NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning
Abstract
1. Introduction
2. Related Work
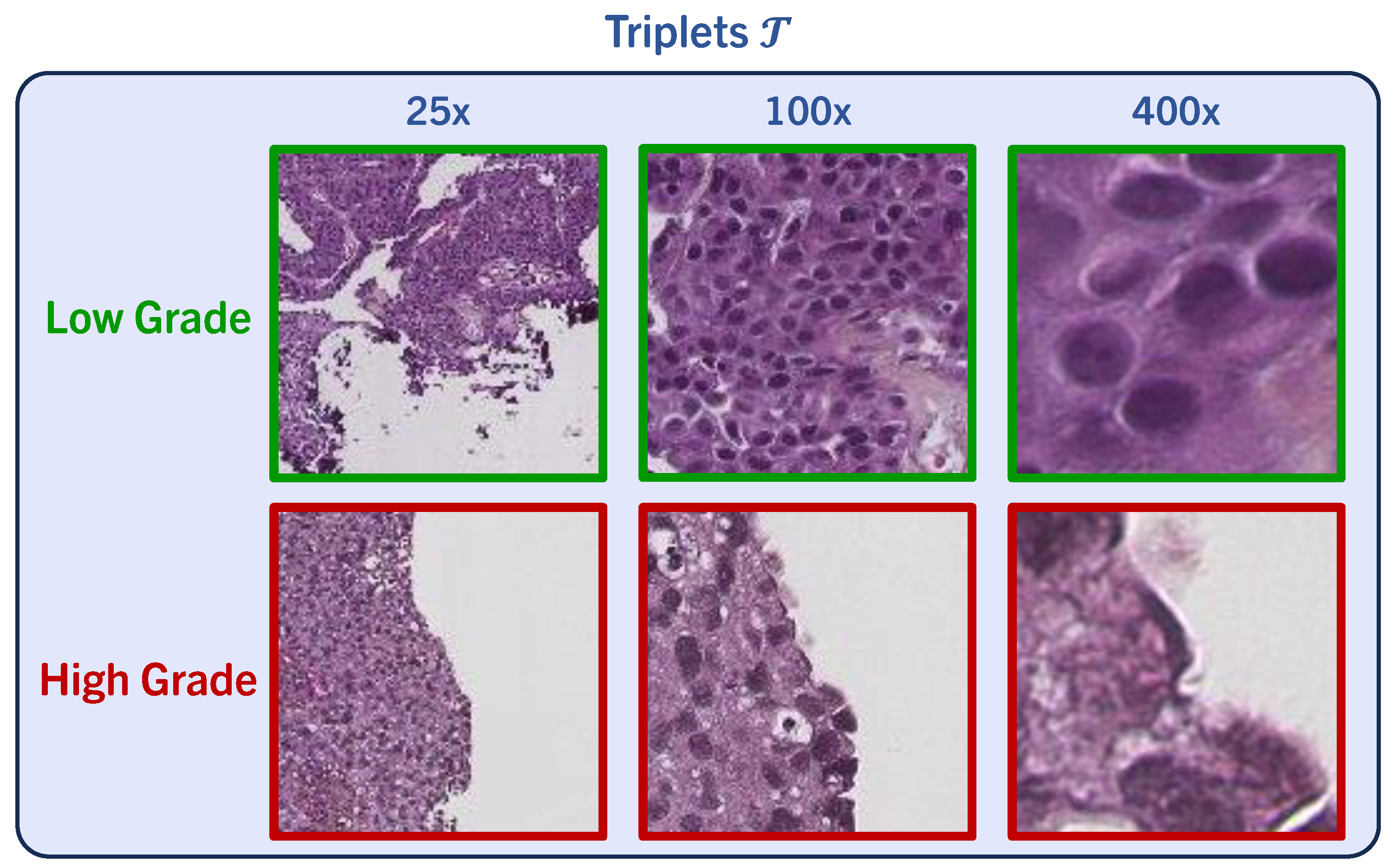
3. Data Material
4. Methods
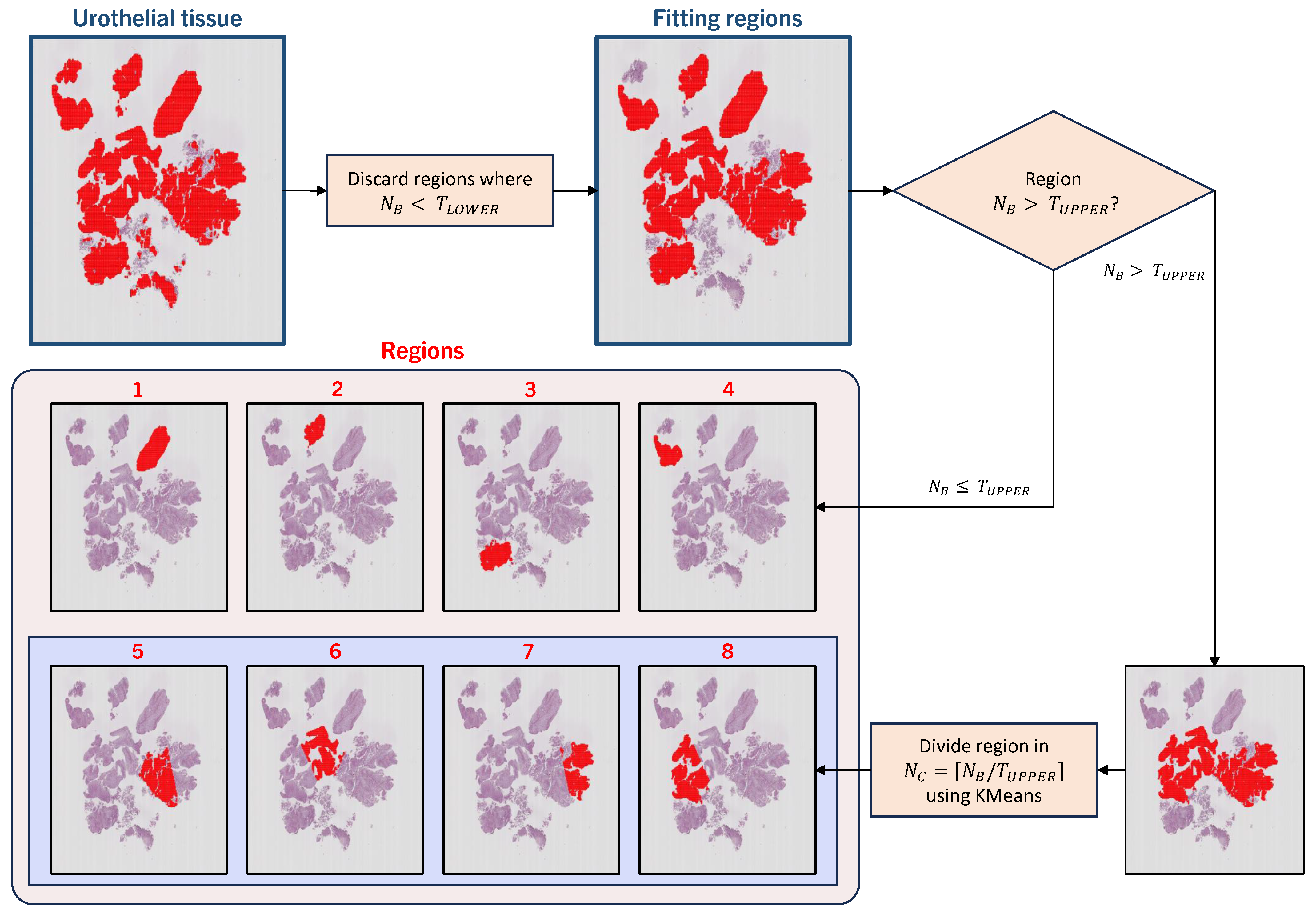
4.1. Automatic Tissue Segmentation and Region Definition
Region Definition
4.2. Multiple-Instance Learning in a WSI Context
Nested Multiple-Instance Architecture
5. Experiments
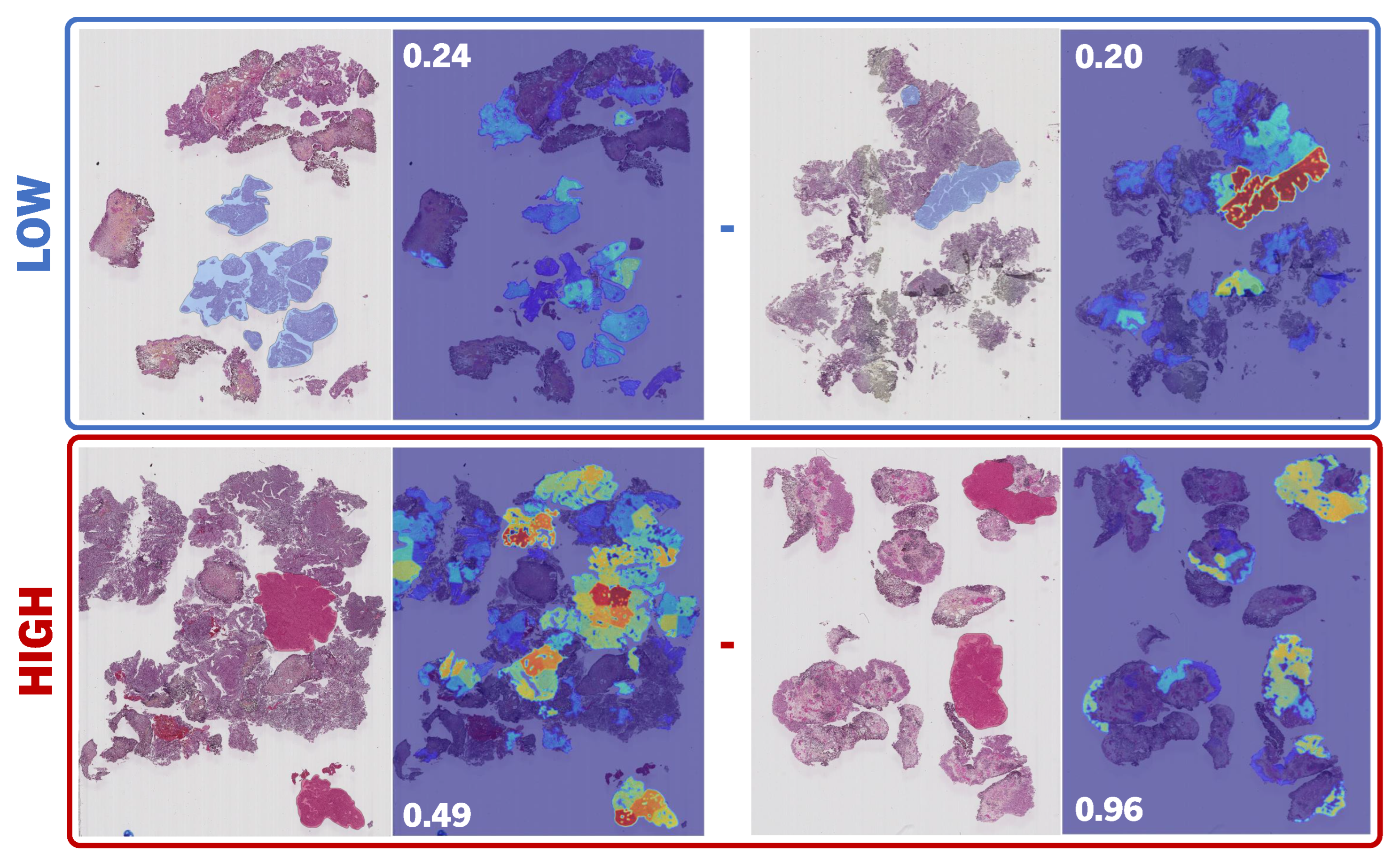
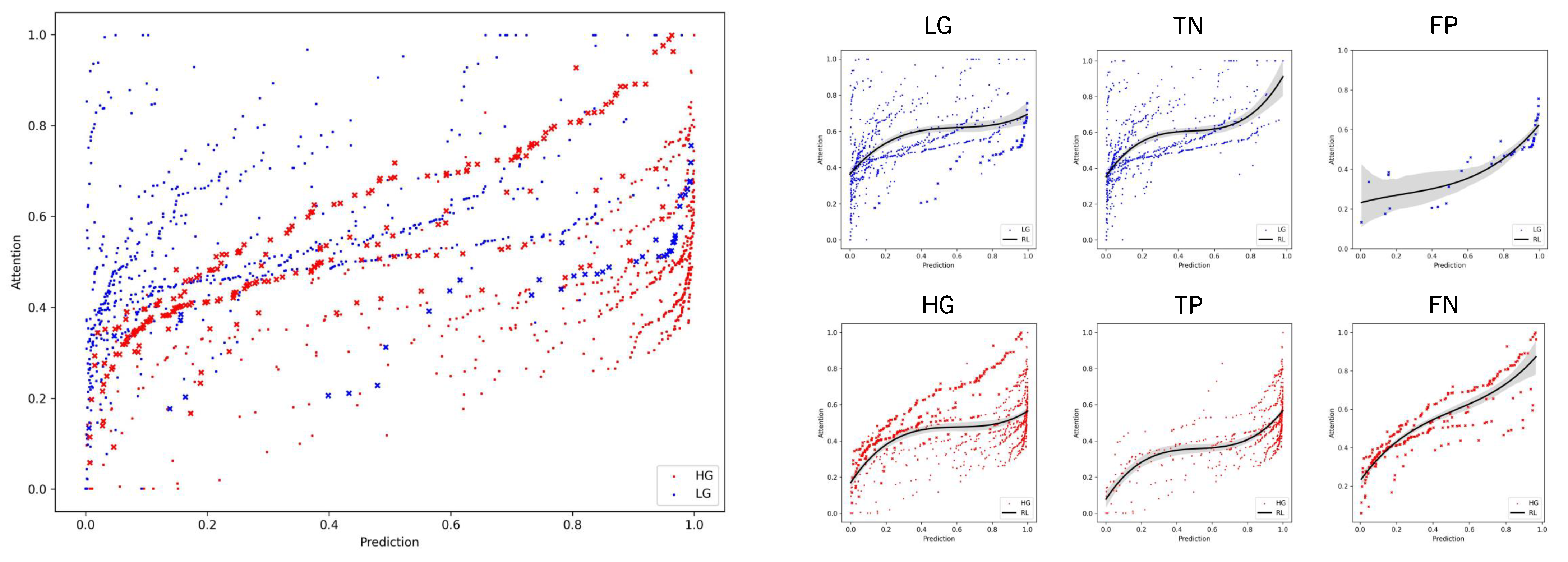
6. Results and Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Teoh, J.Y.C.; Huang, J.; Ko, W.Y.K.; Lok, V.; Choi, P.; Ng, C.F.; Sengupta, S.; Mostafid, H.; Kamat, A.M.; Black, P.C.; et al. Global trends of bladder cancer incidence and mortality, and their associations with tobacco use and gross domestic product per capita. Eur. Urol. 2020, 78, 893–906. [Google Scholar] [CrossRef]
- Burger, M.; Catto, J.W.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.A.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013, 63, 234–241. [Google Scholar] [CrossRef]
- Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Escrig, J.L.D.; Gontero, P.; Liedberg, F.; Masson-Lecomte, A.; Mostafid, A.H.; et al. European Association of Urology guidelines on non–muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ). Eur. Urol. 2022, 81, 75–94. [Google Scholar] [CrossRef]
- Eble, J. World Health Organization classification of tumours. In Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs; IARC Press: Lyon, France, 2004; pp. 68–69. [Google Scholar]
- Tosoni, I.; Wagner, U.; Sauter, G.; Egloff, M.; Knönagel, H.; Alund, G.; Bannwart, F.; Mihatsch, M.J.; Gasser, T.C.; Maurer, R. Clinical significance of interobserver differences in the staging and grading of superficial bladder cancer. BJU Int. 2000, 85, 48–53. [Google Scholar] [CrossRef]
