This section presents the high evaluation of GENSIM, focusing on its effectiveness in simplifying medical texts for elderly users. The experiments are designed to assess both linguistic quality and demographic suitability, comparing GENSIM against a broad range of baseline models across multiple automatic and human evaluation metrics. We report results on three benchmark datasets, analyze component contributions through ablation studies, and demonstrate the model ability to generate outputs that are both medically accurate and cognitively accessible.
4.3. Evaluation Metrics
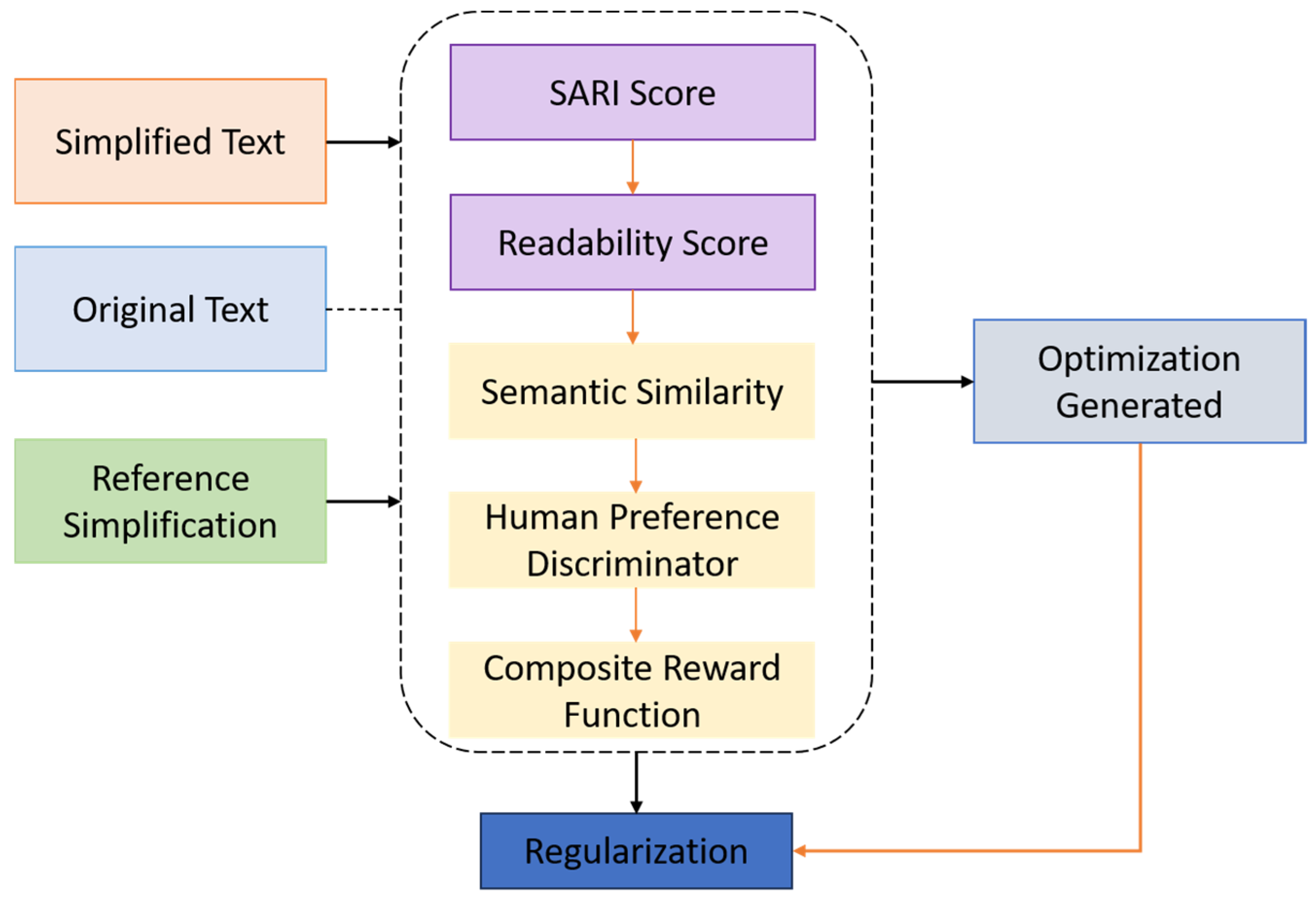
The evaluation of medical text simplification, particularly when targeting elderly populations, necessitates a comprehensive framework that captures not only lexical and syntactic simplification, but also semantic preservation, narrative quality, and demographic suitability. To rigorously assess the performance of GENSIM, we employed a combination of automatic and human-centered evaluation metrics. These metrics span three core dimensions: (i) linguistic simplification, (ii) semantic fidelity, and (iii) usability for elderly users. We utilized four widely adopted automatic metrics to evaluate simplification quality, each capturing a distinct aspect of generation performance.
