In this research, the Results section has been divided into three parts: Wrapper-Based Feature Selection Method and Hyperparameter Output, Ensemble Voting Classifier (Soft and Hard Voting) Analysis, and Diverse Counterfactual Explanations on Misclassified Data. The first part elaborates the best feature set based on the accuracy of each wrapper case using corresponding hyperparameter values for the RF classifier. The second part explains the use of the ensemble voting classifier to obtain better outcomes and analysis. Lastly, the third part analyzes the application of DiCE to the misclassified data obtained from the ensemble classifier. This final subsection mainly explores how much of a patient’s information needs to change in order to result in a different outcome or deviate from the actual outcome.
3.1. Wrapper-Based Feature Selection Method and Hyperparameter Output
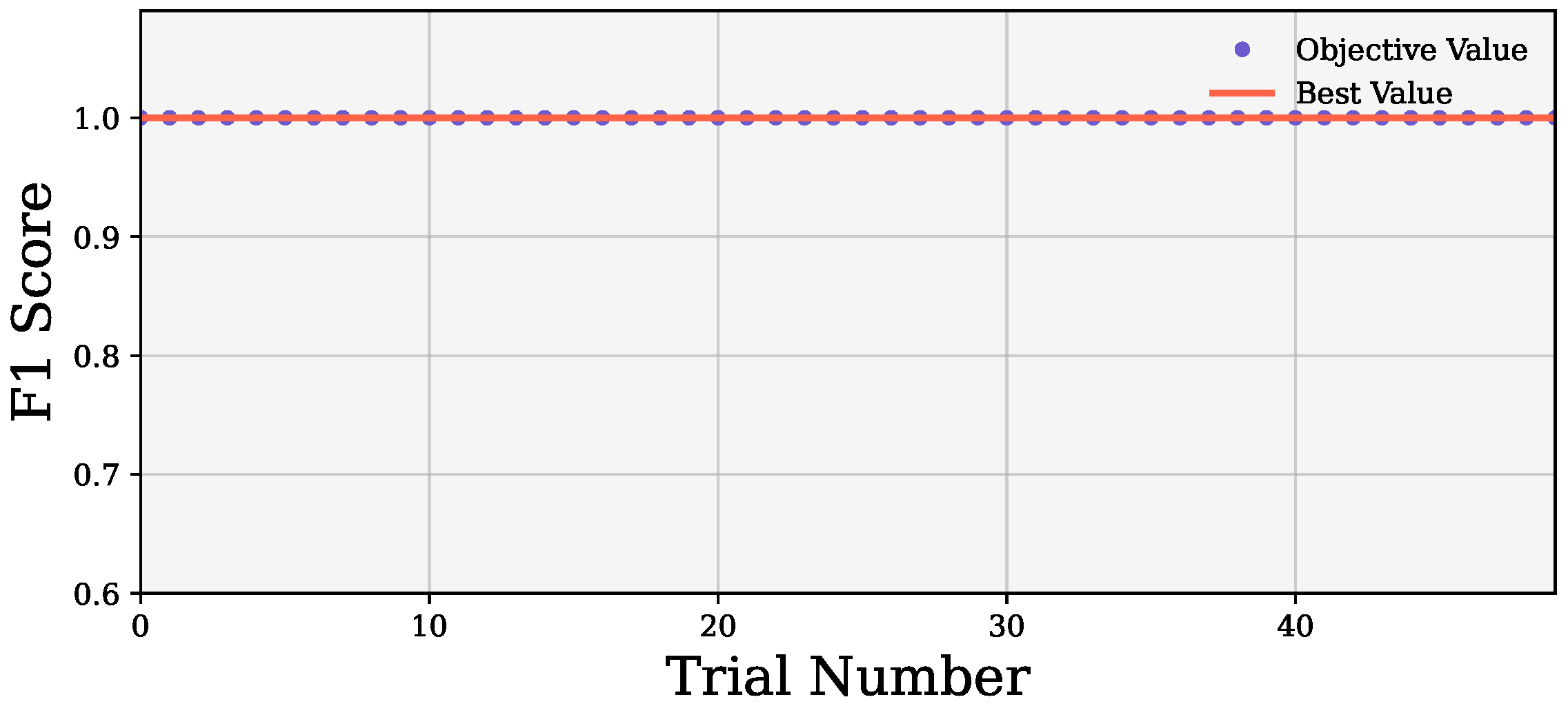
All three wrapper methods were optimized with Optuna before evaluation. We used 50 trials in Optuna to extract the best hyperparameter set. During the optimization, the models gained a perfect cost function of 1 from the first trial, as shown in
Figure 5.
