Integrating Convolutional Neural Networks with Attention Mechanisms for Magnetic Resonance Imaging-Based Classification of Brain Tumors
Abstract
1. Introduction
- This study presents a novel approach that combines hybrid attention with convolution neural networks to improve the efficiency of diagnosing glioma, meningioma, pituitary, and no-tumor cases.
- The objective of this study is to emphasize the effectiveness of the proposed method in comparison to previous studies, showcasing its capacity to provide effective results with fewer resources. Moreover, the method’s capacity for usage in a clinical research context is thoroughly evaluated.
- The findings from this study demonstrate that the proposed method surpasses the previous studies in terms of performance, as demonstrated on the benchmark dataset. Additionally, the study evaluates the prediction competencies of the framework by comparing it to pre-trained models, ultimately improving diagnostics methodologies and clinical necessities.
2. Literature Review
3. Materials and Methods
3.1. Dataset
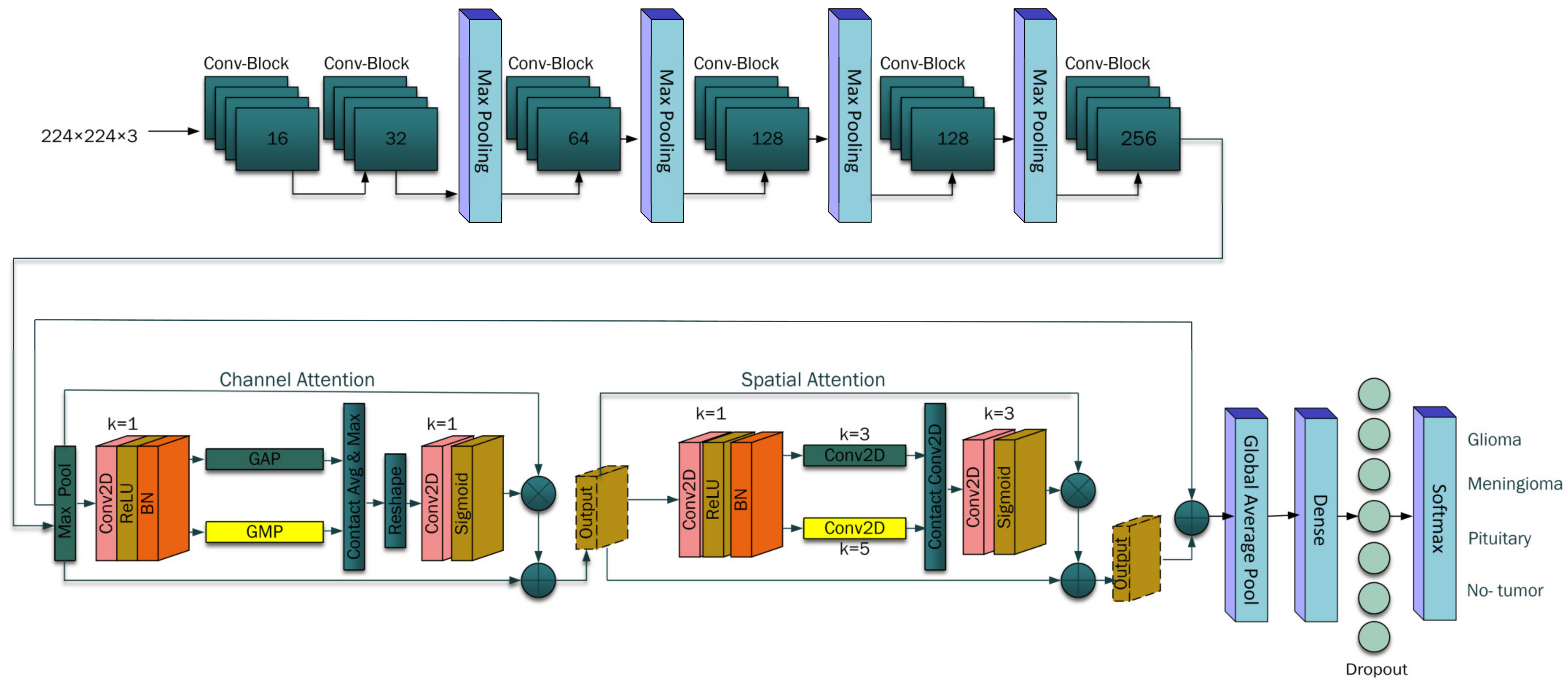
3.2. Proposed Architecture
| Algorithm 1: Pseudo-code for Hybrid Attention Mechanism |
Input:
|
3.3. Activation and Losses Functions
3.4. Optimization Techniques
3.5. Pre-Trained Models
4. Experimental Results
| Algorithm 2: 5-Fold Cross-Validation for Model Evaluation |
|
4.1. Evaluation Matrices
4.2. Confusion Matrices
5. Discussion
| Authors | Dataset | Classes | Methods | Precision | Recall | F1-Score | Accuracy |
|---|---|---|---|---|---|---|---|
| Gumaei et al. [18] | Figshare 3064 Images | 3 | Hybrid PCA-NGIST-RELM | - | - | - | 94.23 |
| Swati et al. [26] | Figshare 3064 Images | 3 | VGG19-Fine tune | 89.52 | - | 91.73 | 94.82 |
| Kaplan et al. [14] | Figshare 3064 Images | 3 | NLBP-αLBP-KNN | - | - | - | 95.56 |
