1. Introduction
Electroretinography (ERG) is a non-invasive form of assessing the functional health of the retina through its response to light stimulation. The stimulation is presented as a series of interval-based light pulses, which trigger varying responses based on the state of retinal adaptation and the wavelength, duration, strength, and stimulating frequency of the light pulse [
1]. Cone photoreceptors, which are responsible for photopic or ’daytime’ vision, require more quanta for activation compared to the rod photoreceptors that function in scotopic or ’night-time’ vision, requiring fewer quanta to activate [
1,
2]. The responses from the photoreceptors and post-receptoral neurons (bipolar, horizontal, amacrine, and ganglion cells) all contribute to the overall size and shape of the recorded one-dimensional (1D) ERG signal [
1,
3].
Several ERG recording methods, including full-field flash, pattern, and multifocal, can be utilized for the early detection and diagnosis of a wide variety of retinal-related diseases, including early diabetic retinopathy, glaucoma, retinal dystrophies, and age-related macular degeneration [
1,
2,
4,
5,
6].
Typically, the full-field ERG (ffERG) signals last about 250 ms and have a frequency range of 0 to 300 Hz [
7]. This test is crucial for assessing the functionality of the retina, which is essential for vision. As illustrated in
Figure 1, the ERG signal primarily consists of two main components: the a-wave and the b-wave. The a-wave is the initial negative deflection in the ERG signal and is generated by the retina’s photoreceptor cells (rods and cones). Following the a-wave, the b-wave is a positive deflection produced by the inner retinal cells, mainly the bipolar and Müller glial cells. These waves are crucial for understanding the retina’s response to light stimuli. The main characteristics of these waves are their amplitudes and time to peak. The amplitude of the a-wave (Va) and the b-wave (Vb) refers to the height of these waves, measured in microvolts (
). These amplitudes reflect the strength of the response generated by the retinal cells. The time to peak of the a-wave (Ta) and the b-wave (Tb) represents the time it takes for these waves to reach their maximum height after the light stimulus, measured in milliseconds (ms) [
1]. These time-domain features are essential for diagnosing various retinal conditions. In addition to the main a- and b-wave components, the ERG signal can also include other components such as the Oscillatory Potentials (OPs) and the Photopic Negative Response (PhNR). The OPs are high-frequency wavelets superimposed on the ascending limb of the b-wave. They are thought to originate from the inner retinal layers, particularly the amacrine cells, and are useful for evaluating inner retinal function. The PhNR is the negative wave following the b-wave peak and is shaped by the retinal ganglion cells. An additional test protocol is the Flicker ERG, recorded using a pulse presented at 30 Hz. The response primarily assesses the cone system’s functionality; it is responsible for color vision and visual acuity under photopic conditions and is useful for diagnosing cone-related disorders [
8].
A number of different ERG signals may be extracted based on the different electrophysiological protocols and clinical applications [
9]. The scotopic 0.01 ERG response is obtained under dark-adapted conditions and is generated by rod photoreceptors with a dominant b-wave and minimal a-wave. The scotopic 2.0 ERG has a stronger flash strength when presented under dark-adapted (DA) conditions and has a mixed rod–cone response. Under light-adapted (LA) conditions, the photopic 2.0 ERG response is a cone-driven response of typically smaller amplitude owing to the fewer cones in the human retina [
1].
Currently, the most widely used form of ERG analysis and feature extraction is time-domain analysis, which involves the identification of the
a- and
b-wave amplitudes and their corresponding time to peaks, usually by algorithms that find the peaks automatically that can then be checked by the clinician [
1,
2]. However, the time-domain features do not fully reveal the underlying energy contributions of the neural generators (photoreceptors, bipolar, amacrine, horizontal, and retinal ganglion cells). So, alternative methods using signal analysis have been explored to deconstruct the signal further [
10]. These methods include Power Spectral Density (PSD) and Fourier Transform, as well as time–frequency-domain methods such as the Short-Time Fourier Transform (STFT), and Continuous and Discrete Wavelet Transforms [
2]. Although these methods have yet to be explored as extensively as the time domain, they offer a more detailed analysis and additional features than those provided by pure time-domain analysis.
Regarding time–frequency analysis, the predominant research has been on Wavelet Transforms, with limited exploration of the Short-Time Fourier Transform (STFT) in analyzing ERG signals. Thus, this study uses STFT as an additional signal analytical approach to the ERG. As referenced in
Section 2, the existing literature has predominantly employed STFT as a complementary technique to Wavelet Transform methods as a comparison. This highlights the opportunity to delve deeper into the potential benefits and insights that STFT could offer in the overall analysis of ERG signals.
