Advancing Cordyceps militaris Industry: Gene Manipulation and Sustainable Biotechnological Strategies
Abstract
1. Introduction
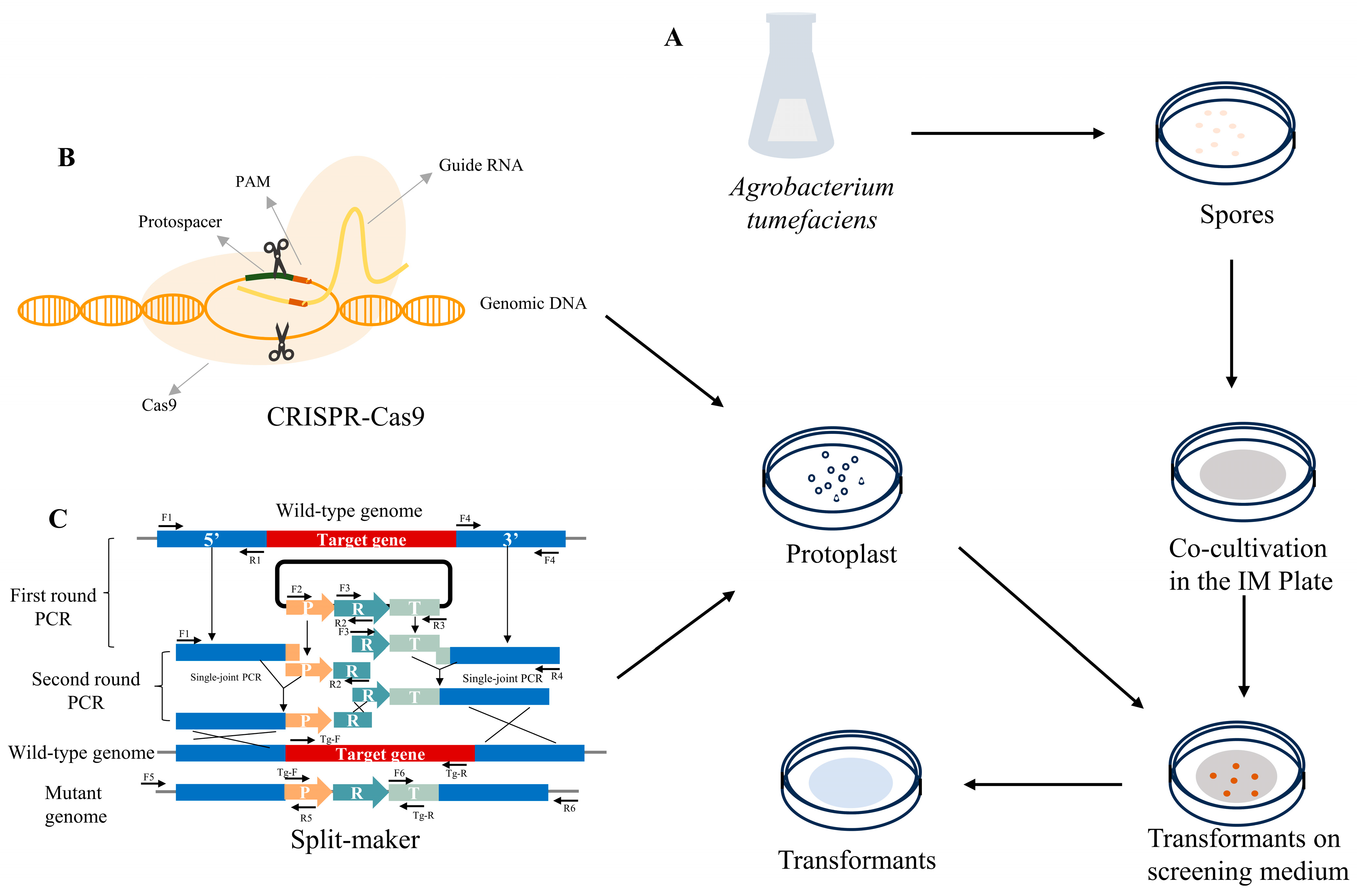
2. The Genetic Manipulation Technology of Cordyceps militaris
2.1. Commonly Used and Efficient Genetic Transformation System
2.1.1. Agrobacterium tumefaciens-Mediated Transformation
2.1.2. Polyethylene Glycol (PEG)-Mediated Protoplast Transformation
2.2. Selective Marker for the Detection of Positive Transformants
2.2.1. Auxotrophic Markers
2.2.2. Drug-Resistance Markers
2.3. Split-Marker Approach
2.4. CRISPR-Cas9 Gene-Editing Technology
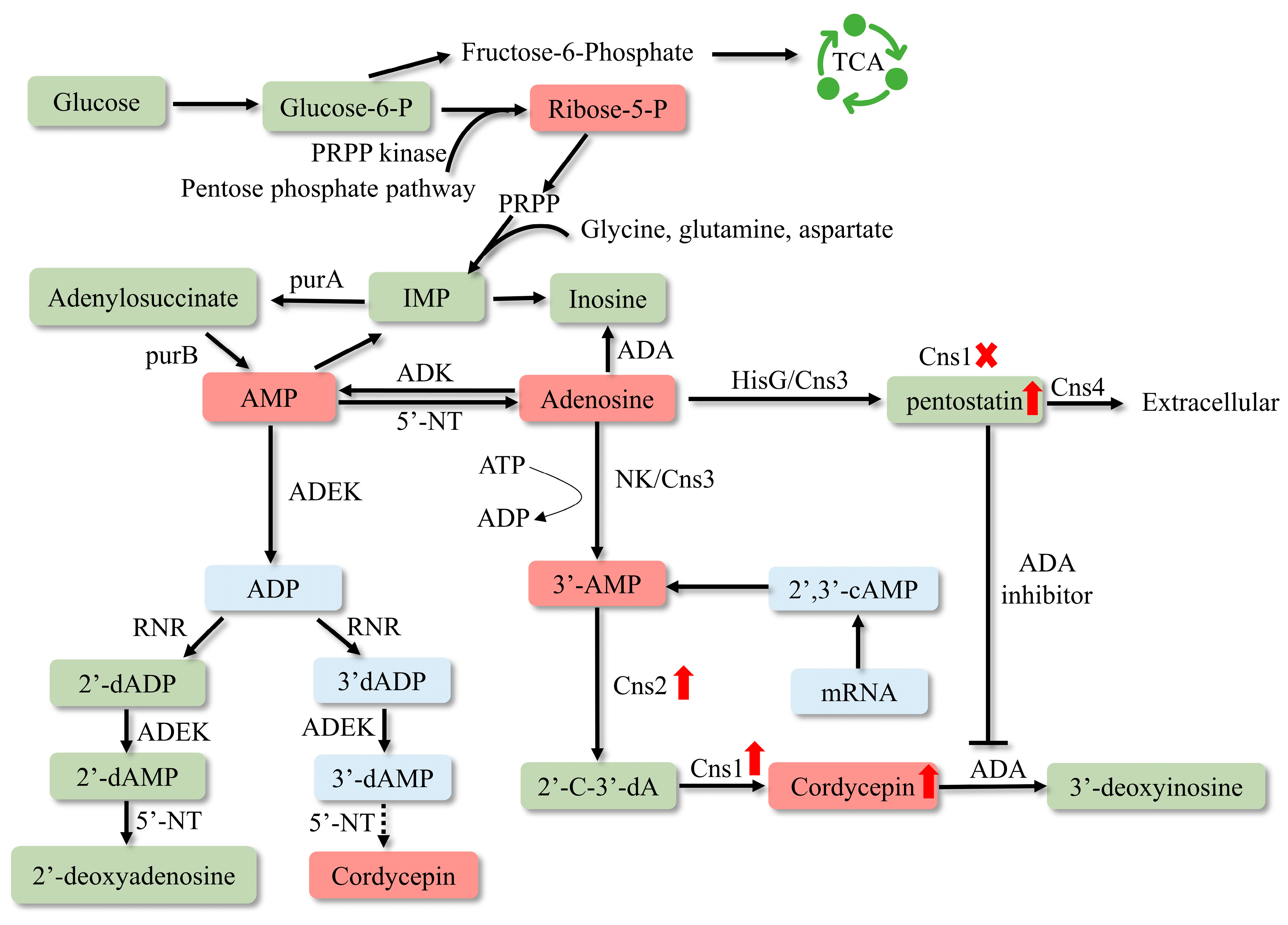
3. Increasing the Content of Secondary Metabolites of C. militaris by Genetic Engineering
3.1. The Enhancement of COR Production
3.2. The Enhancement of Cordyceps Polysaccharides Content
3.3. The Enhancement of Other Active Secondary Metabolites Production
4. Promoting the Growth and Development of C. militaris by Genetic Engineering
| Gene Name | Accession Number | Annotation | Function | Reference |
|---|---|---|---|---|
| Cry-DASH | CCM_00774 | Cryptochrome DASH | Fruiting-body development regulation | [95] |
| WC-1 | AGO64764 | Blue light receptor white collar 1 | Mycelium growth and fruiting-body development regulation | [78] |
| VVD | CCM_04514 | GATA transcription factor LreA | Fruiting-body development regulation | [79] |
| Lec3 | CCM_01589 | Lectin family integral membrane protein, putative | Fruiting-body development regulation | [94] |
| Lec4 | CCM_03832 | Ricin B-related lectin | Fruiting-body development regulation | [38] |
| Hyd1 | CCM_03537 | Hydrophobin 2 | Fruiting-body development regulation | [84] |
| Hyd4 | CCM_07964 | Hydrophobin | Fruiting-body development regulation | [85] |
| fhp | CCM_05119 | Flavohemoprotein | Fruiting-body development regulation | [99] |
| Snf1 | CCM_05552 | Carbon catabolite derepressing protein kinase | Fruiting-body development regulation | [100] |
| Chi1 | CCM_08231 | Class V chitinase, putative | Fruiting-body development regulation | [101] |
| Chi4 | CCM_04817 | Class V chitinase | Fruiting-body development regulation | [101] |
| crf1 | CCM_07998 | Fungal-specific transcription factor | Fruiting-body development regulation | [102] |
5. Breeding for Disease Resistance of C. militaris Using CRISPR/Cas9
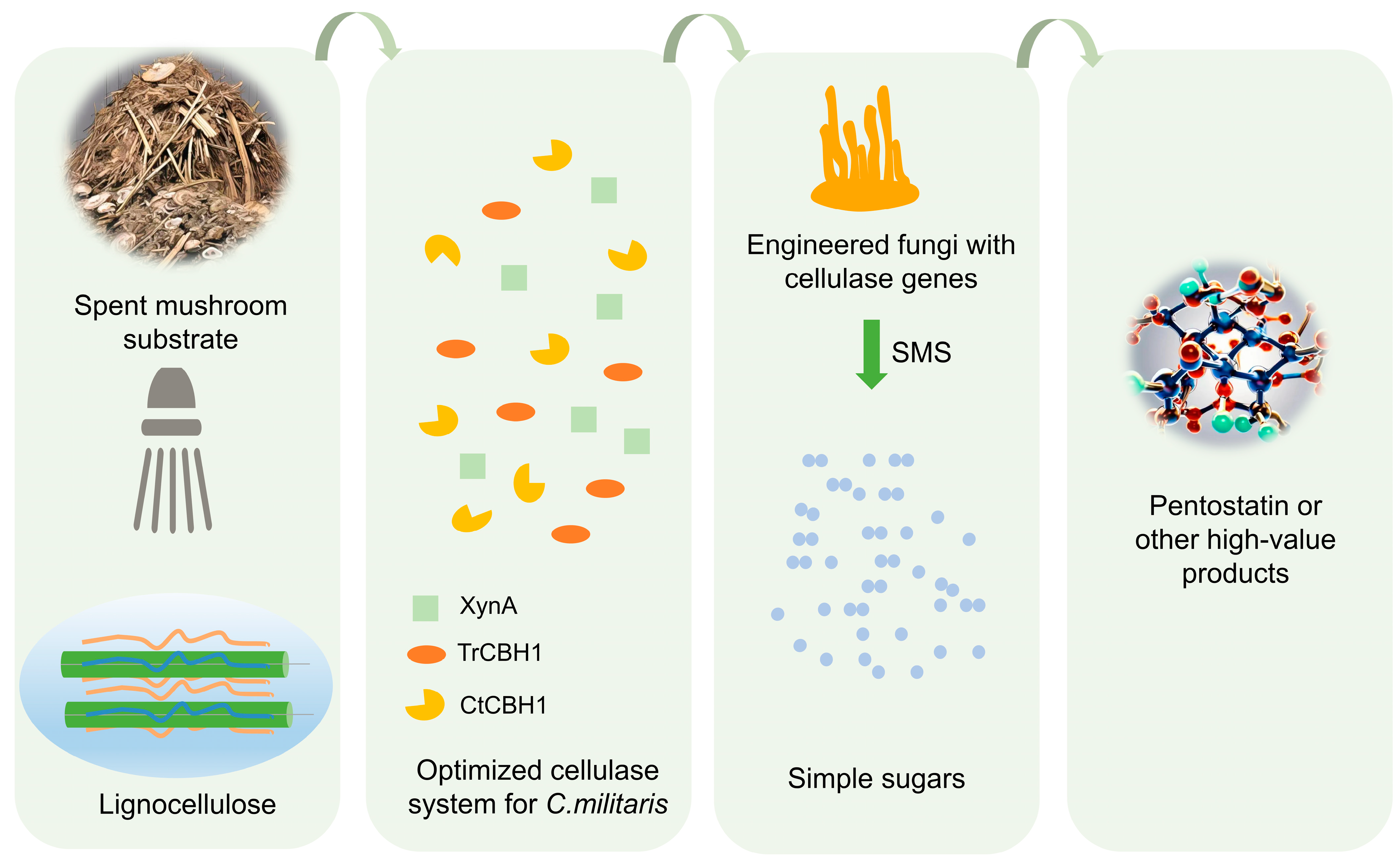
6. Resource Utilization of Agricultural Waste and Sustainable Production of C. militaris
7. Conclusions and Expectation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buenz, E.J.; Bauer, B.A.; Osmundson, T.W.; Motley, T.J. The traditional Chinese medicine Cordyceps sinensis and its effects on apoptotic homeostasis. J. Ethnopharmacol. 2005, 96, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.D. Biotechnological production and applications of Cordyceps militaris, a valued traditional Chinese medicine. Crit. Rev. Biotechnol. 2015, 35, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Jędrejko, K.J.; Lazur, J.; Muszyńska, B. Cordyceps militaris: An Overview of Its Chemical Constituents in Relation to Biological Activity. Foods 2021, 10, 2634. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: Current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Cohen, J.; Asatiani, M.D.; Varshney, V.K.; Yu, H.T.; Yang, Y.C.; Li, Y.H.; Mau, J.L.; Wasser, S.P. Chemical Composition and Nutritional and Medicinal Value of Fruit Bodies and Submerged Cultured Mycelia of Culinary-Medicinal Higher Basidiomycetes Mushrooms. Int. J. Med. Mushrooms 2014, 16, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.A.; Elkhalifa, A.E.O.; Siddiqui, A.J.; Patel, M.; Awadelkareem, A.M.; Snoussi, M.; Ashraf, M.S.; Adnan, M.; Hadi, S. Cordycepin for Health and Wellbeing: A Potent Bioactive Metabolite of an Entomopathogenic Cordyceps Medicinal Fungus and Its Nutraceutical and Therapeutic Potential. Molecules 2020, 25, 2735. