Searching for the Best Machine Learning Algorithm for the Detection of Left Ventricular Hypertrophy from the ECG: A Review
Abstract
1. Introduction
2. Methods
2.1. Literature Search
2.2. Data Extraction and Classification
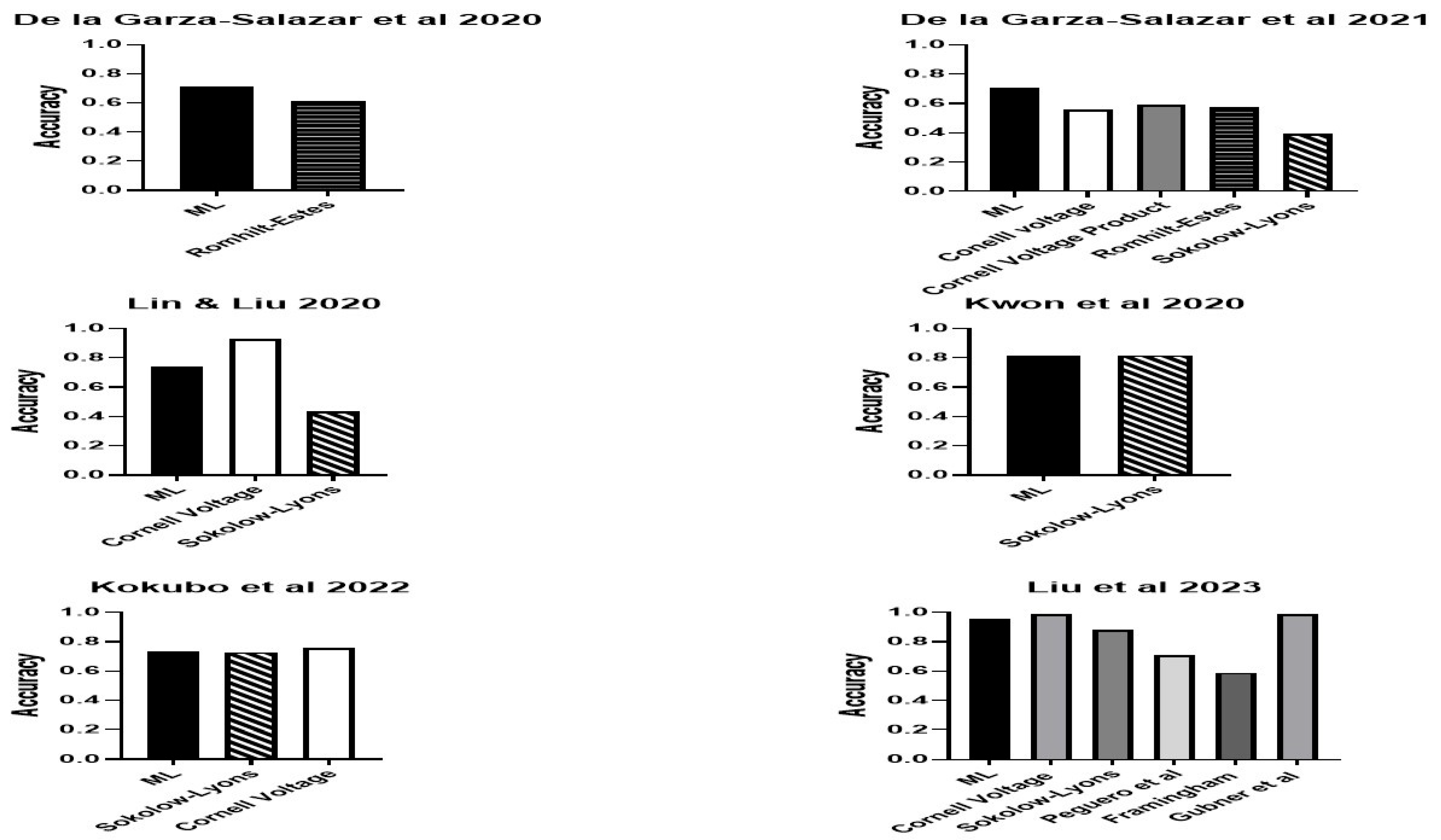
3. Results
4. Discussion
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| SVM | Support vector machine |
| LR | logistic regression |
| RF | Random Forest |
| ENN | Ensemble neural network |
| CNN | Convolutional neural network |
| DNN | Deep neural network |
| AUC | Area under the receiver operating curve |
References
- Levy, D.; Garrison, R.J.; Savage, D.D.; Kannel, W.B.; Castelli, W.P. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 1990, 322, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Giles, W.H.; Croft, J.B. Left ventricular hypertrophy as a predictor of coronary heart disease mortality and the effect of hypertension. Am. Heart J. 2000, 140, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Abdi-Ali, A.; Miller, R.J.H.; Southern, D.; Zhang, M.; Mikami, Y.; Knudtson, M.; Heydari, B.; Howarth, A.G.; Lydell, C.P.; James, M.T.; et al. LV Mass Independently Predicts Mortality and Need for Future Revascularization in Patients Undergoing Diagnostic Coronary Angiography. JACC Cardiovasc. Imaging 2018, 11, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Rabkin, S.W.; Shiekh, I.A.; Wood, D.A. The Impact of Left Ventricular Mass on Diastolic Blood Pressure Targets for Patients with Coronary Artery Disease. Am. J. Hypertens. 2016, 29, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bavishi, C.; Sardar, P.; Agarwal, V.; Krishnamoorthy, P.; Grodzicki, T.; Messerli, F.H. Meta-analysis of left ventricular hypertrophy and sustained arrhythmias. Am. J. Cardiol. 2014, 114, 1049–1052. [Google Scholar] [CrossRef]
- Varvarousis, D.; Kallistratos, M.; Poulimenos, L.; Triantafyllis, A.; Tsinivizov, P.; Giannakopoulos, A.; Kyfnidis, K.; Manolis, A. Cardiac arrhythmias in arterial hypertension. J. Clin. Hypertens. 2020, 22, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Rabkin, S.W. Considerations in understanding the coronary blood flow- left ventricular mass relationship in patients with hypertension. Curr. Cardiol. Rev. 2017, 13, 75–83. [Google Scholar] [CrossRef]
- Yi, S.; Wang, F.; Wan, M.; Yi, X.; Zhang, Y.; Sun, S. Prediction of stroke with electrocardiographic left ventricular hypertrophy in hypertensive patients: A meta-analysis. J. Electrocardiol. 2020, 61, 27–31. [Google Scholar] [CrossRef]
- Sokolow, M.; Lyon, T.P. The ventricular complex in left ventricular hypertrophy as obtained by unipolar precordial and limb leads. Am. Heart J. 1949, 37, 161–186. [Google Scholar] [CrossRef]
- Koito, H.; Spodick, D.H. Accuracy of the RV6:RV5 voltage ratio for increased left ventricular mass. Am. J. Cardiol. 1988, 62, 985–987. [Google Scholar] [CrossRef]