STFT could be selected as the most interpretable of the transformations mentioned above. The spectrogram is a 2D representation of the signal with the time on the horizontal axis and the frequency on the vertical axis, which can be given as follows:
where
is the fast Fourier Transform of spectrum calculation;
corresponds to the representation of the input signal
x with the window function
w (with given length and form) for time position
and frequency position
f. Let us denote that in general the spectrum can be expressed as
[
11].
Equation (
1) provides a linear, unambiguous, and reversible relationship between the input (
x) and output results (
). The power spectrum density for the next processing can be given as
due to the complex origin of the equation [
11].
STFT uses a sliding overlapping window function to convert the signals to a time–frequency domain using the fast Fourier Transform (FFT) algorithm. This produces a
spectrogram representation with the time on the horizontal axis, the frequency on the vertical axis, and the amplitude/power represented as a color map.
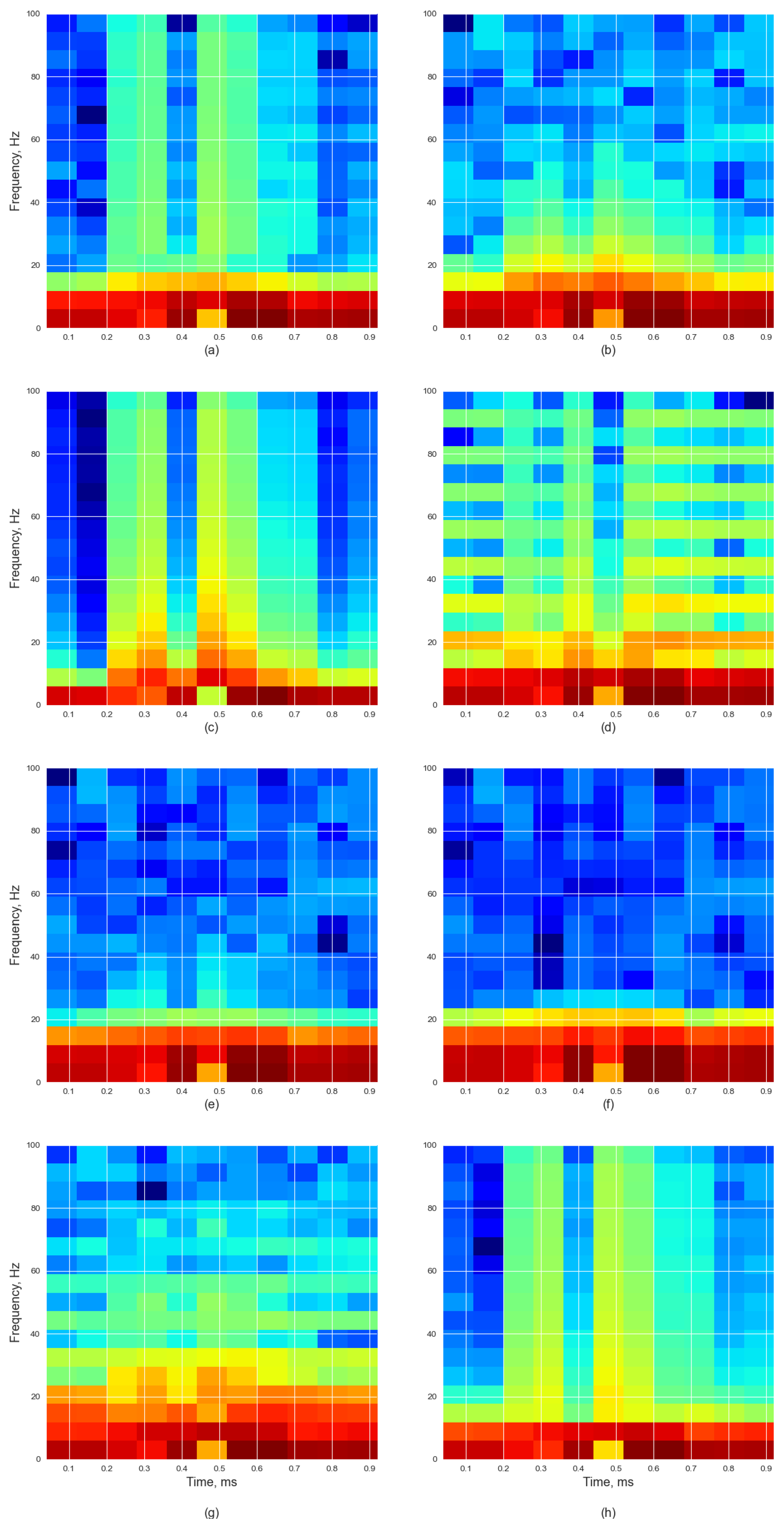
Figure 1 depicts a healthy (top) and an unhealthy (bottom) signal with dystrophy in the time domain and their corresponding spectrogram representations calculated using STFT. We can see that the spectrogram gives us the signal frequency, which is in the range of 0–100 Hz, along with the time the frequency occurs and how much power that frequency contains, with red representing higher power frequencies and blue representing lower power frequencies.