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhu, G.; Yan, W.; Lv, Y.; Wang, X.; Jin, G.; Cui, M.; Lin, Z.; Ren, X. Cordycepin Inhibits Cancer Cell Proliferation and Angiogenesis through a DEK Interaction via ERK Signaling in Cholangiocarcinoma. J. Pharmacol. Exp. Ther. 2020, 373, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Tania, M. Cordycepin and kinase inhibition in cancer. Drug Discov. Today 2023, 28, 103481. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.P.; Qian, Z.Q.; Wu, H. Enhancing cordycepin production in liquid static cultivation of Cordyceps militaris by adding vegetable oils as the secondary carbon source. Bioresour. Technol. 2018, 268, 60–67. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Li, W.J.; Li, Q.P.; Qian, Z.M.; Liu, X.Z.; Dong, C.H. A breakthrough in the artificial cultivation of Chinese cordyceps on a large-scale and its impact on science, the economy, and industry. Crit. Rev. Biotechnol. 2019, 39, 181–191. [Google Scholar] [CrossRef]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, Z.Q.; Xue, Y.P.; Baker, P.J.; Wu, H.; Xu, F.; Teng, Y.; Brathwaite, M.E.; Zheng, Y.G. Biosynthetic Pathway Analysis for Improving the Cordycepin and Cordycepic Acid Production in Hirsutella sinensis. Appl. Biochem. Biotechnol. 2016, 179, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.X.; Tong, X.X.; Yokoyama, W.; Cao, J.; Wang, F.; Peng, C.; Guo, J.L. Overexpression of ribonucleotide reductase small subunit, RNRM, increases cordycepin biosynthesis in transformed Cordyceps militaris. Chin. J. Nat. Med. 2020, 18, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Wei, T.; Ye, Z.W.; Yun, F.; Kang, L.Z.; Tang, H.B.; Guo, L.Q.; Lin, J.F. Efficient CRISPR-Cas9 Gene Disruption System in Edible-Medicinal Mushroom Cordyceps militaris. Front. Microbiol. 2018, 9, 1157. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Xiao, M.; Chai, S.; Zhu, Z.; Wang, Y.; Zhou, Z. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb. Biotechnol. 2021, 14, 2343–2355. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.L.; Wang, X.P.; Liu, M.Q.; Wang, F.; Liu, Q.Z.; Dong, C.H. Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2594–2606. [Google Scholar] [CrossRef]
- Kang, N.; Lee, H.H.; Park, I.; Seo, Y.S. Development of High Cordycepin-Producing Cordyceps militaris Strains. Mycobiology 2017, 45, 31–38. [Google Scholar] [CrossRef]
- Kunhorm, P.; Chueaphromsri, P.; Chaicharoenaudomrung, N.; Noisa, P. Enhancement of cordycepin production from Cordyceps militaris culture by epigenetic modification. Biotechnol. Lett. 2022, 44, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.Y.; Yang, H.; Wang, C.; Liu, H.H.; Lu, X.Y.; Tian, Y. Microbial synthesis of cordycepin, current systems and future perspectives. Trends Food Sci. Technol. 2023, 132, 162–170. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, K.H.; Im, C.H.; Ali, A.; Lee, C.Y.; Kong, W.S.; Ryu, J.S. Identification of Degenerate Nuclei and Development of a SCAR Marker for Flammulina velutipes. PLoS ONE 2014, 9, e107207. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, S.W.; Ryu, J.S.; Lee, C.Y.; Ro, H.S. Isolation of a Variant Strain of Pleurotus eryngii and the Development of Specific DNA Markers to Identify the Variant Strain. Mycobiology 2014, 42, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.P.; Bian, Y.B.; Wang, J.J.; Wang, G.Z.; Ma, X.L.; Xu, Z.Y. Biological and Molecular Characteristics of a Novel Partitivirus Infecting the Edible Fungus Lentinula edodes. Plant Dis. 2017, 101, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Deng, C.H.; Zhang, L.Y.; Hu, K.I. Molecular analysis and biochemical characteristics of degenerated strains of Cordyceps militaris. Arch. Microbiol. 2017, 199, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Yin, J.; Zhang, B.; Li, Z.; Zhao, S.; Gui, Z. Genome-wide analysis of DNA methylation in subcultured Cordyceps militaris. Arch. Microbiol. 2019, 201, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Z.; Dong, C.H.; Liu, X.Z.; Liu, J.K.; Hyde, K.D. Calcarisporium cordycipiticola sp nov., an important fungal pathogen of Cordyceps militaris. Phytotaxa 2016, 268, 135–144. [Google Scholar] [CrossRef]