| Huang et al. [20] | Figshare 3064 Images | 3 | CNNBCN | - | - | - | 95.49 |
| Ghassemi et al. [22] | Figshare 3064 Images | 3 | CNN-based GAN | 95.29 | - | 95.10 | 95.60 |
| Ayadi et al. [23] | Figshare 3064 Images | 3 | DSURF-HOG-SVM | - | 88.84 | 89.37 | 90.27 |
| Noreen et al. [24] | Figshare 3064 Images | 3 | InceptionV3 Ensemble | 93.00 | 92.00 | 92.00 | 94.34 |
| Satyanarayana et al. [27] | Figshare 3064 Images | 3 | AMEA-CNN-MCA | - | - | - | 94.00 |
| Deepak et al. [28] | Figshare 3064 Images | 3 | CNN-MV-KNN | - | - | 95.06 | 95.60 |
| Almalki et al. [49] | Kaggle 2870 Images | 4 | SURF-KAZE-SVM | - | - | - | 95.33 |
| Asiri et al. [51] | Kaggle 2870 Images | 4 | GAN-Softmax | 92.00 | 93.00 | 93.00 | 94.32 |
| Shilaskar et al. [50] | Figshare, SARTAJ, Br35H 7023 Images | 4 | HOG-XG Boost | 92.07 | 91.82. | 91.85 | 92.02 |
| Our work | Figshare, SARTAJ, Br35H, 7023 Images | 4 | CNN-Hybrid Attention | 98.30 | 98.30 | 98.20 | 98.33 |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khazaei, Z.; Goodarzi, E.; Borhaninejad, V.; Iranmanesh, F.; Mirshekarpour, H.; Mirzaei, B.; Naemi, H.; Bechashk, S.M.; Darvishi, I.; Ershad Sarabi, R.; et al. The association between incidence and mortality of brain cancer and human development index (HDI): An ecological study. BMC Public Health 2020, 20, 1696. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. The Global Cancer Observatory—All cancers. Int. Agency Res. Cancer—WHO 2020, 419, 199–200. Available online: https://gco.iarc.fr/today/home (accessed on 12 February 2023).
- Gliomas|Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/gliomas (accessed on 12 February 2023).
- Meningioma|Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/meningioma (accessed on 12 February 2023).