- Netto, G.J.; Amin, M.B.; Berney, D.M.; Compérat, E.M.; Gill, A.J.; Hartmann, A.; Menon, S.; Raspollini, M.R.; Rubin, M.A.; Srigley, J.R.; et al. The 2022 World Health Organization classification of tumors of the urinary system and male genital organs—Part B: Prostate and urinary tract tumors. Eur. Urol. 2022, 82, 469–482. [Google Scholar] [CrossRef]
- Hentschel, A.E.; van Rhijn, B.W.; Bründl, J.; Compérat, E.M.; Plass, K.; Rodríguez, O.; Henríquez, J.D.S.; Hernández, V.; de la Peña, E.; Alemany, I.; et al. Papillary urothelial neoplasm of low malignant potential (PUN-LMP): Still a meaningful histo-pathological grade category for Ta, noninvasive bladder tumors in 2019? In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2020; Volume 38, pp. 440–448. [Google Scholar]
- Jones, T.D.; Cheng, L. Reappraisal of the papillary urothelial neoplasm of low malignant potential (PUNLMP). Histopathology 2020, 77, 525–535. [Google Scholar] [CrossRef]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine learning methods for histopathological image analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Cui, M.; Zhang, D.Y. Artificial intelligence and computational pathology. Lab. Investig. 2021, 101, 412–422. [Google Scholar] [CrossRef]
- Azam, A.S.; Miligy, I.M.; Kimani, P.K.; Maqbool, H.; Hewitt, K.; Rajpoot, N.M.; Snead, D.R. Diagnostic concordance and discordance in digital pathology: A systematic review and meta-analysis. J. Clin. Pathol. 2020, 74, 448–455. [Google Scholar] [CrossRef]
- Kanwal, N.; Pérez-Bueno, F.; Schmidt, A.; Engan, K.; Molina, R. The devil is in the details: Whole slide image acquisition and processing for artifacts detection, color variation, and data augmentation: A review. IEEE Access 2022, 10, 58821–58844. [Google Scholar] [CrossRef]