- Liu, Q.; Dong, C.H. Dual Transcriptomics Reveals Interspecific Interactions between the Mycoparasite Calcarisporium cordycipiticola and Its Host Cordyceps militaris. Microbiol. Spectr. 2023, 11, e04800-22. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Meng, G.; Wang, M.; Li, X.; Liu, M.; Wang, F.; Yang, Y.; Dong, C. Safe-Harbor-Targeted CRISPR/Cas9 System and Cmhyd1 Overexpression Enhances Disease Resistance in Cordyceps militaris. J. Agric. Food Chem. 2023, 71, 15249–15260. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.L.; Huang, C.H.; Cao, L.; Xie, C.H.; Han, R.H. Agrobacterium tumefaciens mediated transformation as a tool for insertional mutagenesis in medicinal fungus Cordyceps militaris. Fungal Biol. 2011, 115, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.L.; Qiu, X.H.; Han, R.C. Identification of the Genes Involved in the Fruiting Body Production and Cordycepin Formation of Cordyceps militaris Fungus. Mycobiology 2015, 43, 37–42. [Google Scholar] [CrossRef]
- Thai, H.D.; Nguyen, B.T.; Nguyen, V.M.; Nguyen, Q.H.; Tran, V.T. Development of a new Agrobacterium-mediated transformation system based on a dual auxotrophic approach in the filamentous fungus Aspergillus oryzae. World J. Microbiol. Biotechnol. 2021, 37, 92. [Google Scholar] [CrossRef]
- Koukaki, M.; Giannoutsou, E.; Karagouni, A.; Diallinas, G. A novel improved method for Aspergillus nidulans transformation. J. Microbiol. Methods 2003, 55, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Hinnen, A.; Hicks, J.B.; Fink, G.R. Transformation of yeast. Proc. Natl. Acad. Sci. USA 1978, 75, 1929–1933. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Ye, Z.; Yun, F.; Lin, J.; Guo, L.; Chen, B.; Mu, Z. Targeted Gene Deletion in Cordyceps militaris Using the Split-Marker Approach. Mol. Biotechnol. 2018, 60, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.W.; Ye, Z.W.; Yu, Y.H.; Lin, J.F.; Guo, L.Q.; Chen, B.X.; Tang, H.B.; Wei, T.; Chen, L.T.; Yun, F. The efficient genetic transformation of Cordycepsmilitaris by using mononuclear protoplasts. Sci. Hortic. 2019, 243, 307–313. [Google Scholar] [CrossRef]
- Zhang, M.; Shan, Y.L.; Gao, H.T.; Wang, B.; Liu, X.; Dong, Y.Y.; Liu, X.M.; Yao, N.; Zhou, Y.G.; Li, X.W.; et al. Expression of a recombinant hybrid antimicrobial peptide magainin II-cecropin B in the mycelium of the medicinal fungus Cordyceps militaris and its validation in mice. Microb. Cell Factories 2018, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Shang, Y.T.; Wang, L.H.; Tian, X.Q.; Tran, V.T.; Yao, L.H.; Zeng, B.; Hu, Z.H. Construction of a new Agrobacterium tumefaciens-mediated transformation system based on a dual auxotrophic approach in Cordyceps militaris. J. Microbiol. Biotechnol. 2024, 34, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Chen, P.; Yang, S.; Zhang, H. Homologous overexpression of genes in Cordyceps militaris improves the production of polysaccharides. Food Res. Int. 2021, 147, 110452. [Google Scholar] [CrossRef]
- Ono, A.; Suzuki, T.; Takeshima, Y.; Kashiwa, T.; Motoyama, T.; Choi, J.H.; Sato, C.; Konno, N.; Miyakawa, H.; Ogata, M.; et al. CmLec4, a lectin from the fungus Cordyceps militaris, controls host infection and fruiting body formation. Int. J. Biol. Macromol. 2022, 215, 303–311. [Google Scholar] [CrossRef]
- Zeng, J.; Zhou, Y.; Lyu, M.; Huang, X.; Xie, M.; Huang, M.; Chen, B.X.; Wei, T. Cordyceps militaris: A novel mushroom platform for metabolic engineering. Biotechnol. Adv. 2024, 74, 108396. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Xue, L.N.; Wei, T.; Wang, N.; Zhong, J.R.; Ye, Z.W.; Guo, L.Q.; Lin, J.F. Multiplex gene precise editing and large DNA fragment deletion by the CRISPR-Cas9-TRAMA system in edible mushroom Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2982–2991. [Google Scholar] [CrossRef]