The STFT spectrogram shows the energies within each frequency band from 0 to 100 Hz. The horizontal axis of the spectrogram denotes time bins (in milliseconds), and the vertical axis represents frequency bands in Hertz (Hz). It should be noted that the black arrows in
Figure 1b show that the spectrogram frequency distribution is from 0 to 50 ms and 0 to 20 Hz (maximum energy), from 60 to 80 ms and 0 to 10 Hz (medium energy), and from 0 to 67 ms and 15 to 30 Hz (low energy). The black arrows in
Figure 1d show that the spectrogram frequency distribution is from 0 to 80 ms and 0 to 15 Hz (maximum energy), from 0 to 80 ms and 15 to 25 Hz (medium energy), and from 25 Hz (low energy). The key difference between the healthy signal (
Figure 1b) and the unhealthy signal (
Figure 1d) is evident in the energy distribution across the frequency bands. The healthy signal shows a more diverse energy spread with the maximum energy occurring at higher frequencies (0–20 Hz) compared to the unhealthy signal, where the maximum energy is concentrated at lower frequencies (0–15 Hz). This indicates that unhealthy signals tend to have more energy concentrated in the lower frequency bands, suggesting a potential marker for identifying signal health.
This study compared various window functions for optimal feature extraction using STFT and spectrogram generation to classify the signals and determine which window yielded the best features for ERG signal classification. Several combinations of window function, window size, and window overlap were used to extract spectrogram images to train deep learning (DL) models, and manual feature extraction, which was used to train classical machine learning (ML) models. The results from both approaches were compared to determine which window yielded the best signal classification and whether DL had an advantage over the classical ML approaches. The main contributions of this study were the use of different window parameter combinations for feature extraction and the application of DL for classifying the extracted spectrogram images.
The paper is organized as follows:
Section 2 reviews relevant studies and those employing STFT for feature extraction in similar fields.
Section 3 presents the materials and methods used for the study, which include the ERG signal database and the pipeline for signal processing, feature extraction using the STFT, model building, and evaluation.
Section 4 and
Section 5 describe the results obtained from the analyses using multiple evaluation metrics and discuss the outcomes, and, finally,
Section 6 concludes with the implications of the findings for the analysis of the ERG and future directions.
2. Related Works
ERG analysis methods can be divided broadly into three different approaches: time-domain analysis, which involves analyzing the amplitudes and time to peaks of the signal; frequency-domain analysis, which involves studying the frequencies of the signal; and time-frequency analysis, which involves studying the signal’s frequencies at the time they occur along with their power and nonlinear methods [
2]. Time-domain analysis, for the most part, is the most popular method used in the literature because it is fast and usually provides differences in amplitude or time to peak when there is a retinal disease. However, subtle or early functional changes may not be evident in time-domain analysis initially, such as in diabetes and glaucoma, thus the application of signal analysis may improve earlier diagnosis in both. In addition, signal analysis may also support classification between groups in early neurological disorders [
10].
Several studies have used frequency-domain methods to analyze ERG signals. These methods provide a different perspective on signal analysis by providing spectral information unavailable in the time domain. Most studies in the frequency domain use the FT with the FFT algorithm [
12] to convert the signal into the frequency domain before analyzing it. A few other methods, namely Power Spectral Density (PSD) and Linear Prediction (LP), have also been used. In [
7,
13], Gur et al. were able to find similarities between corneal and non-corneal ERG signals by using FFT and LP to identify specific frequencies in normal corneal ERGs under different conditions. After studying the Oscillatory Potentials (OPs) from the ERG signals of diabetic patients using the FFT, Vander Torren et al. [
14] concluded that it was possible to express OPs quantitatively even in pathologies. Similarly, by studying photopic and scotopic ERGs in the Fourier spectrum and comparing them to the time domain, Li et al. [
15] were also able to highlight differences in the dominant frequency and power between the scotopic and photopic ERGs. In a different study, Sieving et al. [
16] used discrete Fourier Transform (DFT) to study Flicker ERGs cycle by cycle, extracting real-time harmonic components.