SARI measures the quality of simplification by computing the n-gram overlap for three operations—additions, deletions, and copying—between the model output, the reference simplification(s), and the source sentence. Unlike BLEU, which rewards only surface similarity, SARI is specifically designed for simplification and correlates well with human judgments. Higher SARI scores indicate better simplification balance:
where
is the original sentence,
is the system output, and
is the reference.
FKGL quantifies the reading level required to comprehend a text, based on sentence length and word syllable count. It is particularly useful for aligning output complexity with age-based readability guidelines. Outputs with FKGL ≤ 6 are considered suitable for the general public, and particularly for older adults:
BERTScore uses contextual embeddings from a pretrained language model to compute semantic similarity between the generated and reference sentences. It measures how well the meaning is preserved, even when surface word forms differ significantly:
Although originally designed for machine translation, BLEU is included to report n-gram overlap between system outputs and references. It offers an upper bound on fluency and lexical matching, though it often undervalues simplification effectiveness due to its reliance on exact phrase matches.
Recognizing the inherent limitations of relying solely on automated evaluation metrics, we implemented a structured human evaluation protocol involving both domain experts—such as medical educators and geriatric healthcare professionals—and senior citizen volunteers. A randomly selected subset of 200 samples from each test corpus was independently rated by annotators using a 5-point Likert scale. The evaluation focused on three principal dimensions: the degree to which the simplified outputs preserved the critical semantic content of the original sentences, the ease with which the content could be understood based on vocabulary, sentence structure, and grammatical clarity, and the extent to which the outputs met the communicative needs and preferences of adults aged 65 and older, particularly in terms of narrative tone, empathetic phrasing, and explanatory clarity. Inter-annotator agreement, assessed via Cohen’s
κ, achieved a mean value of 0.81, indicating substantial reliability. Outputs that were identified as exhibiting over-simplification—characterized by loss of essential information or hallucinated content—were documented separately and excluded from the final average scoring to ensure fidelity and rigor in assessment
Table 3.
This dual-pronged evaluation framework ensures that the performance of GENSIM is assessed both algorithmically and experientially, enabling robust benchmarking across general NLP goals and user-centered healthcare communication outcomes. Although the use of automatic metrics can give different viewpoints, they all have certain disadvantages in the case of simplification. As an illustration, BLEU can lower the value of paraphrased outputs, SARI can assign a negative score to outputs that have only partial overlaps, and FKGL may fail to recognize semantic adequacy. We decided to use several metrics together and add human evaluation to decrease the possible errors.
4.4. Results
To comprehensively evaluate the effectiveness of GENSIM in simplifying medical texts for elderly readers, we compared it against SOTA competitive models encompassing both traditional and state-of-the-art simplification approaches. These models span multiple categories: rule-based, statistical, neural encoder–decoder, pretrained transformer-based models, and reinforcement learning-enhanced systems. All models were evaluated on the SimpleDC, PLABA, and NIH-SeniorHealth test sets using the evaluation framework.
Table 4 presents a detailed comparison of GENSIM against competitive baseline models across four core metrics: SARI, FKGL, BERTScore, and BLEU. These metrics collectively evaluate simplification quality in terms of structural modification (SARI), readability (FKGL), semantic fidelity (BERTScore), and surface fluency (BLEU). All models were evaluated on the same test sets to ensure consistency and comparability.