- Rachmawati, R.; Kinoshita, H.; Nihira, T. Establishment of transformation system in Cordyceps militaris by using integration vector with benomyl resistance gene. In Proceedings of the 3rd International Conference on Sustainable Future for Human Security (SUSTAIN), Kyoto Univ, Kyoto, Japan, 3–5 November 2012; pp. 142–149. [Google Scholar]
- Cunningham, K.G.; Manson, W.; Spring, F.S.; Hutchinson, S.A. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature 1950, 166, 949. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal Cordycepin Biosynthesis Is Coupled with the Production of the Safeguard Molecule Pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489.e4. [Google Scholar] [CrossRef]
- Vongsangnak, W.; Raethong, N.; Mujchariyakul, W.; Nguyen, N.N.; Leong, H.W.; Laoteng, K. Genome-scale metabolic network of Cordyceps militaris useful for comparative analysis of entomopathogenic fungi. Gene 2017, 626, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.-d.; Wang, W.; Zhong, J.-J. Enhancement of cordycepin production in submerged cultures of Cordyceps militaris by addition of ferrous sulfate. Biochem. Eng. J. 2012, 60, 30–35. [Google Scholar] [CrossRef]
- Kato, T.; Nishimura, K.; Suparmin, A.; Ikeo, K.; Park, E.Y. Effects of Cordycepin in Cordyceps militaris during Its Infection to Silkworm Larvae. Microorganisms 2021, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Z.S.; Bao, D.P.; Li, B.; Yin, X.; Wu, Y.Y.; Chen, H.Y.; Tang, G.R.; Li, N.Y.; Zou, G. Improving Hypoxia Adaption Causes Distinct Effects on Growth and Bioactive Compounds Synthesis in an Entomopathogenic Fungus Cordyceps militaris. Front. Microbiol. 2021, 12, 698436. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, P.; Xu, L.; Xu, D.; Hu, W.; Cheng, Y.; Yang, S. Construction of Cordycepin High-Production Strain and Optimization of Culture Conditions. Curr. Microbiol. 2022, 80, 12. [Google Scholar] [CrossRef] [PubMed]
- Jeennor, S.; Anantayanon, J.; Panchanawaporn, S.; Chutrakul, C.; Vongsangnak, W.; Laoteng, K. Efficient de novo production of bioactive cordycepin by Aspergillus oryzae using a food-grade expression platform. Microb. Cell Fact. 2023, 22, 253. [Google Scholar] [CrossRef]
- Wang, H.; Fu, X.; Zuo, X.; Zhang, C.; Lu, W. Overproduction of cordycepin in Saccharomyces cerevisiae by cordycepin synthase screening and metabolic engineering. AIChE J. 2024, 70, e18361. [Google Scholar] [CrossRef]
- Duan, X.Y.; Tian, Y.; Song, Z.Q.; Song, L.P.; Lin, W.B.; Wang, C.; Yang, H.; Lu, X.Y.; Ji, X.J.; Liu, H.H. High-level de novo biosynthesis of cordycepin by systems metabolic engineering in Yarrowia lipolytica. Bioresour. Technol. 2022, 363, 127862. [Google Scholar] [CrossRef]
- Song, Z.Q.; Lin, W.B.; Duan, X.Y.; Song, L.P.; Wang, C.; Yang, H.; Lu, X.Y.; Ji, X.J.; Tian, Y.; Liu, H.H. Increased Cordycepin Production in Yarrowia lipolytica Using Combinatorial Metabolic Engineering Strategies. Acs Synth. Biol. 2023, 12, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Methacanon, P.; Madla, S.; Kirtikara, K.; Prasitsil, M. Structural elucidation of bioactive fungi-derived polymers. Carbohydr. Polym. 2005, 60, 199–203. [Google Scholar] [CrossRef]
- Ujita, M.; Inoue, R.; Makino, Y.; Katsuno, Y.; Okumura, H. Binding specificity of the recombinant cytoplasmic domain of Cordyceps militaris β-1,3-glucan synthase catalytic subunit. Biosci. Biotechnol. Biochem. 2011, 75, 171–174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, N.; Ge, X.; Yin, X.; Yang, L.; Chen, L.; Shao, R.; Xu, W. A review on polysaccharide biosynthesis in Cordyceps militaris. Int. J. Biol. Macromol. 2024, 260, 129336. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bai, Y.; Dai, R.; Guo, X.; Liu, Z.H.; Yuan, S. Improved Polysaccharide Production by Homologous Co-overexpression of Phosphoglucomutase and UDP Glucose Pyrophosphorylase Genes in the Mushroom Coprinopsis cinerea. J. Agric. Food Chem. 2018, 66, 4702–4709. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhong, Y.; Ding, L.; Liu, X.; Xu, S.; Guo, D.; Blennow, A.; Xue, J. Biosynthesis, structure and functionality of starch granules in maize inbred lines with different kernel dehydration rate. Food Chem. 2022, 368, 130796. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zhao, F.; Han, S.; Zhang, Y. Enhanced rhamnolipids production in Pseudomonas aeruginosa SG by selectively blocking metabolic bypasses of glycosyl and fatty acid precursors. Biotechnol. Lett. 2020, 42, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, F.; Xu, Z.; Ding, Z. Bioactive Mushroom Polysaccharides: A Review on Monosaccharide Composition, Biosynthesis and Regulation. Molecules 2017, 22, 955. [Google Scholar] [CrossRef] [PubMed]