- Pituitary Tumors—Symptoms and Causes—Mayo Clinic. 2018. Available online: https://www.mayoclinic.org/diseases-conditions/pituitary-tumors/symptoms-causes/syc-20350548 (accessed on 18 March 2023).
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2020, 131, 244–260. [Google Scholar] [CrossRef]
- Kang, S.H.; Lee, Y. Motion Artifact Reduction Using U-Net Model with Three-Dimensional Simulation-Based Datasets for Brain Magnetic Resonance Images. Bioengineering 2024, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.; Ma, Y.; Ullah, I.; Ghadi, Y.Y.; Khan, M.Z.; Khan, M.A.; Abdusalomov, A.; Alqahtani, F.; Shehata, A.M. Brain Tumor Classification from MRI Using Image Enhancement and Convolutional Neural Network Techniques. Brain Sci. 2023, 13, 1320. [Google Scholar] [CrossRef] [PubMed]
- Ukwuoma, C.C.; Qin, Z.; Heyat, M.B.B.; Akhtar, F.; Smahi, A.; Jackson, J.K.; Furqan Qadri, S.; Muaad, A.Y.; Monday, H.N.; Nneji, G.U. Automated Lung-Related Pneumonia and COVID-19 Detection Based on Novel Feature Extraction Framework and Vision Transformer Approaches Using Chest X-ray Images. Bioengineering 2022, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Battineni, G.; Chintalapudi, N.; Hossain, M.A.; Losco, G.; Ruocco, C.; Sagaro, G.G.; Traini, E.; Nittari, G.; Amenta, F. Artificial Intelligence Models in the Diagnosis of Adult-Onset Dementia Disorders: A Review. Bioengineering 2022, 9, 370. [Google Scholar] [CrossRef] [PubMed]
- Altini, N.; Brunetti, A.; Puro, E.; Taccogna, M.G.; Saponaro, C.; Zito, F.A.; De Summa, S.; Bevilacqua, V. NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering 2022, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Chen, S.; Jiang, N.; Hu, H. An Effective WSSENet-Based Similarity Retrieval Method of Large Lung CT Image Databases. KSII Trans. Internet Inf. Syst. 2022, 16, 2359–2376. [Google Scholar] [CrossRef]
- Deng, X.; Liu, E.; Li, S.; Duan, Y.; Xu, M. Interpretable Multi-Modal Image Registration Network Based on Disentangled Convolutional Sparse Coding. IEEE Trans. Image Process. 2023, 32, 1078–1091. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, K.; Kaya, Y.; Kuncan, M.; Ertunç, H.M. Brain tumor classification using modified local binary patterns (LBP) feature extraction methods. Med. Hypotheses 2020, 139, 109696. [Google Scholar] [CrossRef]
- El-Shafai, W.; Mahmoud, A.A.; El-Rabaie, E.S.M.; Taha, T.E.; Zahran, O.F.; El-Fishawy, A.S.; Soliman, N.F.; Alhussan, A.A.; Abd El-Samie, F.E. Hybrid Segmentation Approach for Different Medical Image Modalities. Comput. Mater. Contin. 2022, 73, 3455–3472. [Google Scholar] [CrossRef]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Yang, J.; Yang, B.; Yin, Z.; Liu, M.; Yin, L.; Zheng, W. Analysis and Design of Surgical Instrument Localization Algorithm. C.—Comput. Model. Eng. Sci. 2022, 137, 669–685. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A Hybrid Feature Extraction Method with Regularized Extreme Learning Machine for Brain Tumor Classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Srujan, K.S.; Shivakumar, S.; Sitnur, K.; Garde, O.; Pk, P. Brain Tumor Segmentation and Classification using CNN model. Int. Res. J. Eng. Technol. 2020, 7, 4077–4080. [Google Scholar]
- Huang, Z.; Du, X.; Chen, L.; Li, Y.; Liu, M.; Chou, Y.; Jin, L. Convolutional Neural Network Based on Complex Networks for Brain Tumor Image Classification with a Modified Activation Function. IEEE Access 2020, 8, 89281–89290. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Automated Categorization of Brain Tumor from MRI Using CNN features and SVM. J. Ambient Intell. Humaniz. Comput. 2020, 12, 8357–8369. [Google Scholar] [CrossRef]