GENSIM achieves the highest performance across all major dimensions. With a SARI score of 47.1, it significantly outperforms the next best model, indicating a superior ability to modify sentences in a way that aligns with human references while preserving key information. Its FKGL score of 4.8 places it well below the recommended sixth-grade threshold, demonstrating exceptional readability for elderly users. Importantly, GENSIM maintains a BERTScore of 0.892, reflecting strong semantic retention, and a BLEU score of 63.2, suggesting high fluency and lexical alignment with human-authored simplifications. In contrast, models such as Alkaldi et al. [
13], MUSS [
20], and GPT-3.5 with Persona Prompt perform reasonably well in one or two dimensions but fall short in balancing all four. For instance, GPT-3.5 + Persona Prompt achieves relatively strong SARI (42.4) and BERTScore (0.880), yet its FKGL (6.0) is less favorable for elderly comprehension. Similarly, Khan et al. [
15] approach GENSIM in SARI and BERTScore but underperform on readability.
Baseline transformer models such as BART-PLABA [
16], [
17] demonstrate acceptable fluency (BLEU ~57–58) but generate outputs with higher FKGL (6.9–7.2), indicating reduced accessibility for older adults. RLHF-enhanced models such as ACCESS + RLHF (Khan et al. [
15]) show improvement over purely supervised variants but still lag behind GENSIM in SARI and FKGL, emphasizing the importance of age-targeted architectural tuning. Earlier approaches like EditNTS [
22], SIMPLER [
23], and rule-based systems provide decent simplification on paper but fail to preserve semantic meaning and fluency, as reflected in low BERTScore. Moreover, Longformer-based models such as those by Guo et al. [
25] and Sun et al. [
27]—though capable of handling long documents—underperform across all dimensions, with notably poor FKGL and BERTScore, reflecting their limitations in tailoring output to elderly users.
To better understand the contribution of each core component of the GENSIM architecture, we conducted a series of ablation experiments. These experiments systematically removed or replaced architectural modules and training strategies, allowing us to quantify their individual and synergistic impact on performance. All variants were trained and evaluated under identical conditions using the SimpleDC test set, with results reported in
Table 5.
To further assess the robustness of GENSIM across diverse areas of medicine, we conducted a stratified evaluation of its outputs on subdomains represented in the PLABA and NIH-SeniorHealth corpora, including oncology, cardiology, immunology, and geriatrics. This analysis aimed to determine whether GENSIM’s simplification quality is consistent across domains that vary in terminology density, syntactic complexity, and relevance to elderly health contexts.
Table 5 details the results for each subdomain. GENSIM kept the FKGL value lower than or equal to 6 for all subdomains, thus ensuring the texts are readable for the elderly. The materials of oncology and cardiology came out with a bit higher semantic fidelity (BERTScore 0.896 and 0.894, respectively) than that of immunology (0.885), in which more considerable terminology density had a more significant influence on simplification accuracy. Texts related to geriatrics, which were mostly extracted from the NIH-SeniorHealth corpus, indicated the closest conformity to elderly readability preferences by having the lowest FKGL (4.6) together with the highest semantic retention. These outcomes corroborate that the GENSIM model is universally applicable to different subdomains with minimal changes in its results. The most significant thing is that the model stable performance in generating outputs that fall under the sixth-grade readability level indicates that it can be used extensively in diverse healthcare communication scenarios.
The results help to support the framework’s strength and real-world value when the range of biomedical content can be very diverse.