- Cheah, I.K.; Halliwell, B. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2012, 1822, 784–793. [Google Scholar] [CrossRef]
- Beelman, R.B.; Kalaras, M.D.; Phillips, A.T.; Richie, J.P., Jr. Is ergothioneine a ‘longevity vitamin’ limited in the American diet? J. Nutr. Sci. 2020, 9, e52. [Google Scholar] [CrossRef]
- Ames, B.N. Prolonging healthy aging: Longevity vitamins and proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 10836–10844. [Google Scholar] [CrossRef] [PubMed]
- Tsiantas, K.; Tsiaka, T.; Koutrotsios, G.; Siapi, E.; Zervakis, G.I.; Kalogeropoulos, N.; Zoumpoulakis, P. On the Identification and Quantification of Ergothioneine and Lovastatin in Various Mushroom Species: Assets and Challenges of Different Analytical Approaches. Molecules 2021, 26, 1832. [Google Scholar] [CrossRef] [PubMed]
- Kamide, T.; Takusagawa, S.; Tanaka, N.; Ogasawara, Y.; Kawano, Y.; Ohtsu, I.; Satoh, Y.; Dairi, T. High Production of Ergothioneine in Escherichia coli using the Sulfoxide Synthase from Methylobacterium strains. J. Agric. Food Chem. 2020, 68, 6390–6394. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kawano, Y.; Satoh, Y.; Dairi, T.; Ohtsu, I. Gram-scale fermentative production of ergothioneine driven by overproduction of cysteine in Escherichia coli. Sci. Rep. 2019, 9, 1895. [Google Scholar] [CrossRef] [PubMed]
- Osawa, R.; Kamide, T.; Satoh, Y.; Kawano, Y.; Ohtsu, I.; Dairi, T. Heterologous and High Production of Ergothioneine in Escherichia coli. J. Agric. Food Chem. 2018, 66, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- van der Hoek, S.A.; Darbani, B.; Zugaj, K.E.; Prabhala, B.K.; Biron, M.B.; Randelovic, M.; Medina, J.B.; Kell, D.B.; Borodina, I. Engineering the Yeast Saccharomyces cerevisiae for the Production of L- (+) -Ergothioneine. Front. Bioeng. Biotechnol. 2019, 7, 262. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Pan, H.Y.; Guo, L.Q.; Lin, J.F.; Liao, H.L.; Li, H.Y. Successful biosynthesis of natural antioxidant ergothioneine in Saccharomyces cerevisiae required only two genes from Grifola frondosa. Microb. Cell Fact. 2020, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Takusagawa, S.; Satoh, Y.; Ohtsu, I.; Dairi, T. Ergothioneine production with Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2019, 83, 181–184. [Google Scholar] [CrossRef]
- van der Hoek, S.A.; Rusnák, M.; Jacobsen, I.H.; Martínez, J.L.; Kell, D.B.; Borodina, I. Engineering ergothioneine production in Yarrowia lipolytica. FEBS Lett. 2022, 596, 1356–1364. [Google Scholar] [CrossRef]
- Pluskal, T.; Ueno, M.; Yanagida, M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system. PLoS ONE 2014, 9, e97774. [Google Scholar] [CrossRef]
- Chen, B.X.; Xue, L.N.; Wei, T.; Ye, Z.W.; Li, X.H.; Guo, L.Q.; Lin, J.F. Enhancement of ergothioneine production by discovering and regulating its metabolic pathway in Cordyceps militaris. Microb. Cell Fact. 2022, 21, 169. [Google Scholar] [CrossRef] [PubMed]
- Kot, A.M.; Błażejak, S.; Gientka, I.; Kieliszek, M.; Bryś, J. Torulene and torularhodin: “new” fungal carotenoids for industry? Microb. Cell Factories 2018, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.-H.; Hao, Y.-F.; Li, Y.-M.; Liang, Y.-J.; Jiang, J.-G. Inhibiting Lycopene Cyclases to Accumulate Lycopene in High β-Carotene-Accumulating Dunaliella bardawil. Food Bioprocess. Technol. 2016, 9, 1002–1009. [Google Scholar] [CrossRef]