- Kvikstad, V.; Mangrud, O.M.; Gudlaugsson, E.; Dalen, I.; Espeland, H.; Baak, J.P.; Janssen, E.A. Prognostic value and reproducibility of different microscopic characteristics in the WHO grading systems for pTa and pT1 urinary bladder urothelial carcinomas. Diagn. Pathol. 2019, 14, 90. [Google Scholar] [CrossRef]
- van der Kwast, T.; Liedberg, F.; Black, P.C.; Kamat, A.; van Rhijn, B.W.; Algaba, F.; Berman, D.M.; Hartmann, A.; Lopez-Beltran, A.; Samaratunga, H.; et al. International Society of Urological Pathology expert opinion on grading of urothelial carcinoma. Eur. Urol. Focus 2022, 8, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Berbís, M.A.; McClintock, D.S.; Bychkov, A.; Van der Laak, J.; Pantanowitz, L.; Lennerz, J.K.; Cheng, J.Y.; Delahunt, B.; Egevad, L.; Eloy, C.; et al. Computational pathology in 2030: A Delphi study forecasting the role of AI in pathology within the next decade. EBioMedicine 2023, 88, 104427. [Google Scholar] [CrossRef]
- Ciompi, F.; Geessink, O.; Bejnordi, B.E.; De Souza, G.S.; Baidoshvili, A.; Litjens, G.; Van Ginneken, B.; Nagtegaal, I.; Van Der Laak, J. The importance of stain normalization in colorectal tissue classification with convolutional networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI), Melbourne, Australia, 18–21 April 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 160–163. [Google Scholar]
- Fuster, S.; Khoraminia, F.; Kiraz, U.; Kanwal, N.; Kvikstad, V.; Eftestøl, T.; Zuiverloon, T.C.; Janssen, E.A.; Engan, K. Invasive cancerous area detection in Non-Muscle invasive bladder cancer whole slide images. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–5. [Google Scholar]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A.; González, F. High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: Application to invasive breast cancer detection. PLoS ONE 2018, 13, e0196828. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Reis, S.; Gazinska, P.; Hipwell, J.H.; Mertzanidou, T.; Naidoo, K.; Williams, N.; Pinder, S.; Hawkes, D.J. Automated classification of breast cancer stroma maturity from histological images. IEEE Trans. Biomed. Eng. 2017, 64, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Dundar, M.M.; Badve, S.; Bilgin, G.; Raykar, V.; Jain, R.; Sertel, O.; Gurcan, M.N. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans. Biomed. Eng. 2011, 58, 1977–1984. [Google Scholar] [CrossRef]
- Andreassen, C.; Fuster, S.; Hardardottir, H.; Janssen, E.A.; Engan, K. Deep Learning for Predicting Metastasis on Melanoma WSIs. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–5. [Google Scholar]
- Tabatabaei, Z.; Colomer, A.; Moll, J.O.; Naranjo, V. Toward More Transparent and Accurate Cancer Diagnosis With an Unsupervised CAE Approach. IEEE Access 2023, 11, 143387–143401. [Google Scholar] [CrossRef]
- Deng, R.; Liu, Q.; Cui, C.; Yao, T.; Long, J.; Asad, Z.; Womick, R.M.; Zhu, Z.; Fogo, A.B.; Zhao, S.; et al. Omni-seg: A scale-aware dynamic network for renal pathological image segmentation. IEEE Trans. Biomed. Eng. 2023, 70, 2636–2644. [Google Scholar] [CrossRef]
- Jiao, P.; Zheng, Q.; Yang, R.; Ni, X.; Wu, J.; Chen, Z.; Liu, X. Prediction of HER2 Status Based on Deep Learning in H&E-Stained Histopathology Images of Bladder Cancer. Biomedicines 2024, 12, 1583. [Google Scholar] [CrossRef] [PubMed]
- Dietterich, T.G.; Lathrop, R.H.; Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 1997, 89, 31–71. [Google Scholar] [CrossRef]
- Maron, O.; Lozano-Pérez, T. A framework for multiple-instance learning. Adv. Neural Inf. Process. Syst. 1997, 10, 570–576. [Google Scholar]
- Carbonneau, M.A.; Cheplygina, V.; Granger, E.; Gagnon, G. Multiple instance learning: A survey of problem characteristics and applications. Pattern Recognit. 2018, 77, 329–353. [Google Scholar] [CrossRef]