Table 3 shows the selected hyperparameter set. Optuna returned the best value of the number of estimators of 144, min and max group of 5, and remove proportion of 0.5391963650868431.
3.1.1. Cross-Validation Performance
In addition, we observed the 5-fold cross-validation mean result before feature selection. In this context, we have used the hyperparameters for the RF classifier selected by the Optuna optimizer. In the 5-fold cross-validation context, the RF classifier achieved a mean accuracy of 73.99%, a mean F1-score of 67.34%, mean precision of 70.48%, and recall of 68.11%. The results are presented in
Table 4.
3.1.2. Wrapper-Based Feature Selection Performance
The parameters achieved from the Optuna result were used for each wrapper-method feature selection case and effectively determined the best features. Using the selected features, we evaluated the classification metrics of accuracy, F1-score, precision, and recall for SFS, SBS, and EFS.
(i) SFS: The SFS wrapper method selected a feature subset containing Age, BMI, Glucose, Adiponectin, and Resistin, which enabled the RF classifier to achieve an accuracy of 85.71% and F1-score of 83.87%. The model also secured a high precision of 92.85% and a recall of 76.47%, indicating the usefulness of SFS in carrying more informative features. These results reveal that SFS contributes to improved classification performance through optimal feature selection.
(ii) SBS: SBS chose a feature subset including Age, Glucose, HOMA, Adiponectin, and Resistin; however, the resulting model performance declined. The RF classifier achieved only 68.57% accuracy and 68.57% F1-score, with a precision of 66.66% and recall of 70.58%. This outcome suggests that SBS may retain less pertinent features.
(iii) EFS: EFS selected a similar optimal feature subset as SFS, resulting in corresponding classification outcomes. The RF classifier achieved 85.71% accuracy, 83.87% F1-score, 92.85% precision, and 76.47% recall using the selected features. Among the three methods (SFS, SBS, and EFS), Age, Glucose, Adiponectin, and Resistin emerged as common features, indicating their significant impact on the models. The results of each wrapper-method are presented in