- Linden, H.; Ballario, P.; Macino, G. Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 1997, 22, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Talora, C.; Franchi, L.; Linden, H.; Ballario, P.; Macino, G. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. Embo J. 1999, 18, 4961–4968. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Verma, S.; Corrochano, L.M. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet. Biol. 2010, 47, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Guo, M.M.; Yang, H.J.; Guo, S.P.; Dong, C.H. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, F.; Yang, Y.; Wang, Y.; Dong, C. CmVVD is involved in fruiting body development and carotenoid production and the transcriptional linkage among three blue-light receptors in edible fungus Cordyceps militaris. Environ. Microbiol. 2020, 22, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.W.; Zhao, Y.; Tang, H.B.; Ye, Z.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Transcriptome Analysis of Cordyceps militaris Reveals Genes Associated with Carotenoid Synthesis and Identification of the Function of the Cmtns Gene. Front. Microbiol. 2019, 10, 2105. [Google Scholar] [CrossRef]
- Rottenberg, M.E.; Masocha, W.; Ferella, M.; Petitto-Assis, F.; Goto, H.; Kristensson, K.; McCaffrey, R.; Wigzell, H. Treatment of African trypanosomiasis with cordycepin and adenosine deaminase inhibitors in a mouse model. J. Infect. Dis. 2005, 192, 1658–1665. [Google Scholar] [CrossRef]
- Zou, G.; Li, B.; Wang, Y.; Yin, X.; Gong, M.; Shang, J.J.; Wei, Y.J.; Li, X.L.; Bao, D.P. Efficient conversion of spent mushroom substrate into a high value-added anticancer drug pentostatin with engineered Cordyceps militaris. Green. Chem. 2021, 23, 10030–10038. [Google Scholar] [CrossRef]
- Lian, T.T.; Yang, T.; Liu, G.J.; Sun, J.D.; Dong, C.H. Reliable reference gene selection for Cordyceps militaris gene expression studies under different developmental stages and media. Fems Microbiol. Lett. 2014, 356, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, F.; Liu, M.; Dong, C. Hydrophobin CmHYD1 Is Involved in Conidiation, Infection and Primordium Formation, and Regulated by GATA Transcription Factor CmAreA in Edible Fungus, Cordyceps militaris. J. Fungi 2021, 7, 674. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, M.; Dong, C. Hydrophobin Gene Cmhyd4 Negatively Regulates Fruiting Body Development in Edible Fungi Cordyceps militaris. Int. J. Mol. Sci. 2023, 24, 4586. [Google Scholar] [CrossRef] [PubMed]
- vanWetter, M.A.; Schuren, F.H.J.; Schuurs, T.A.; Wessels, J.G.H. Targeted mutation of the SC3 hydrophobin gene of Schizophyllum commune affects formation of aerial hyphae. Fems Microbiol. Lett. 1996, 140, 265–269. [Google Scholar] [CrossRef]
- Sammer, D.; Krause, K.; Gube, M.; Wagner, K.; Kothe, E. Hydrophobins in the Life Cycle of the Ectomycorrhizal Basidiomycete Tricholoma vaccinum. PLoS ONE 2016, 11, e0167773. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X.; Chen, R.L.; Yan, J.J.; Long, Y.; Tong, Z.J.; Song, H.B.; Xie, B.G. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.D.; Ainouz, I.L.; Deoliveira, J.T.A.; Cavada, B.S. Plant-lectins, chemical and biological aspects. Mem. Do Inst. Oswaldo Cruz 1991, 86, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhao, C.G.; Tong, X.; Qi, Y.P. A lectin with mycelia differentiation and antiphytovirus activities from the edible mushroom Agrocybe aegerita. J. Biochem. Mol. Biol. 2003, 36, 214–222. [Google Scholar] [CrossRef]
- Nagata, Y.; Yamashita, M.; Honda, H.; Akabane, J.; Uehara, K.; Saito, A.; Sumisa, F.; Nishibori, K.; Oodaira, Y. Characterization, occurrence, and molecular cloning of a lectin from Grifola frondosa Jacalin-related lectin of fungal origin. Biosci. Biotechnol. Biochem. 2005, 69, 2374–2380. [Google Scholar] [CrossRef]
- Jung, E.C.; Kim, K.D.; Bae, C.H.; Kim, J.C.; Kim, D.K.; Kim, H.H. A mushroom lectin from ascomycete Cordyceps militaris. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2007, 1770, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Wang, H.; Ng, T.B. A haemagglutinin from the medicinal fungus Cordyceps militaris. Biosci. Rep. 2009, 29, 321–327. [Google Scholar] [CrossRef]