- Ghassemi, N.; Shoeibi, A.; Rouhani, M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control 2020, 57, 101678. [Google Scholar] [CrossRef]
- Ayadi, W.; Charfi, I.; Elhamzi, W.; Atri, M. Brain tumor classification based on hybrid approach. Vis. Comput. 2020, 38, 107–117. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Alassafi, M.O. Brain Tumor Classification Based on Fine-Tuned Models and the Ensemble Method. Comput. Mater. Contin. 2021, 67, 3967–3982. [Google Scholar] [CrossRef]
- Ahmad, B.; Sun, J.; You, Q.; Palade, V.; Mao, Z. Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 2022, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, G.; Appala Naidu, P.; Subbaiah Desanamukula, V.; Satish kumar, K.; Chinna Rao, B. A mass correlation based deep learning approach using deep Convolutional neural network to classify the brain tumor. Biomed. Signal Process. Control 2023, 81, 104395. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor categorization from imbalanced MRI dataset using weighted loss and deep feature fusion. Neurocomputing 2023, 520, 94–102. [Google Scholar] [CrossRef]
- Rezaei, K.; Agahi, H.; Mahmoodzadeh, A. A Weighted Voting Classifiers Ensemble for the Brain Tumors Classification in MR Images. IETE J. Res. 2020, 68, 3829–3842. [Google Scholar] [CrossRef]
- Yadav, S. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification. In Proceedings of the 2016 IEEE 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016. [Google Scholar] [CrossRef]
- Robbins, H.; Monro, S. A Stochastic Approximation Method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference for Learning Representations ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Nickparvar, M. Brain Tumor MRI Dataset. 2021. Available online: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset (accessed on 10 May 2023).
- Cheng, J. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 10 May 2023).
- Brain Tumor Classification (MRI)|Kaggle. Available online: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri (accessed on 10 July 2023).
- Br35H :: Brain Tumor Detection 2020. Available online: https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection?select=no (accessed on 10 May 2023).
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, ICML 2015, Lille, France, 6–11 July 2015; Volume 1, pp. 448–456. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve Restricted Boltzmann machines. In Proceedings of the ICML 2010—27th International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Computer Vision—ECCV 2018, Proceedings of the 15th European Conference, Munich, Germany, 8–14 September 2018; Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2018; Volume 11211, pp. 3–19. [Google Scholar]
- Bin Tufail, A.; Ullah, I.; Rehman, A.U.; Khan, R.A.; Khan, M.A.; Ma, Y.K.; Hussain Khokhar, N.; Sadiq, M.T.; Khan, R.; Shafiq, M.; et al. On Disharmony in Batch Normalization and Dropout Methods for Early Categorization of Alzheimer’s Disease. Sustainability 2022, 14, 4695. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: https://www.deeplearningbook.org (accessed on 10 February 2022).
- Moradi, R.; Berangi, R.; Minaei, B. A Survey of Regularization Strategies for Deep Models; Springer: Amsterdam, The Netherlands, 2020; Volume 53, ISBN 0123456789. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 299, 345–350. [Google Scholar] [CrossRef]
- ReduceLROnPlateau. Available online: https://keras.io/api/callbacks/reduce_lr_on_plateau/ (accessed on 24 May 2023).