Using Welch’s Power Spectral Density (PSD), Karimi et al. [
17] were able to find significant differences in the frequency components in the scotopic and photopic ERGs of patients with and without retinitis pigmentosa. To search for signs of retinal pathologies in patients with stage I and II open-angle glaucoma, Zueva et al. [
18] analyzed the frequency responses from Flicker and pattern ERGs by decomposing them into a Fourier Series.
While the frequency-domain methods mentioned above provide spectral information about ERG signals, they need to improve significantly regarding temporal information, which is crucial for ERG analysis. Time - frequency domain methods offer a way to obtain both spectral and temporal information from the signals and represent it in a or format. Unlike the classical FT, STFT allows us to visualize the signal’s frequencies, the time window at which they occur, and how strong that frequency is at that point in time. This allows us to extract multi-dimensional features that are otherwise not accessible in the time domain or frequency domain alone.
To the best of our knowledge and findings, virtually all time-frequency ERG analysis studies are based on Continuous and Discrete Wavelet Transforms, except very few studies that included STFT as part of the analysis. In [
19], STFT was one of the time–frequency methods used along with Continuous Wavelet Transform (CWT) and Discrete Wavelet Transform (DWT) to analyze the photopic ERG signals obtained from a healthy subject. In [
20], STFT was applied along with CWT and DWT to analyze the effects of obesity on ERG signals. Three different responses (cone, rod, and maximal combined) were analyzed, and features were extracted using STFT, CWT, and DWT, after which the results from these methods were compared. In [
21], STFT and DWT were used to determine the frequency components of the three photopic and Flicker 30Hz ERG signals of patients with Central Retinal Vein Occlusion (CRVO). More recently, ref. [
8] used CWT to manually extract features from adult and pediatric signals, which were used to train a Decision Tree classifier by combining time-domain features (a- and b-wave amplitudes and implicit times) with the wavelet features. In a similar study to this paper, ref. [
22] compared several mother wavelet combinations to determine which combination would better classify pediatric ERG signals.
It is worth noting that a significant drawback of STFT is its time–frequency resolution trade-off, which stems from the uncertainty principle (Gabor limit in signal processing) [