- Bao, D.P.; Ma, Y.W.; Gong, M.; Li, Y.; Gao, Y.N.; Yang, R.H.; Yang, R.F.; Mao, W.J.; Wang, Y. Sequence analysis and heterologous expression of lectin-like gene CMLec3 from the medicinal fungus Cordyceps militaris. Mycoscience 2018, 59, 119–123. [Google Scholar] [CrossRef]
- Wang, F.; Song, X.H.; Dong, X.M.; Zhang, J.J.; Dong, C.H. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biotechnol. 2017, 101, 4645–4657. [Google Scholar] [CrossRef]
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700. [Google Scholar] [CrossRef]
- Tagua, V.G.; Pausch, M.; Eckel, M.; Gutiérrez, G.; Miralles-Durán, A.; Sanz, C.; Eslava, A.P.; Pokorny, R.; Corrochano, L.M.; Batschauer, A. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 15130–15135. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Dunlap, J.C.; Loros, J.J. Fungal Light Sensing at the Bench and Beyond. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 96, pp. 1–51. [Google Scholar]
- Lou, H.W.; Zhao, Y.; Chen, B.X.; Yu, Y.H.; Tang, H.B.; Ye, Z.W.; Lin, J.F.; Guo, L.Q. Cmfhp Gene Mediates Fruiting Body Development and Carotenoid Production in Cordyceps militaris. Biomolecules 2020, 10, 410. [Google Scholar] [CrossRef]
- Wāng, Y.; Wang, R.; Wáng, Y.; Li, Y.; Yang, R.H.; Gong, M.; Shang, J.J.; Zhang, J.S.; Mao, W.J.; Zou, G.; et al. Diverse function and regulation of CmSnf1 in entomopathogenic fungus Cordyceps militaris. Fungal Genet. Biol. 2020, 142, 103415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Yin, Y.Y.; Cui, Y.; Zhang, Y.X.; Liu, B.Y.; Ma, Y.C.; Liu, Y.N.; Liu, G.Q. Chitinase Is Involved in the Fruiting Body Development of Medicinal Fungus Cordyceps militaris. Life 2023, 13, 764. [Google Scholar] [CrossRef]
- He, R.; Zhang, L.; Lan, J.; Mei, S.; Li, Y. Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris. Biology 2022, 11, 1535. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, Y.Y.; Zhang, X.L.; Li, K.; Li, X.; Wang, F.; Xu, F.X.; Dong, C.H. Infection Process and Genome Assembly Provide Insights into the Pathogenic Mechanism of Destructive Mycoparasite Calcarisporium cordycipiticola with Host Specificity. J. Fungi 2021, 7, 918. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Kong, X.; Lu, Z.; Xiao, M.; Chen, M.; Zhu, L.; Shen, Y.; Hu, X.; Song, S. Para-aminobenzoic acid (PABA) synthase enhances thermotolerance of mushroom Agaricus bisporus. PLoS ONE 2014, 9, e91298. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, P.; Iotti, M.; Zeppa, S.D.; Lancellotti, E.; Amicucci, A.; Zambonelli, A. Morphological and functional changes in mycelium and mycorrhizas of Tuber borchii due to heat stress. Fungal Ecol. 2017, 29, 20–29. [Google Scholar] [CrossRef]
- Jiaojiao, Z.; Fen, W.; Kuanbo, L.; Qing, L.; Ying, Y.; Caihong, D. Heat and light stresses affect metabolite production in the fruit body of the medicinal mushroom Cordyceps militaris. Appl. Microbiol. Biotechnol. 2018, 102, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Sanmiya, K.; Suzuki, K.; Egawa, Y.; Shono, M. Mitochondrial small heat-shock protein enhances thermotolerance in tobacco plants. FEBS Lett. 2004, 557, 265–268. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, S.; Luo, Y.; Ma, C.; Gong, Y.; Zhou, Y.; Gao, S.; Huang, Z.; Yan, L.; Hu, Y.; et al. The heat shock protein 40 LeDnaJ regulates stress resistance and indole-3-acetic acid biosynthesis in Lentinula edodes. Fungal Genet. Biol. 2018, 118, 37–44. [Google Scholar] [CrossRef]
- Ling, Y.Y.; Ling, Z.L.; Zhao, R.L. Construction of a heat-resistant strain of Lentinus edodes by fungal Hsp20 protein overexpression and genetic transformation. Front. Microbiol. 2022, 13, 1009885. [Google Scholar] [CrossRef]
- Leong, Y.K.; Ma, T.W.; Chang, J.S.; Yang, F.C. Recent advances and future directions on the valorization of spent mushroom substrate (SMS): A review. Bioresour. Technol. 2022, 344, 126157. [Google Scholar] [CrossRef]