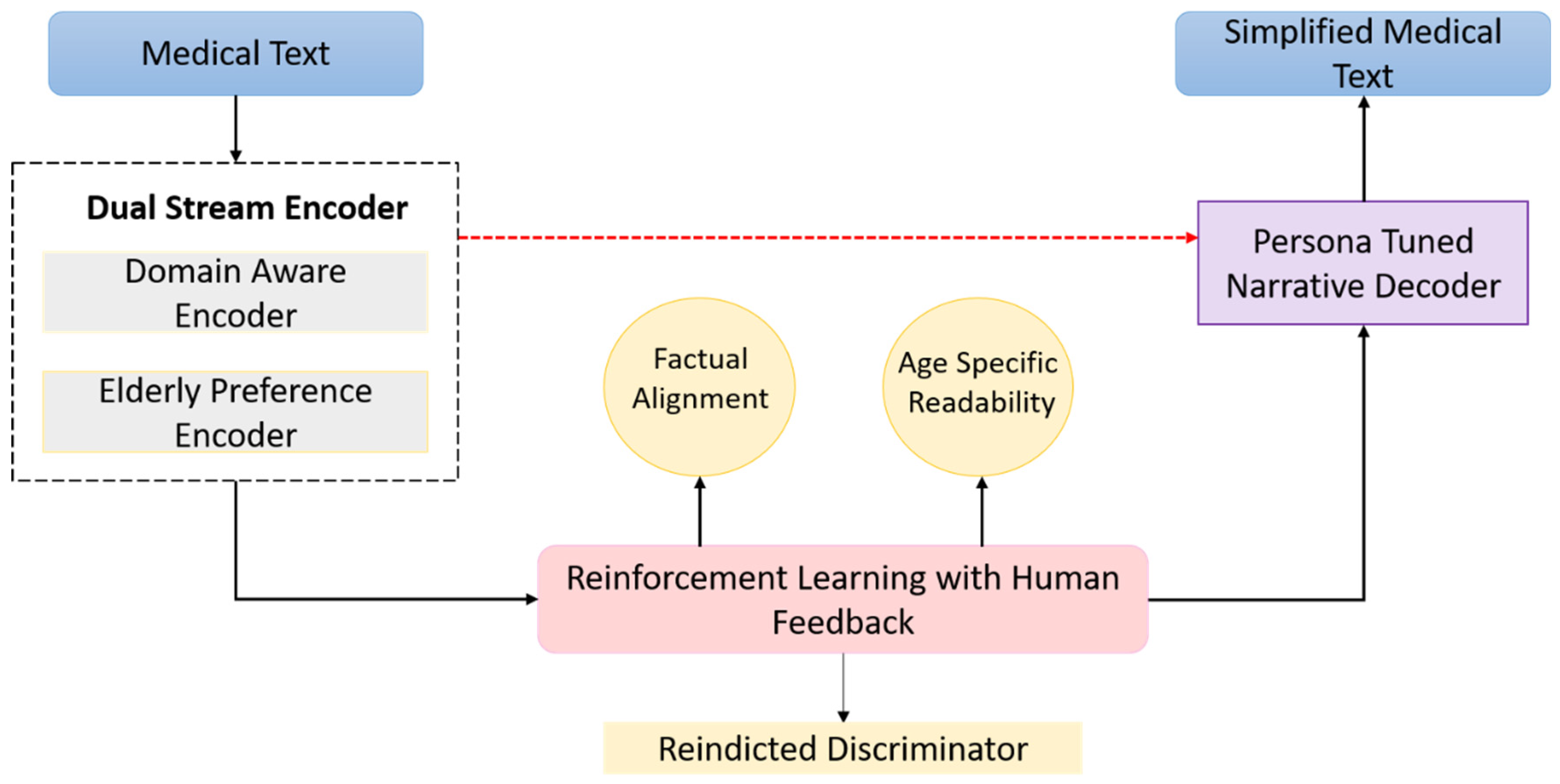
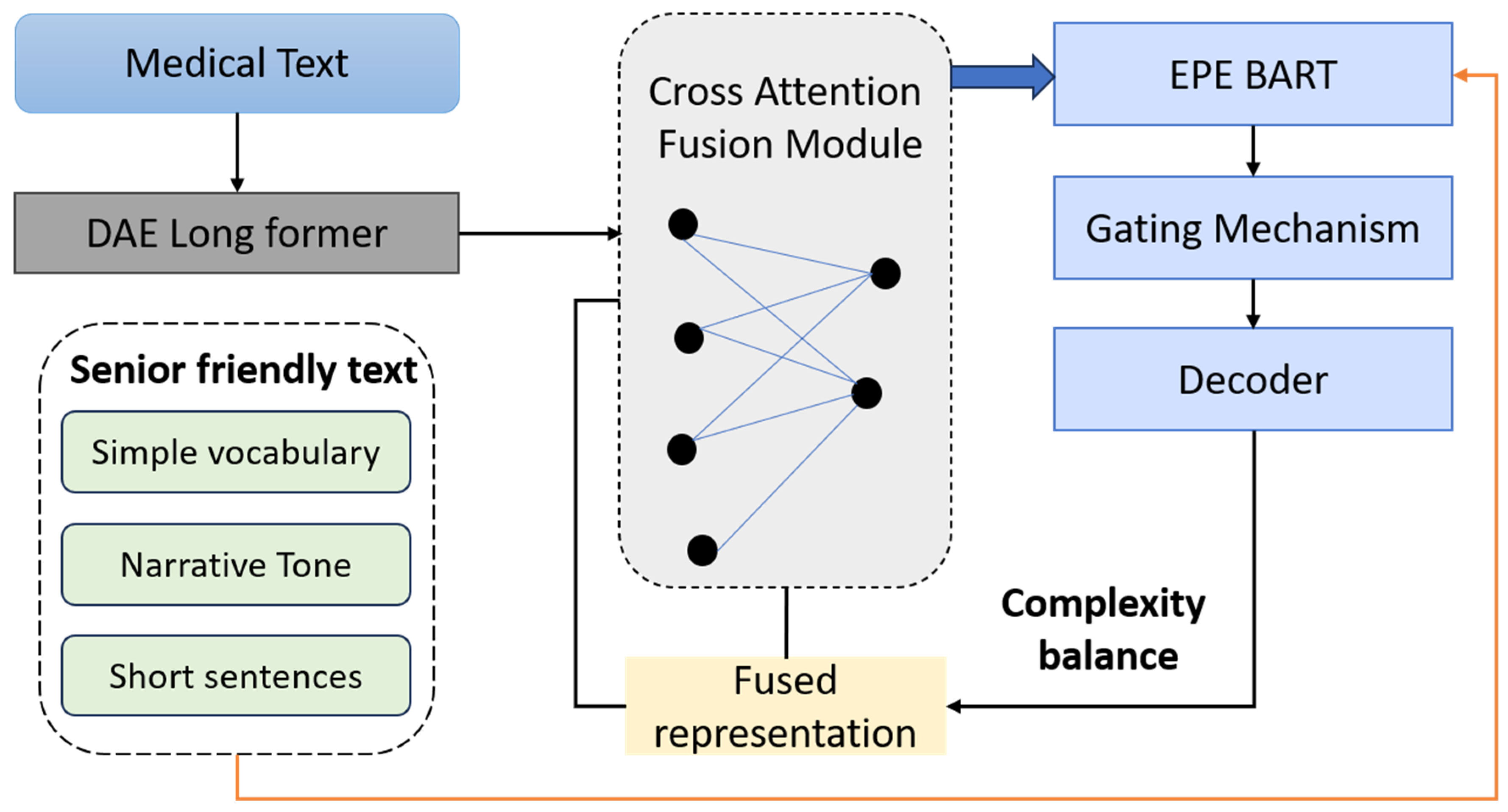
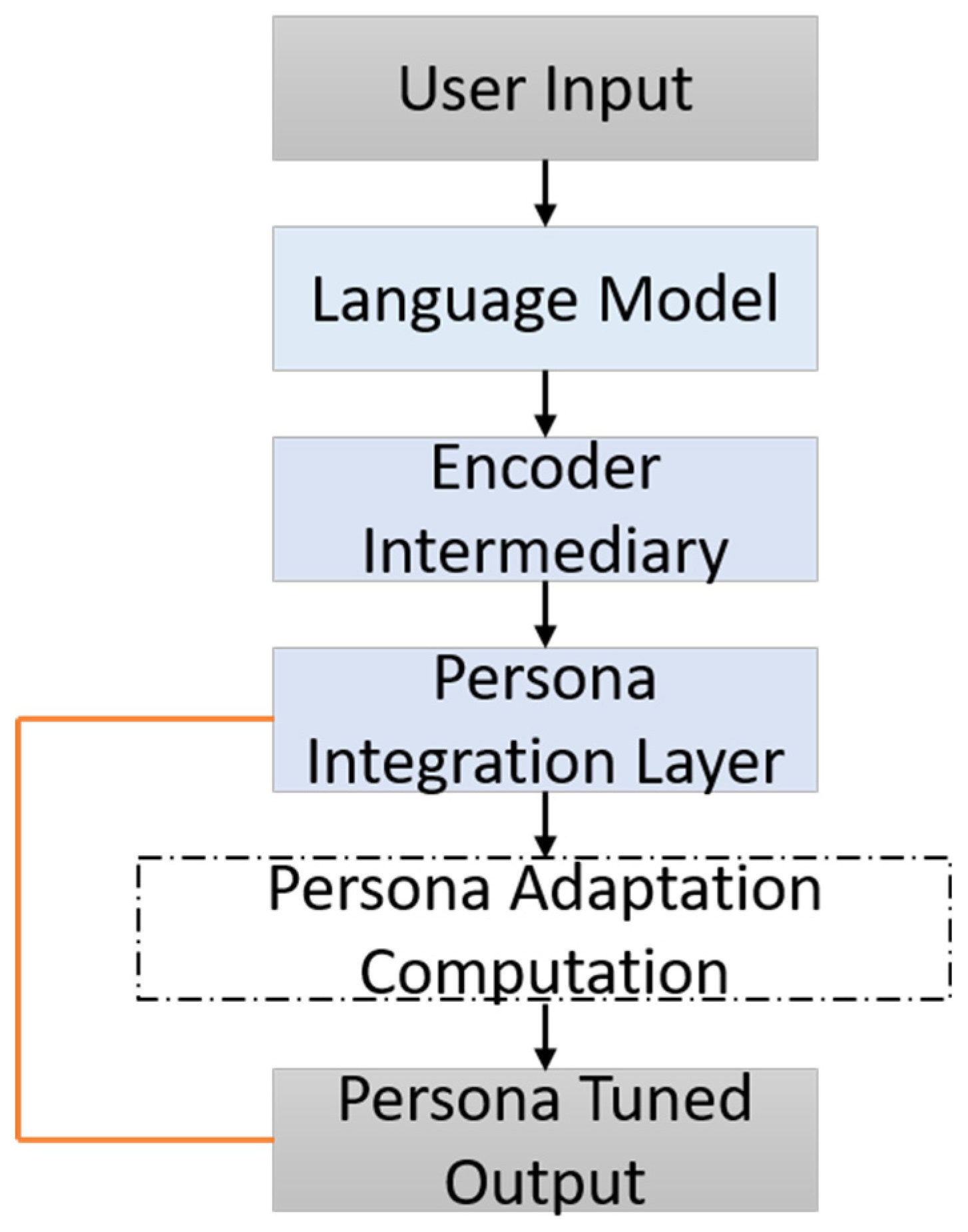
The ablation study concentrated on five principal architectural and training components integral to the GENSIM framework. First, the Dual-Stream Encoder, comprising the DAE and the EPE, operates through a cross-attentive fusion mechanism to integrate clinical accuracy with age-adapted linguistic style. Second, the RLHF module serves to optimize the generation process by aligning the model’s output with both readability constraints and semantic preservation through composite reward functions. Third, the PTND enhances stylistic adaptability by leveraging fine-tuning on elderly-specific corpora and employing conditioning cues tailored for senior-friendly expression. Fourth, the Discriminator Reward Signal introduces a human-aligned preference model that quantifies the readability and acceptability of generated texts, serving as a critical feedback signal in the RLHF loop. Finally, the use of Persona Embeddings allows for fine-grained control over narrative tone, syntactic structure, and simplification strategies, thereby enabling the model to adapt dynamically to diverse communicative goals specific to the elderly population
Table 6.