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the Proceedings—30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Computer Vision—ECCV 2016, Proceedings of the 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2016; Volume 9908, pp. 630–645. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Ting, K.M. Confusion Matrix. In Encyclopedia of Machine Learning and Data Mining; Springer: Boston, MA, USA, 2017; p. 260. [Google Scholar] [CrossRef]
- Almalki, Y.E.; Ali, M.U.; Ahmed, W.; Kallu, K.D.; Zafar, A.; Alduraibi, S.K.; Irfan, M.; Basha, M.A.A.; Alshamrani, H.A.; Alduraibi, A.K. Robust Gaussian and Nonlinear Hybrid Invariant Clustered Features Aided Approach for Speeded Brain Tumor Diagnosis. Life 2022, 12, 1084. [Google Scholar] [CrossRef] [PubMed]
- Shilaskar, S.; Mahajan, T.; Bhatlawande, S.; Chaudhari, S.; Mahajan, R.; Junnare, K. Machine Learning based Brain Tumor Detection and Classification using HOG Feature Descriptor. In Proceedings of the International Conference on Sustainable Computing and Smart Systems (ICSCSS 2023), Coimbatore, India, 14–16 June 2023; pp. 67–75. [Google Scholar]
- Asiri, A.A.; Shaf, A.; Ali, T.; Aamir, M.; Usman, A.; Irfan, M.; Alshamrani, H.A.; Mehdar, K.M.; Alshehri, O.M.; Alqhtani, S.M. Multi-Level Deep Generative Adversarial Networks for Brain Tumor Classification on Magnetic Resonance Images. Intell. Autom. Soft Comput. 2023, 36, 127–143. [Google Scholar] [CrossRef]







| Models | Parameters | Precision | Recalls | F1-Score | Accuracy | Training Time(s) |
|---|---|---|---|---|---|---|
| Xception | 22,963,756 | 92.35 | 92.20 | 92.25 | 92.64 | 1228.13 |
| ResNet50V2 | 25,667,076 | 90.00 | 90.05 | 90.10 | 90.39 | 614.07 |
| DenseNet201 | 20,293,188 | 92.95 | 92.75 | 92.85 | 93.20 | 1274.99 |
| ResNet101V2 | 44,728,836 | 86.10 | 86.15 | 86.15 | 86.51 | 1035.39 |
| DenseNet169 | 14,351,940 | 94.90 | 95.00 | 94.90 | 95.29 | 964.36 |
| Proposed method without Attention | 829,172 | 96.85 | 96.75 | 96.80 | 96.97 | 423.99 |
| Proposed method with Attention | 928,688 | 98.30 | 98.30 | 98.20 | 98.33 | 460.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasheed, Z.; Ma, Y.-K.; Ullah, I.; Al-Khasawneh, M.; Almutairi, S.S.; Abohashrh, M. Integrating Convolutional Neural Networks with Attention Mechanisms for Magnetic Resonance Imaging-Based Classification of Brain Tumors. Bioengineering 2024, 11, 701. https://doi.org/10.3390/bioengineering11070701
Rasheed Z, Ma Y-K, Ullah I, Al-Khasawneh M, Almutairi SS, Abohashrh M. Integrating Convolutional Neural Networks with Attention Mechanisms for Magnetic Resonance Imaging-Based Classification of Brain Tumors. Bioengineering. 2024; 11(7):701. https://doi.org/10.3390/bioengineering11070701
Chicago/Turabian StyleRasheed, Zahid, Yong-Kui Ma, Inam Ullah, Mahmoud Al-Khasawneh, Sulaiman Sulmi Almutairi, and Mohammed Abohashrh. 2024. "Integrating Convolutional Neural Networks with Attention Mechanisms for Magnetic Resonance Imaging-Based Classification of Brain Tumors" Bioengineering 11, no. 7: 701. https://doi.org/10.3390/bioengineering11070701
APA StyleRasheed, Z., Ma, Y.-K., Ullah, I., Al-Khasawneh, M., Almutairi, S. S., & Abohashrh, M. (2024). Integrating Convolutional Neural Networks with Attention Mechanisms for Magnetic Resonance Imaging-Based Classification of Brain Tumors. Bioengineering, 11(7), 701. https://doi.org/10.3390/bioengineering11070701