- Martín, C.; Zervakis, G.I.; Xiong, S.; Koutrotsios, G.; Strætkvern, K.O. Spent substrate from mushroom cultivation: Exploitation potential toward various applications and value-added products. Bioengineered 2023, 14, 2252138. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, M.; Chen, J.; Li, S.; Shao, Y.; Yang, J.; Li, J. Characteristics of bio-oil produced by the pyrolysis of mixed oil shale semi-coke and spent mushroom substrate. Fuel 2017, 200, 218–224. [Google Scholar] [CrossRef]
- Lam, S.S.; Lee, X.Y.; Nam, W.L.; Phang, X.Y.; Liew, R.K.; Yek, P.N.; Ho, Y.L.; Ma, N.L.; Rosli, M.H. Microwave vacuum pyrolysis conversion of waste mushroom substrate into biochar for use as growth medium in mushroom cultivation. J. Chem. Technol. Biotechnol. 2019, 94, 1406–1415. [Google Scholar] [CrossRef]
- Pérez-Chávez, A.M.; Mayer, L.; Albertó, E. Mushroom cultivation and biogas production: A sustainable reuse of organic resources. Energy Sustain. Dev. 2019, 50, 50–60. [Google Scholar] [CrossRef]
- Wan Mahari, W.A.; Peng, W.; Nam, W.L.; Yang, H.; Lee, X.Y.; Lee, Y.K.; Liew, R.K.; Ma, N.L.; Mohammad, A.; Sonne, C.; et al. A review on valorization of oyster mushroom and waste generated in the mushroom cultivation industry. J. Hazard. Mater. 2020, 400, 123156. [Google Scholar] [CrossRef] [PubMed]
- Jasiūnas, L.; Pedersen, T.H.; Toor, S.S.; Rosendahl, L.A. Biocrude production via supercritical hydrothermal co-liquefaction of spent mushroom compost and aspen wood sawdust. Renew. Energy 2017, 111, 392–398. [Google Scholar] [CrossRef]
- Ryden, P.; Efthymiou, M.N.; Tindyebwa, T.A.M.; Elliston, A.; Wilson, D.R.; Waldron, K.W.; Malakar, P.K. Bioethanol production from spent mushroom compost derived from chaff of millet and sorghum. Biotechnol. Biofuels 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Raethong, N.; Thananusak, R.; Cheawchanlertfa, P.; Prabhakaran, P.; Rattanaporn, K.; Laoteng, K.; Koffas, M.; Vongsangnak, W. Functional genomics and systems biology of Cordyceps species for biotechnological applications. Curr. Opin. Biotechnol. 2023, 81, 102939. [Google Scholar] [CrossRef]

| Strains | Method | Selection Marker | Target Gene | Transformation Efficiency | References |
|---|---|---|---|---|---|
| JM4 | Agrobacterium tumefaciens-mediated transformation (ATMT) | Hygromycin B | - | 30–600 cfu/ 1 × 105 spores | [28] |
| HF 374-1, HF 432-1, HF 432-2, HF 438, and CM 001-5 | Protoplast-mediated transformation (PMT) | Benomyl | LaeA (global regulator) | 7 cfu/μg | [41] |
| CM10 | Split-Marker | Basta | Tns (encoding a terpenoid synthase) | 4.53 cfu/μg | [33] |
| CM10 | CRISPR-Cas9 | Basta | pyrG (encoding orotic acid-5′-monophosphate decarboxylase) | 1.7 cfu/μg | [14] |
| CM15 | CRISPR-Cas9 | Basta | Cns1 (oxidoreductase domain-containing protein) | 5 cfu/μg | [40] |
| CGMCC 3.16323 | CRISPR-Cas9 | hygromycin | Cmwc-1(blue light receptor white collar 1) | 5.5 cfu/μg | [16] |
| Cmvvd (GATA transcription factor LreA) | 8.8 cfu/μg | ||||
| CGMCC 3.16323 | CRISPR-Cas9 | hygromycin | Hyd1 (Hydrophobin 2) | - | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Wu, Y.; Song, J.; Ma, M.; Xiao, Y.; Zeng, B. Advancing Cordyceps militaris Industry: Gene Manipulation and Sustainable Biotechnological Strategies. Bioengineering 2024, 11, 783. https://doi.org/10.3390/bioengineering11080783
Hu Y, Wu Y, Song J, Ma M, Xiao Y, Zeng B. Advancing Cordyceps militaris Industry: Gene Manipulation and Sustainable Biotechnological Strategies. Bioengineering. 2024; 11(8):783. https://doi.org/10.3390/bioengineering11080783
Chicago/Turabian StyleHu, Yan, Yijian Wu, Jiayi Song, Maomao Ma, Yunzhu Xiao, and Bin Zeng. 2024. "Advancing Cordyceps militaris Industry: Gene Manipulation and Sustainable Biotechnological Strategies" Bioengineering 11, no. 8: 783. https://doi.org/10.3390/bioengineering11080783
APA StyleHu, Y., Wu, Y., Song, J., Ma, M., Xiao, Y., & Zeng, B. (2024). Advancing Cordyceps militaris Industry: Gene Manipulation and Sustainable Biotechnological Strategies. Bioengineering, 11(8), 783. https://doi.org/10.3390/bioengineering11080783