- Mercan, E.; Aksoy, S.; Shapiro, L.G.; Weaver, D.L.; Brunyé, T.T.; Elmore, J.G. Localization of diagnostically relevant regions of interest in whole slide images: A comparative study. J. Digit. Imaging 2016, 29, 496–506. [Google Scholar] [CrossRef]
- Tibo, A.; Frasconi, P.; Jaeger, M. A network architecture for multi-multi-instance learning. In Proceedings of the Machine Learning and Knowledge Discovery in Databases: European Conference (ECML PKDD), Skopje, Macedonia, 18–22 September 2017; Springer: Cham, Switzerland, 2017; pp. 737–752. [Google Scholar]
- Tibo, A.; Jaeger, M.; Frasconi, P. Learning and interpreting multi-multi-instance learning networks. J. Mach. Learn. Res. 2020, 21, 7890–7949. [Google Scholar]
- Fuster, S.; Eftestøl, T.; Engan, K. Nested multiple instance learning with attention mechanisms. In Proceedings of the 2022 21st IEEE International Conference on Machine Learning and Applications (ICMLA), Nassau, Bahamas, 12–14 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 220–225. [Google Scholar]
- Khoraminia, F.; Fuster, S.; Kanwal, N.; Olislagers, M.; Engan, K.; van Leenders, G.J.; Stubbs, A.P.; Akram, F.; Zuiverloon, T.C. Artificial Intelligence in Digital Pathology for Bladder Cancer: Hype or Hope? A Systematic Review. Cancers 2023, 15, 4518. [Google Scholar] [CrossRef]
- Wetteland, R.; Kvikstad, V.; Eftestøl, T.; Tøssebro, E.; Lillesand, M.; Janssen, E.A.; Engan, K. Automatic diagnostic tool for predicting cancer grade in bladder cancer patients using deep learning. IEEE Access 2021, 9, 115813–115825. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Xiong, L.; Yan, D.; Xia, L.; Yuan, J.; Wang, J.; Jiao, P.; et al. Accurate Diagnosis and Survival Prediction of Bladder Cancer Using Deep Learning on Histological Slides. Cancers 2022, 14, 5807. [Google Scholar] [CrossRef]