- Crow, R.S.; Prineas, R.J.; Rautaharju, P.; Hannan, P.; Liebson, P.R. Relation between electrocardiography and echocardiography for left ventricular mass in mild systemic hypertension (results from Treatment of Mild Hypertension Study). Am. J. Cardiol. 1995, 75, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Labib, S.B.; Anderson, K.M.; Christiansen, J.C.; Kannel, W.B.; Castelli, W.P. Determinants of sensitivity and specificity of electrocardiographic criteria for left ventricular hypertrophy. Circulation 1990, 81, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Casale, P.N.; Devereux, R.B.; Kligfield, P.; Eisenberg, R.R.; Miller, D.H.; Chaudhary, B.S.; Phillips, M.C. Electrocardiographic detection of left ventricular hypertrophy: Development and prospective validation of improved criteria. J. Am. Coll. Cardiol. 1985, 6, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Peguero, J.G.; Lo Presti, S.; Perez, J.; Issa, O.; Brenes, J.C.; Tolentino, A. Electrocardiographic criteria for the diagnosis of left ventricular hypertrophy. J. Am. Coll. Cardiol. 2017, 69, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Pewsner, D.; Jüni, P.; Egger, M.; Battaglia, M.; Sundström, J.; Bachmann, L.M. Accuracy of electrocardiography in diagnosis of left ventricular hypertrophy in arterial hypertension: Systematic review. Br. Med. J. Br. Med. J. Publ. Group 2007, 335, 711. [Google Scholar] [CrossRef] [PubMed]
- Rautaharju, P.M.; Soliman, E.Z. Electrocardiographic left ventricular hypertrophy and the risk of adverse cardiovascular events: A critical appraisal. J. Electrocardiol. 2014, 47, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Fagard, R.H.; Staessen, J.A.; Thijs, L.; Celis, H.; Birkenhäger, W.H.; Bulpitt, C.J.; de Leeuw, P.W.; Leonetti, G.; Sarti, C.; Tuomilehto, J.; et al. Prognostic significance of electrocardiographic voltages and their serial changes in elderly with systolic hypertension. Hypertension 2004, 44, 459–464. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kannel, W.B.; Gordon, T.; Castelli, W.P.; Margolis, J.R. Electrocardiographic left ventricular hypertrophy and risk of coronary heart disease: The Framingham Study. Ann. Intern. Med. Am. Coll. Physicians 1970, 72, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Rabkin, S.W.; Mathewson, F.A.L.; Tate, R.B. The electrocardiogram in apparently healthy men and the risk of sudden death. Br. Heart J. 1982, 47, 546–552. [Google Scholar] [CrossRef]
- De Bacquer, D.; De Backer, G.; Kornitzer, M.; Blackburn, H. Prognostic value of ECG findings for total, cardiovascular disease, and coronary heart disease death in men and women. Heart 1998, 80, 570–577. [Google Scholar] [CrossRef]
