagging |
| Research (exosome-treated) | Significant recovery with respect to positive control | Longer fibers, fewer disruptions and abnormal clusters than positive control | Markedly improved elasticity with respect to positive control | Reduced wrinkles, firmer skin, less aging than positive control |
Table 15.
Overall histological assessment of collagen and elastin from murine models at 6 weeks.
Table 15.
Overall histological assessment of collagen and elastin from murine models at 6 weeks.
| Criterion | Negative Control | Positive Control | Research Group |
|---|
| Collagen Density | High | Sharply reduced | Well recovered |
| Collagen Structure | Well organized arrangement | Disrupted, disordered | Well recovered, minimal disruptions |
| Elastin Density | High | Sharply reduced | Well recovered |
| Elastin Structure | Intact | Fragmented, deformed | Fewer disruptions, better arrangement |
| Skin Elasticity | Excellent | Sharply reduced, sagging | Markedly recovered |
| Inflammation Status | None | Mild inflammation, fibrosis | Significantly reduced inflammation |
| Wrinkles | None | Pronounced | Significantly reduced |
| Scarring/Fibrosis | None | Small scars, fibrosis | No scars, good tissue recovery |