The full GENSIM model outperforms all ablated variants across all metrics, confirming the importance of each design element. Notably, removing the EPE leads to a substantial increase in FKGL, validating its role in tailoring linguistic complexity to elderly users. Moreover, removing the entire dual-stream encoder and relying solely on the domain encoder (DAE) results in the steepest drop in SARI and BERTScore, showing that semantic-narrative alignment from both encoder branches is critical for balance between simplicity and meaning preservation. Eliminating the RLHF training phase results in noticeable degradation across all metrics, particularly in SARI and FKGL, demonstrating the importance of reward-based optimization in capturing broader simplification goals that supervised learning fails to generalize. Replacing the discriminator reward with Zipf-frequency heuristics results in lower BERTScore and SARI, indicating that human-aligned feedback is superior to frequency-based lexical simplification alone.
The Persona-Tuned Decoder contributes significantly to both FKGL and BLEU, suggesting that stylistic conditioning contributes not just to readability, but also to fluency and cohesion. The loss of persona embeddings or narrative structuring leads to outputs that are more technical and less approachable, corroborated by increased FKGL and decreased BLEU. To address the limitations inherent in purely automated metrics, a structured human evaluation was conducted involving domain experts—including medical educators and geriatric health professionals—as well as senior citizen volunteers. A randomly selected subset of 200 samples from each test corpus was assessed using a 5-point Likert scale across three core dimensions: semantic adequacy, linguistic accessibility, and demographic fit. Faithfulness measured the degree to which the output preserved essential medical information such as terminology, risk disclosures, and procedural guidance. Simplicity captured perceived ease of comprehension, focusing on vocabulary, sentence construction, and grammatical clarity. Usefulness evaluated alignment with elderly communication needs, emphasizing narrative tone, supportive language, and explanatory structure. Inter-annotator agreement, computed using Cohen’s κ, achieved a mean score of 0.81, indicating substantial reliability
Table 7.
We engaged human rater groups to implement our human evaluation system that included: (i) medical educators and health professionals specialized in geriatrics (N = 6), (ii) biomedical informatics graduate students trained in health literacy (N = 4), and (iii) senior citizen volunteers aged 65+ recruited from community centers (N = 8). Annotators prepared for the task by evaluating sample sentences and using a shared rubric before rating. Each group independently rated 200 test sentences per dataset, which were randomly selected and stratified, balanced across complexity levels. For measuring consistency, we calculated Cohen’s κ for the overlapping subsets: κ = 0.84 for domain experts, κ = 0.77 for senior volunteers, and κ = 0.81 overall, denoting substantial agreement. Although the number of 200 samples per dataset is limited, this size was enough to indicate the trends across semantic adequacy, readability, and demographic suitability. The human evaluation in large-scale and multi-center settings is still an important direction for future work.
The results demonstrate that GENSIM significantly surpasses previous state-of-the-art systems, including ACCESS with RLHF and GPT-3.5 with persona prompts, particularly in SARI and FKGL—metrics most reflective of simplification quality and age-appropriate readability. While certain baseline models such as DRESS and PGN yield modest FKGL scores, their performance on SARI and BERTScore reveals deficits in semantic preservation or tendencies toward oversimplification. GPT-based systems, although fluent, exhibit inconsistency in applying simplification heuristics unless explicitly guided by structured persona cues. Rule-based systems, though achieving low FKGL scores, suffer from semantic dilution and reduced fluency, as evidenced by their lower BERTScore and BLEU metrics
Table 8.
Along with aggregate metrics, we show qualitative examples that are domain- and difficulty-stratified. Samples of sentences were taken from the test sets that are held out to represent (i) high-jargon oncology/cardiology statements, (ii) instruction-style dosage/procedure text, and (iii) common elder-relevant conditions that need plain, declarative phrasing. We removed those that are too simple and the over-simplification instances that are identified by human review. Short evaluation notes for each example that are based on (a) automatic scores and (b) a two-rater consensus regarding faithfulness, simplicity, and usefulness for 65+ are reported. This constitutes a clear connection between what was simplified and why it is indicative of real clinical communication requirements.