- Jansen, I.; Lucas, M.; Bosschieter, J.; de Boer, O.J.; Meijer, S.L.; van Leeuwen, T.G.; Marquering, H.A.; Nieuwenhuijzen, J.A.; de Bruin, D.M.; Savci-Heijink, C.D. Automated detection and grading of non–muscle-invasive urothelial cell carcinoma of the bladder. Am. J. Pathol. 2020, 190, 1483–1490. [Google Scholar] [CrossRef]
- Spyridonos, P.; Petalas, P.; Glotsos, D.; Cavouras, D.; Ravazoula, P.; Nikiforidis, G. Comparative evaluation of support vector machines and probabilistic neural networks in superficial bladder cancer classification. J. Comput. Methods Sci. Eng. 2006, 6, 283–292. [Google Scholar]
- Zhang, Z.; Xie, Y.; Xing, F.; McGough, M.; Yang, L. Mdnet: A semantically and visually interpretable medical image diagnosis network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6428–6436. [Google Scholar]
- Zhang, Z.; Chen, P.; McGough, M.; Xing, F.; Wang, C.; Bui, M.; Xie, Y.; Sapkota, M.; Cui, L.; Dhillon, J.; et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 2019, 1, 236–245. [Google Scholar] [CrossRef]
- Ilse, M.; Tomczak, J.; Welling, M. Attention-based deep multiple instance learning. In Proceedings of the International Conference on Machine Learning (PMLR), Stockholm, Sweden, 10–15 July 2018; pp. 2127–2136. [Google Scholar]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Wetteland, R.; Engan, K.; Eftestøl, T.; Kvikstad, V.; Janssen, E.A. A multiscale approach for whole-slide image segmentation of five tissue classes in urothelial carcinoma slides. Technol. Cancer Res. Treat. 2020, 19, 1533033820946787. [Google Scholar] [CrossRef]
- Dalheim, O.N.; Wetteland, R.; Kvikstad, V.; Janssen, E.A.M.; Engan, K. Semi-supervised Tissue Segmentation of Histological Images. In Proceedings of the Colour and Visual Computing Symposium, Gjøvik, Norway, 16–17 September 2020; Volume 2688, pp. 1–15. [Google Scholar]