Table 4.
3.2. Ensemble Voting Classifier (Soft and Hard Voting) Analysis
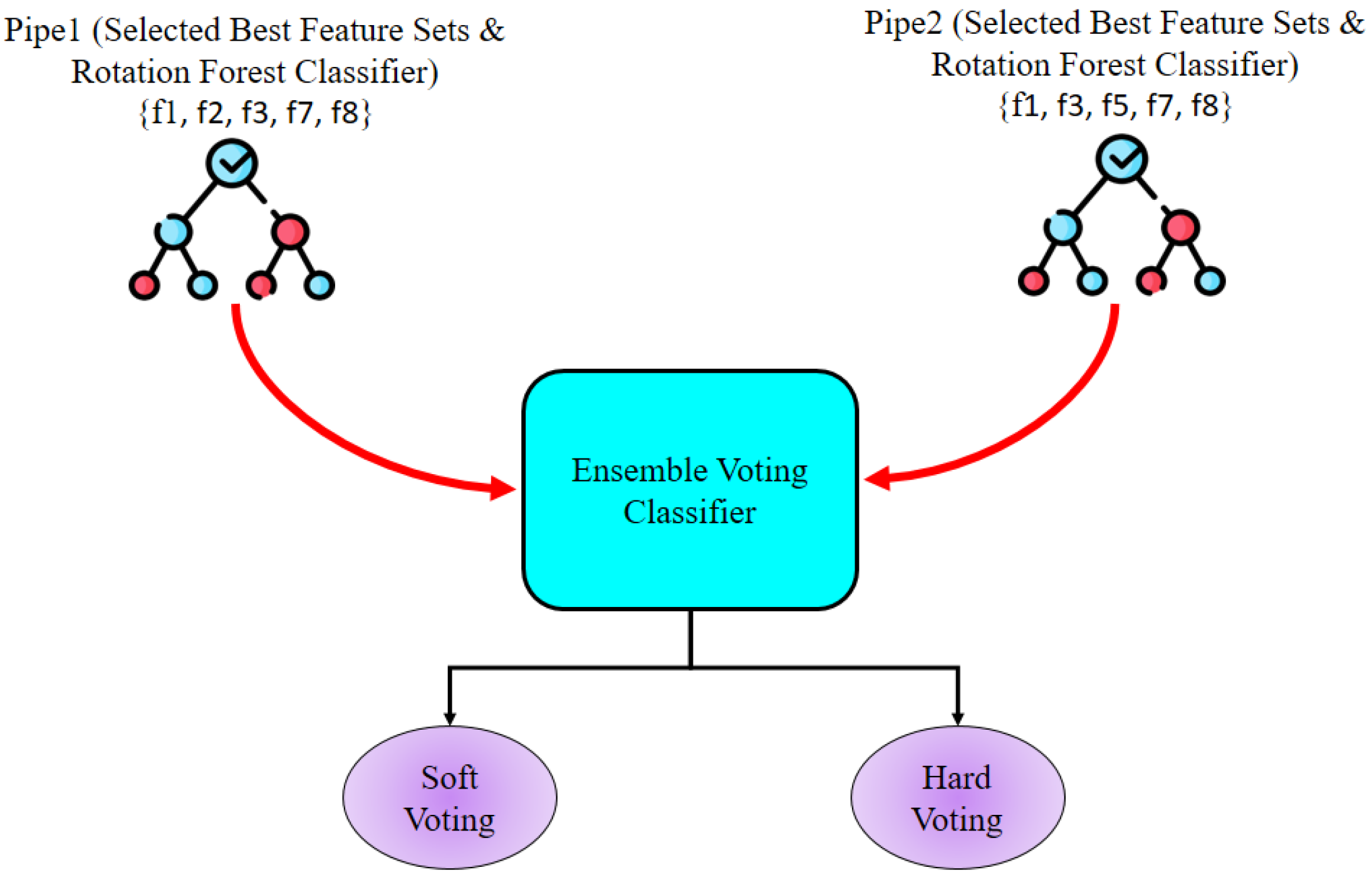
In this research, ensemble learning techniques are used to intensify the classification performance by leveraging the strengths of multiple RF classifiers. Two distinct feature subsets were selected from the three wrapper-based feature selection techniques. Because SFS and EFS provided the same results and same feature subset, one feature subset was chosen from both and another was taken from SBS. For this ensemble learning approach, two pipelines were created, denoted Pipe1 and Pipe2, with each pipeline containing a distinct feature subset and RF classifier. The RF classifiers were parameterized using the optimal hyperparameters, and the Pipe1 and Pipe2 Ensemble Voting Classifier approaches were employed. Finally, soft and hard voting strategies were applied. The overall Ensemble Voting Classifier system is represented in
Figure 6. For soft and hard voting, both cases assigned higher influence to Pipe1 before training.
Table 5 presents the comparative performance of the ensemble methods for hard voting and soft voting in terms of the most important evaluation metrics for both the training and test cases. In the training case, soft voting and hard voting both yielded 100% accuracy, F1-score, precision, and recall, which indicates that the classifier model performed better on seen data. In the test case, the hard voting method performed better with an accuracy of 85.71%, F1-score of 83.87%, and recall of 76.47%. It also showed better precision of 92.86%, indicating a lower rate of false positives. Soft voting continued to be competitive, with 80.00% accuracy, 77.42% F1-score, 85.71% precision, and 70.59% recall. These outcomes generally point towards hard voting offering more stable classification ability.