23]. This means that it is impossible to achieve a high resolution for both the time and frequency components of the signal simultaneously; hence, a compromise has to be made between the two. Thus, the larger the window size, the better the frequency resolution and the lower the time resolution, and the smaller the window size, the better the time resolution, but the lower the frequency resolution.
Table 1 provides several frequency and time–frequency domain methods used for analyzing ERG signals previously, as well as the types of ERG signals used for the studies.
The analyses presented in
Table 1 indicate a prevalent preference among researchers for the FT in frequency-domain analysis and the Wavelet Transform in time–frequency-domain analysis. This inclination towards the Wavelet Transform in time–frequency-domain analysis may stem from considering the time–frequency resolution trade-off. As different mother wavelets in Wavelet Analysis can impact signals uniquely, the STFT also exhibits variations in signal representation based on factors such as the chosen window function, window size, and overlaps between windows during signal processing. This warrants further investigation to ascertain the efficacy of STFT in ERG signal analysis, given its nuanced response to different signal characteristics.
5. Discussion
STFT is a method based on the FT proposed as a solution to the lack of temporal information from the classical FT method. The use of windows that slide across the signal helps extract the spectral and temporal information of the signal and present it in the form of a spectrogram. However, since it is impossible to obtain high temporal and spectral resolution simultaneously, it is necessary to determine which window, window size, and overlap between the sliding window would yield the most optimal features of the ERG signal for classification. As shown in
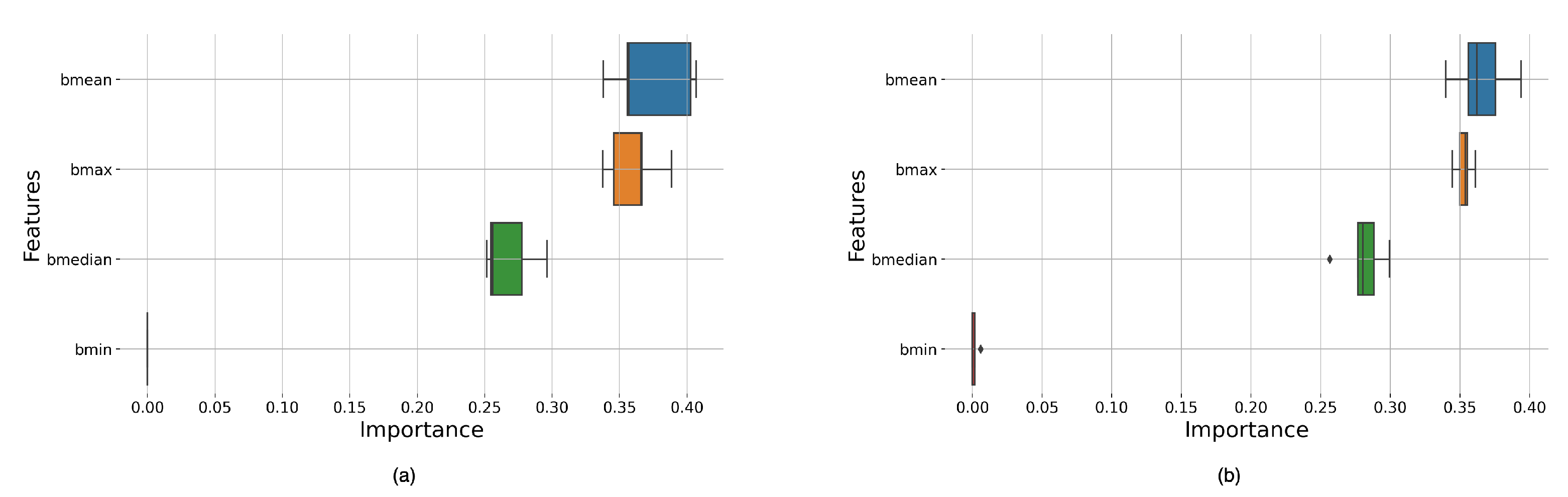
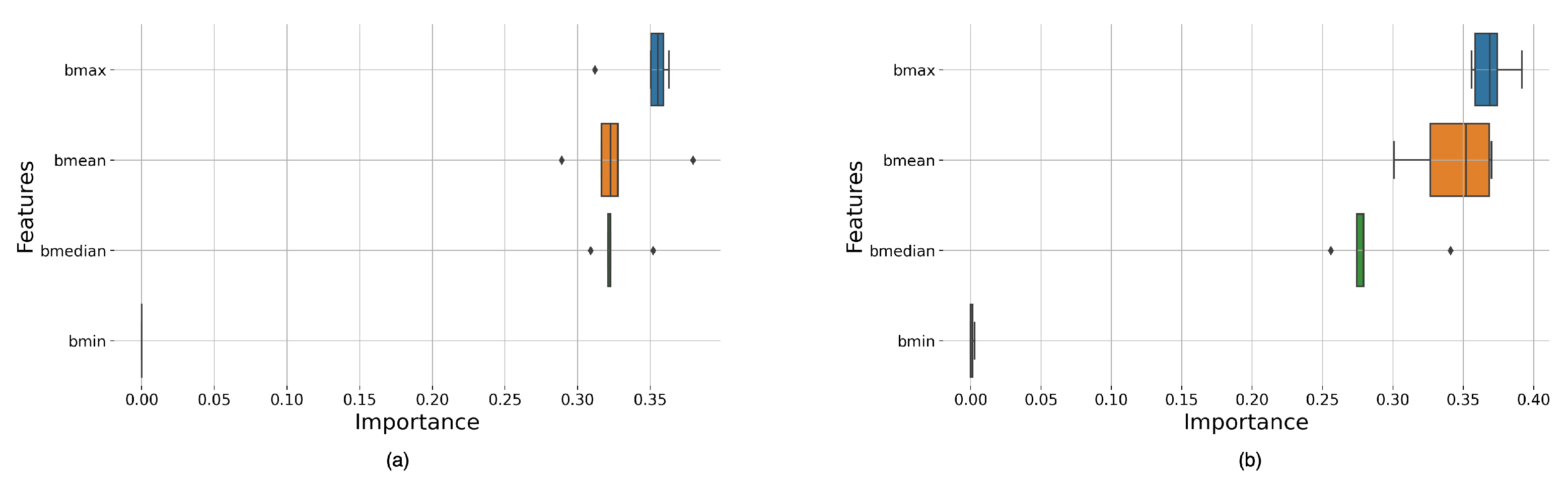
Appendix A, there was very little difference in the results for the window functions studied. However, we can see significant differences in performance depending on window size and overlap.
The classifiers with the best results were those with larger window sizes; given that larger window sizes provided better frequency resolutions, this could indicate that signals with higher frequency resolutions produce the most optimal features.
We can also observe that the RF-based models outperformed the DT-based models. This was expected given that RF uses an ensemble of DTs, hence being able to capitalize on multiple trees rather than a single individual tree to make its predictions.