- Fuster, S.; Khoraminia, F.; Eftestøl, T.; Zuiverloon, T.C.; Engan, K. Active Learning Based Domain Adaptation for Tissue Segmentation of Histopathological Images. In Proceedings of the 2023 31st European Signal Processing Conference (EUSIPCO), Helsinki, Finland, 4–8 September 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1045–1049. [Google Scholar]
- Kvikstad, V. Better Prognostic Markers for Nonmuscle Invasive Papillary Urothelial Carcinomas. Ph.D. Thesis, Universitetet i Stavanger, Stavanger, Norway, 2022. [Google Scholar]
- Wetteland, R.; Engan, K.; Eftestøl, T. Parameterized extraction of tiles in multilevel gigapixel images. In Proceedings of the 2021 12th International Symposium on Image and Signal Processing and Analysis (ISPA), Zagreb, Croatia, 13–15 September 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 78–83. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems 25, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Abraham, N.; Khan, N.M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI), Venice, Italy, 8–11 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 683–687. [Google Scholar]
- Bergsma, W. A bias-correction for Cramér’s V and Tschuprow’s T. J. Korean Stat. Soc. 2013, 42, 323–328. [Google Scholar] [CrossRef]
- Akoglu, H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef]


| Subset | Low-Grade | High-Grade |
|---|---|---|
| Train | 124 (0) | 96 (0) |
| Validation | 17 (0) | 13 (0) |
| Test | 28 (7) | 22 (7) |
| Model | Accuracy | Precision | Recall | F1 Score | AUC | |
|---|---|---|---|---|---|---|
| AbMILMONO | 0.68 (0.07) | 0.71 (0.07) | 0.68 (0.06) | 0.67 (0.07) | 0.36 (0.12) | 0.81 (0.07) |
| AbMILDI | 0.79 (0.09) | 0.80 (0.10) | 0.78 (0.09) | 0.78 (0.09) | 0.57 (0.18) | 0.85 (0.13) |