- Lonn, E.; Mathew, J.; Pogue, J.; Johnstone, D.; Danisa, K.; Bosch, J.; Baird, M.; Dagenais, G.; Sleight, P.; Yusuf, S.; et al. Relationship of electrocardiographic left ventricular hypertrophy to mortality and cardiovascular morbidity in high-risk patients. Eur. J. Cardiovasc. Prev. Rehabil. 2003, 10, 420–428. [Google Scholar] [CrossRef]
- Hsieh, B.P.; Pham, M.X.; Froelicher, V.F. Prognostic value of electrocardiographic criteria for left ventricular hypertrophy. Am. Heart J. 2005, 150, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.M.; Vander Zwaag, R.V.; el-Zeky, F.; Ramanathan, K.B.; Mirvis, D.M. Left ventricular hypertrophy: Effect on survival. J. Am. Coll. Cardiol. 1993, 22, 508–513. [Google Scholar] [CrossRef]
- Hawkins, N.M.; Wang, D.; McMurray, J.J.V.; Pfeffer, M.A.; Swedberg, K.; Granger, C.B.; Yusuf, S.; Pocock, S.J.; Ostergren, J.; Michelson, E.L.; et al. Prevalence and prognostic implications of electrocardiographic left ventricular hypertrophy in heart failure: Evidence from the CHARM programme. Heart 2007, 93, 59–64. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; He, T.; Ding, Y.; Yang, L.; Jiang, X.; Huang, L. Predictive value of electrocardiographic left ventricular hypertrophy in the general population: A meta-analysis. J. Electrocardiol. 2020, 62, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Schlant, R.C.; Adolph, R.J.; DiMarco, J.P.; Dreifus, L.S.; Dunn, M.I.; Fisch, C.; Garson, A., Jr.; Haywood, L.J.; Levine, H.J.; Murray, J.A. Guidelines for electrocardiography. A report of the American College of Cardiology/American Heart Association Task Force on Assessment of Diagnostic and Therapeutic Cardiovascular Procedures (Committee on Electrocardiography). Circulation Am. Heart Assoc. 1992, 85, 1221–1228. [Google Scholar] [CrossRef]
- Unger, T.; Borghi, C.; Charchar, F.; Khan, N.A.; Poulter, N.R.; Prabhakaran, D.; Ramirez, A.; Schlaich, M.; Stergiou, G.S.; Tomaszewski, M.; et al. 2020 International Society of Hypertension global hypertension practice guidelines. J. Hypertens. 2020, 38, 982–1004. [Google Scholar] [CrossRef]
- Okin, P.M.; Roman, M.J.; Devereux, R.B.; Kligfield, P. Electrocardiographic identification of increased left ventricular mass by simple voltage-duration products. J. Am. Coll. Cardiol. 1995, 25, 417–423. [Google Scholar] [CrossRef]
- Romhilt, D.W.; Bove, K.E.; Norris, R.J.; Conyers, E.; Conradi, S.; Rowlands, D.T.; Scott, R. A critical appraisal of the electrocardiographic criteria for the diagnosis of left ventricular hypertrophy. Circ. Am. Heart Assoc. 1969, 40, 185–196. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Lin, G.-M.; Liu, K. An Electrocardiographic System with Anthropometrics via Machine Learning to Screen Left Ventricular Hypertrophy among Young Adults. IEEE J. Transl. Eng. Health Med. 2020, 8, 1800111. [Google Scholar] [CrossRef] [PubMed]