The soft and hard voting strategies were also used to explore their effectiveness and performance in terms of how much they varied. We obtained two identical feature subsets among the three wrapper-based feature selection techniques. In this case, we chose unique feature subsets as well as an identical one which was used to make the pipeline. We created two pipelines, as the two feature subsets are identical; if had we created three pipelines, then these two would be more influential, and the results might vary according to the identical features. To avoid this, we selected only one out of the two identical feature subsets.
Figure 7 presents the confusion matrices for both soft and hard voting ensemble classifiers in distinguishing between the Non-BC (Non-Breast Cancer) and BC (Breast Cancer) classes for both the training and test cases. In the training case, both soft voting and hard voting correctly classified BC and Non-BC, indicating that the ensemble voting classifier models were able to effectively fit the training data without any misclassifications. The hard voting method correctly classified more cases (17 BC and 13 Non-BC) than the soft voting model (16 BC and 12 Non-BC). Additionally, by attaining reduced misclassification rates, the hard voting classifier showed enhanced overall prediction reliability. These results are supported by the superior performance metrics of the hard voting method.
Figure 8 illustrates the Receiver Operating Characteristic (ROC) curves for the soft and hard voting ensemble classifiers. The Area Under the Curve (AUC) for soft voting is slightly higher, designating a better overall ranking performance. Although hard voting yields higher accuracy, soft voting demonstrates stronger discriminative ability across all threshold levels. The ROC curves for both soft and hard voting cases show that soft voting achieves a higher AUC of 88% compared to hard voting with an AUC of 85%, which highlights the former’s better discriminatory power. The ROC curves were developed using the test cases. The performance gap between soft and hard voting may have occurred due to their different mechanisms, as hard voting predicts the final class by majority vote whereas soft voting predicts the final class using the average predicted probabilities. This suggests that soft voting was able to generalize better under varied classification thresholds.
The misclassified examples for the soft and hard voting ensemble classifiers are shown in
Table 6. Key characteristics such as Age, BMI, Glucose, HOMA, Adiponectin, Resistin, and the label transitions shown in the “Label Pair” column have been included in each table. Hard voting classifies five cases incorrectly, while soft voting classifies seven cases incorrectly. Both cases struggle to correctly identify class 1 (Non-BC), which they tend to predict as class 2 (BC). Notably, the instances indexed 18, 24, 30, 31, and 114 were misclassified by both classifiers. The reduced number of errors demonstrates the relatively higher robustness of hard voting in classification accuracy.
Table 7 presents a comparative performance analysis between the proposed models and other classifiers. We compared our work with the XGBoost, LightGBM, and CatBoost classifier models. The Extreme Gradient Boosting (XGBoost) classifier model yielded promising results, obtaining an accuracy of 82.86%, and the model maintained scores across key evaluation metrics, with F1-score, precision, and recall all at 83.33%, highlighting its reliable and balanced predictions. The Light Gradient-Boosting Machine (LightGBM) classifier achieved an accuracy of 80%, F1-score of 78.79%, precision of 81.25%, and recall 76.47%. In addition, the CatBoost classifier achieved an accuracy of 77.14%, F1-score of 75%, precision of 80%, and recall of 70.59%.
In this context, Our Work-1, using the RF classifier 5-fold cross-validation technique, achieved a moderate accuracy of 75.43% and moderate precision of 75%, whereas its F1-score and recall results were 71.61% and 70%, respectively. The Our Work-2 model applies SFS/EFS (RF) and SBS (RF); the former demonstrated excellent performance, with accuracy of 85.71%, precision of 83.87%, recall of 92.86%, and an F1-score of 76.47%, while the latter produced slightly worse results, with an accuracy of 68.57%, precision of 68.57%, recall of 66.66%, and F1-score of 70.58%. The Our Work-3 model utilizes two ensemble voting classifiers; hard voting yielded results similar to those of SFS/EFS, with an accuracy of 85.71%, precision of 83.87%, recall of 92.86%, and F1-score of 76.47%, while soft voting showed a medium level of performance, with accuracy of 80%, precision of 77.42%, recall of 85.71%, and an F1-score of 70.59%.