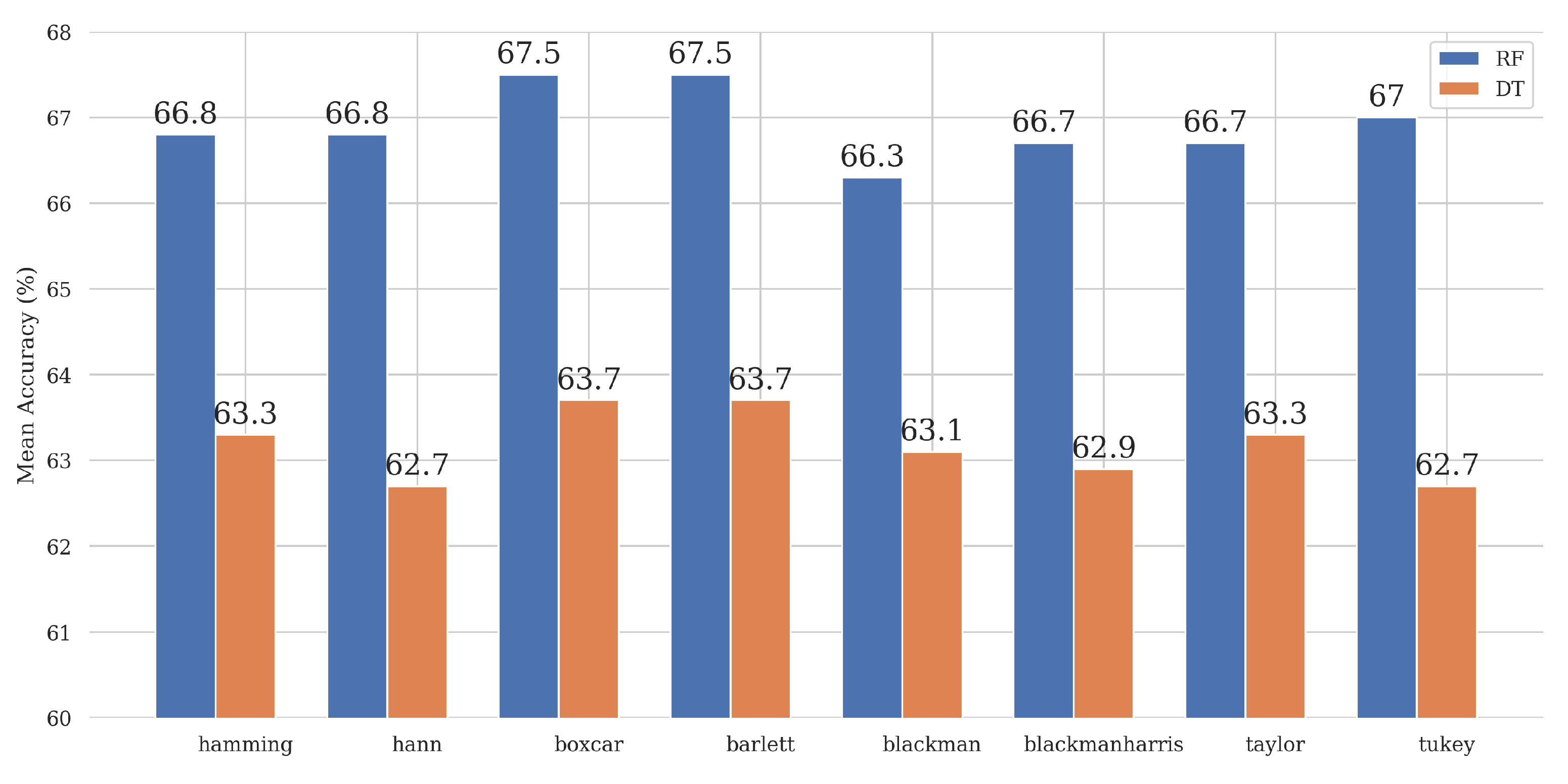
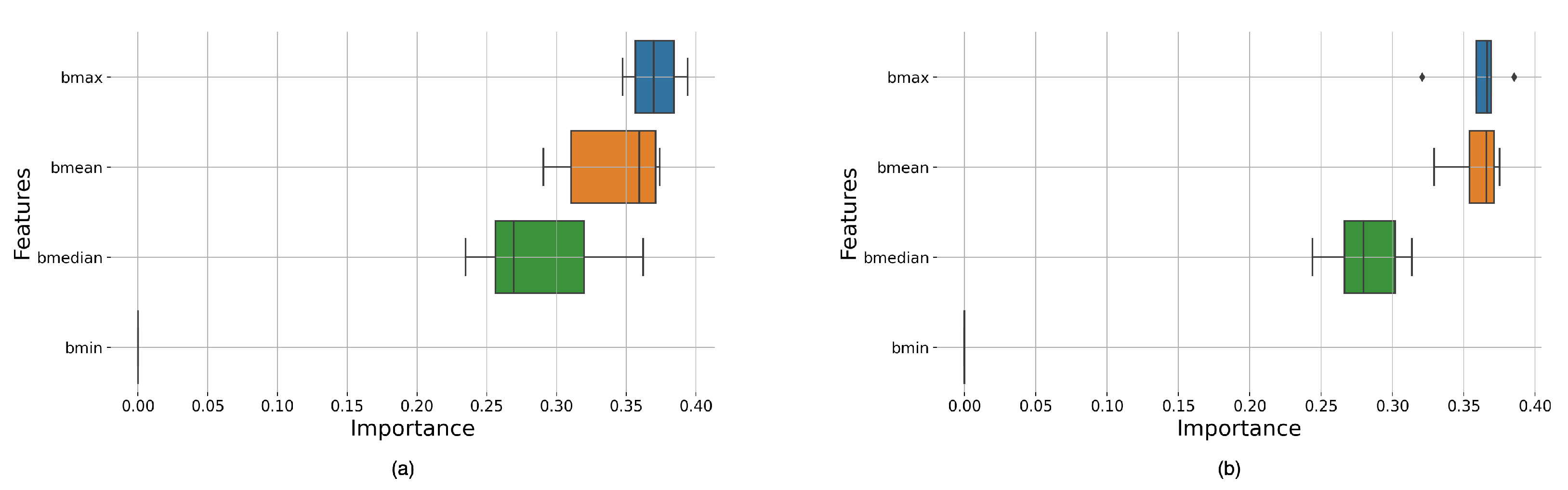
Table A3 shows that Boxcar and Bartlett have the highest mean scores and the most significant variance, AUC 69.3% and 64.4%, due to these windows having multiple high-value score classifiers. As reported in
Table A1 and
Table A2, it can be seen that all windows have nearly identical scores, with Boxcar and Bartlett having better classification Accuracy with higher scores than the others at 70.8%, suggesting that these windows might have slightly better effects on the extracted features than the rest. Thus, the models’ results and performance regarding the window functions were similar. However, there were differences in the metrics regarding the window size and overlaps. One plausible explanation for this was that the window function itself did not affect the signal significantly as much as the size and overlap of the windows do because the latter two determine the signal’s resolution. This effect is described in
Appendix A where all windows have the same maximum value for each metric; however, the Boxcar window, which is a square window and does not change anything in the signal, has the highest mean and variance because it has multiple window sizes with the maximum value metrics. On the other hand, the Bartlett and Boxcar window functions have the best performance among the analyzed window types. The Bartlett window, with an almost triangular shape, is known for being used to prevent the generation of too many oscillations in the frequency domain [
28]. The results of using the window are the same as for the basic Boxcar window. This was likely to be due to the relatively small and straightforward (interpreted) feature space. The STD analysis in