- Sparapani, R.; Dabbouseh, N.M.; Gutterman, D.; Zhang, J.; Chen, H.; Bluemke, D.A.; Lima, J.A.C.; Burke, G.L.; Soliman, E.Z. Detection of Left Ventricular Hypertrophy Using Bayesian Additive Regression Trees: The MESA. J. Am. Heart Assoc. 2019, 8, e009959. [Google Scholar] [CrossRef] [PubMed]
- De la Garza-Salazar, F.; Romero-Ibarguengoitia, M.E.; Rodriguez-Diaz, E.A.; Azpiri-Lopez, J.R.; Gonzalez-Cantu, A. Improvement of electrocardiographic diagnostic accuracy of left ventricular hypertrophy using a Machine Learning approach. PLoS ONE 2020, 15, e0232657. [Google Scholar] [CrossRef]
- Kwon, J.-M.; Jeon, K.-H.; Kim, H.M.; Kim, M.J.; Lim, S.M.; Kim, K.-H.; Song, P.S.; Park, J.; Choi, R.K.; Oh, B.-H. Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. EP Eur. 2020, 22, 412–419. [Google Scholar] [CrossRef] [PubMed]
- De la Garza Salazar, F.; Romero Ibarguengoitia, M.E.; Azpiri López, J.R.; González Cantú, A. Optimizing ECG to detect echocardiographic left ventricular hypertrophy with computer-based ECG data and machine learning. PLoS ONE 2021, 16, e0260661. [Google Scholar] [CrossRef]
- Khurshid, S.; Friedman, S.; Pirruccello, J.P.; Di Achille, P.; Diamant, N.; Anderson, C.D.; Ellinor, P.T.; Batra, P.; Ho, J.E.; Philippakis, A.A.; et al. Deep Learning to Predict Cardiac Magnetic Resonance-Derived Left Ventricular Mass and Hypertrophy From 12-Lead ECGs. Circ. Cardiovasc. Imaging 2021, 14, e012281. [Google Scholar] [CrossRef]
- Sabovcik, F.; Cauwenberghs, N.; Kouznetsov, D.; Haddad, F.; Alonso-Betanzos, A.; Vens, C.; Kuznetsova, T. Applying machine learning to detect early stages of cardiac remodelling and dysfunction. Eur. Hear. J. Cardiovasc. Imaging 2021, 22, 1208–1217. [Google Scholar] [CrossRef]
- Angelaki, E.; Marketou, M.E.; Barmparis, G.D.; Patrianakos, A.; Vardas, P.E.; Parthenakis, F.; Tsironis, G.P. Detection of abnormal left ventricular geometry in patients without cardiovascular disease through machine learning: An ECG-based approach. J. Clin. Hypertens. 2021, 23, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.Y.; Sng, G.; Ho, W.H.; Hankun, W.; Sia, C.-H.; Lee, J.S.; Shen, X.; Tan, B.Y.; Lee, E.C.; Dalakoti, M.; et al. Machine learning versus classical electrocardiographic criteria for echocardiographic left ventricular hypertrophy in a pre-participation cohort. Kardiol. Pol. 2021, 79, 654–661. [Google Scholar]
- Zhao, X.; Huang, G.; Wu, L.; Wang, M.; He, X.; Wang, J.-R.; Zhou, B.; Liu, Y.; Lin, Y.; Liu, D.; et al. Deep learning assessment of left ventricular hypertrophy based on electrocardiogram. Front. Cardiovasc. Med. 2022, 9, 952089. [Google Scholar] [CrossRef]
- Sammani, A.; Jansen, M.; de Vries, N.M.; de Jonge, N.; Baas, A.F.; Te Riele, A.S.J.M.; Asselbergs, F.W.; Oerlemans, M.I.F.J. Automatic Identification of Patients with Unexplained Left Ventricular Hypertrophy in Electronic Health Record Data to Improve Targeted Treatment and Family Screening. Front. Cardiovasc. Med. 2022, 9, 768847. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, T.; Kodera, S.; Sawano, S.; Katsushika, S.; Nakamoto, M.; Takeuchi, H.; Kimura, N.; Shinohara, H.; Matsuoka, R.; Nakanishi, K.; et al. Automatic Detection of Left Ventricular Dilatation and Hypertrophy from Electrocardiograms Using Deep Learning. Int. Heart J. 2022, 63, 939–947. [Google Scholar] [CrossRef]