| AbMILTRI | 0.82 (0.07) | 0.82 (0.07) | 0.82 (0.07) | 0.82 (0.07) | 0.64 (0.14) | 0.91 (0.04) |
| MEANTRI | 0.81 (0.03) | 0.83 (0.03) | 0.80 (0.03) | 0.80 (0.03) | 0.61 (0.05) | 0.92 (0.03) |
| MAXTRI | 0.80 (0.06) | 0.80 (0.06) | 0.78 (0.06) | 0.79 (0.07) | 0.58 (0.13) | 0.85 (0.06) |
| NMGradMONO | 0.68 (0.09) | 0.71 (0.08) | 0.69 (0.08) | 0.68 (0.09) | 0.37 (0.16) | 0.80 (0.06) |
| NMGradDI | 0.83 (0.03) | 0.85 (0.03) | 0.82 (0.03) | 0.82 (0.03) | 0.65 (0.06) | 0.91 (0.04) |
| NMGradTRI | 0.86 (0.03) | 0.87 (0.02) | 0.85 (0.04) | 0.85 (0.03) | 0.71 (0.06) | 0.94 (0.01) |
| Wetteland [35] | 0.90 (-) | 0.87 (-) | 0.80 (-) | 0.83 (-) | - | - |
| Jansen [37] | 0.74 (-) | - | 0.71 (-) | - | 0.48 (0.14) | - |
| Zhang [40] | 0.95 (-) | - | - | - | - | 0.95 (-) |
| Output | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Attention | 0.76 | 0.81 | 0.69 | 0.75 |
| Prediction | 0.89 | 0.83 | 0.91 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuster, S.; Kiraz, U.; Eftestøl, T.; Janssen, E.A.M.; Engan, K. NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning. Bioengineering 2024, 11, 909. https://doi.org/10.3390/bioengineering11090909
Fuster S, Kiraz U, Eftestøl T, Janssen EAM, Engan K. NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning. Bioengineering. 2024; 11(9):909. https://doi.org/10.3390/bioengineering11090909
Chicago/Turabian StyleFuster, Saul, Umay Kiraz, Trygve Eftestøl, Emiel A. M. Janssen, and Kjersti Engan. 2024. "NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning" Bioengineering 11, no. 9: 909. https://doi.org/10.3390/bioengineering11090909
APA StyleFuster, S., Kiraz, U., Eftestøl, T., Janssen, E. A. M., & Engan, K. (2024). NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning. Bioengineering, 11(9), 909. https://doi.org/10.3390/bioengineering11090909