Table A3 also shows that the smallest values are obtained for the Hamming and Hann window function cases for the RF decision algorithm and the Hann and Tukey window function for the DT algorithm.
This is different from DL methods. We assume that it is about automatic feature extraction.
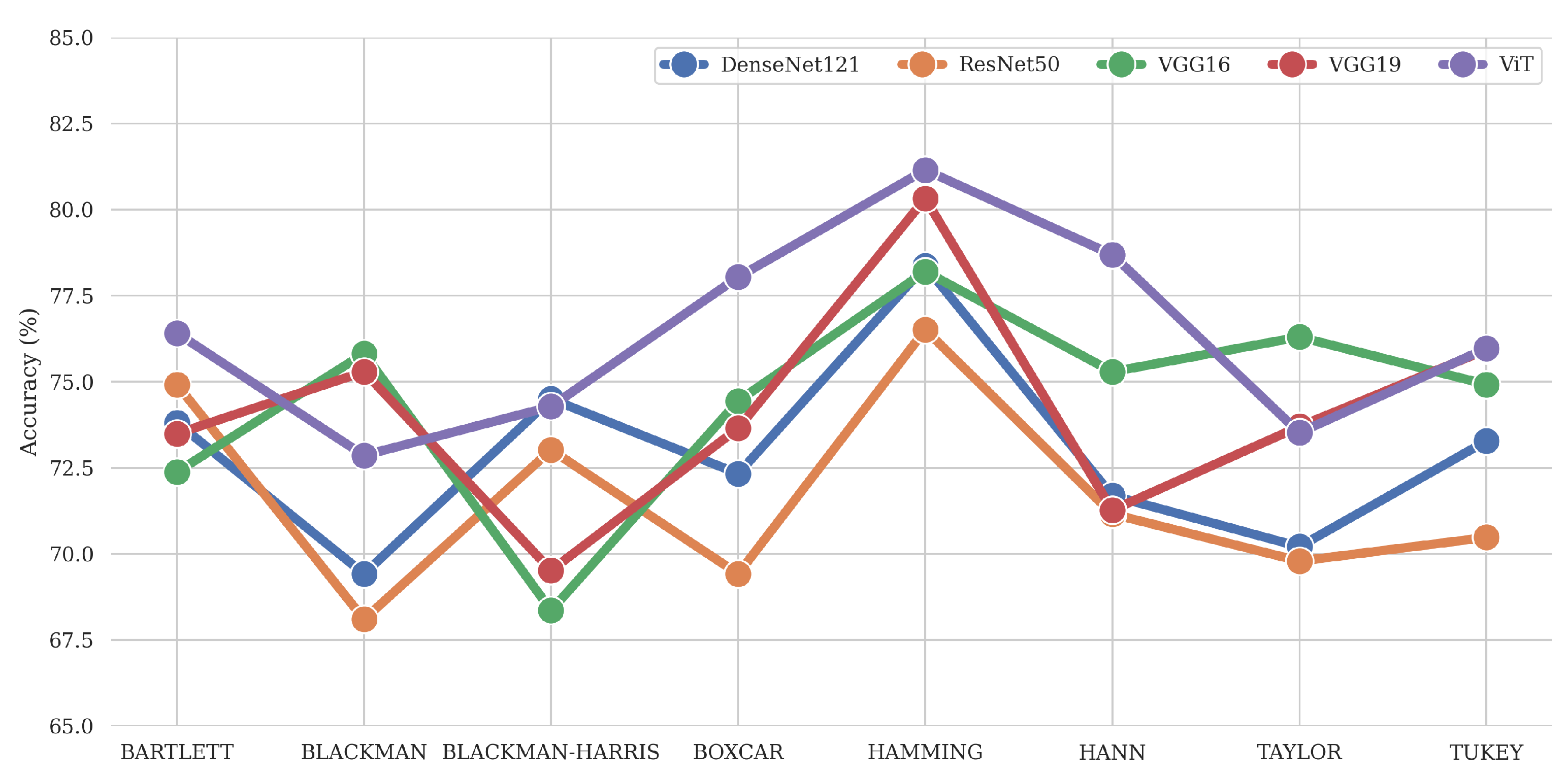
Figure 6 shows that the difference between the windowed features is more significant than the manual feature extraction approach. DL methods extract more features from the signal than can be extracted manually. Due to this, the average metrics values are higher than for the manual feature extraction cases. Comparing both approaches, we can conclude the perceptiveness of the modern DL approaches. However, we must also note that manual feature extraction with STFT can be considered the most explainable approach. Given that DL architectures do a better job at learning and extracting features at a wider scale, they can still be used as feature extractors alongside a classical model for the final classification. This will be explored in future research as it provides the potential to expand the feature space without the need for manual feature extraction.
6. Conclusions
This study investigated various window functions for STFT calculation (and spectrogram generation) to classify ERG signals. The spectrogram images were extracted using several combinations of well-known window functions, window sizes, and window overlap values, and the manual features were extracted to train the classical ML model using the same methods. Based on the comparison of the results of the two approaches, DL can be recommended. In terms of Accuracy, the ViT Small architecture with the Hamming window showed the best performance among the combinations of DL models with window types (81%). However, if manual feature extraction is required, a RF with a rectangular Boxcar window or Bartlett window can be recommended as an alternative to the DL approach. In the study, the mean Accuracy in these cases was 67.5%.
The results of the analysis of ERG using Short-Time Fourier Transform and ML techniques are, of course, dependent on the size of the dataset used for training, thus necessitating a large original sample. To address this limitation, expanding dataset volumes and promoting open data sharing within the electrophysiology community could enhance the diversity and representation of synthetic waveforms. Although these preliminary results have been generated with a relatively small sample set, it is one of the largest in the world by data quantity [
38]. Moreover, we are actively developing larger synthetic datasets to support clinical studies [
45].
Another limitation of this study was the feature space used in the ML approach; the analysis only used four features: the minimum, maximum, median, and mean brightness of the spectrogram; this could be a reason why there was little to no difference between the windows, this limitation was not encountered in the DL approach as the automatic feature extraction of DL architectures gave it access to a more prominent feature space. Hence, in future studies, we will look at expanding the feature space for the ML classifiers, as expanding the feature space for manual feature extraction approaches contributes to improved Accuracy and other metrics while maintaining the overall explainability of the system. This solution is essential for the developed algorithm to be easily understandable in medical applications.