- Naderi, H.; Ramírez, J.; van Duijvenboden, S.; Pujadas, E.R.; Aung, N.; Wang, L.; Anwar Ahmed Chahal, C.; Lekadir, K.; Petersen, S.E.; Munroe, P.B. Predicting left ventricular hypertrophy from the 12-lead electrocardiogram in the UK Biobank imaging study using machine learning. Eur. Heart J. Digit. Health 2023, 4, 316–324. [Google Scholar] [CrossRef]
- Liu, C.-W.; Wu, F.-H.; Hu, Y.-L.; Pan, R.-H.; Lin, C.-H.; Chen, Y.-F.; Tseng, G.-S.; Chan, Y.-K.; Wang, C.-L. Left ventricular hypertrophy detection using electrocardiographic signal. Sci. Rep. 2023, 13, 2556. [Google Scholar] [CrossRef]
- Boser, B.; Guyon, I.; Vapnik, V. A training algorithm for optimal margin classifiers. In Proceedings of the fifth Annual Workshop on Computational Learning Theory—COLT ’92, Pittsburgh, PA, USA, 27–29 July 1992; ACM Press: New York, NY, USA, 1992; pp. 144–152. [Google Scholar]
- Golpour, P.; Ghayour-Mobarhan, M.; Saki, A.; Esmaily, H.; Taghipour, A.; Tajfard, M.; Ghazizadeh, H.; Moohebati, M.; Ferns, G.A. Comparison of Support Vector Machine, Naïve Bayes and Logistic Regression for Assessing the Necessity for Coronary Angiography. Int. J. Environ. Res. Public Health 2020, 17, 6449. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Uddin, S.; Khan, A.; Hossain, M.E.; Moni, M.A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 281. [Google Scholar] [CrossRef]
- Hong, S.; Zhou, Y.; Shang, J.; Xiao, C.; Sun, J. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comput. Biol. Med. 2020, 122, 103801. [Google Scholar] [CrossRef] [PubMed]
- Andaur Navarro, C.L.; Damen, J.A.A.; Takada, T.; Nijman, S.W.J.; Dhiman, P.; Ma, J.; Collins, G.S.; Bajpai, R.; Riley, R.D.; Moons, K.G.M.; et al. Risk of bias in studies on prediction models developed using supervised machine learning techniques: Systematic review. BMJ 2021, 375, n2281. [Google Scholar] [CrossRef]
- Collins, G.S.; Dhiman, P.; Andaur Navarro, C.L.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef]
- Wallace, M.L.; Mentch, L.; Wheeler, B.J.; Tapia, A.L.; Richards, M.; Zhou, S.; Yi, L.; Redline, S.; Buysse, D.J. Use and misuse of random forest variable importance metrics in medicine: Demonstrations through incident stroke prediction. BMC Med. Res. Methodol. 2023, 23, 144. [Google Scholar] [CrossRef] [PubMed]
- Rabkin, S.W.; Zhou, J. Estimating left ventricular mass from the electrocardiogram across the spectrum of LV mass from normal to increased LV mass in an older aged group. Cardiol. Res. Pract. 2024, 6634222. [Google Scholar] [CrossRef] [PubMed]
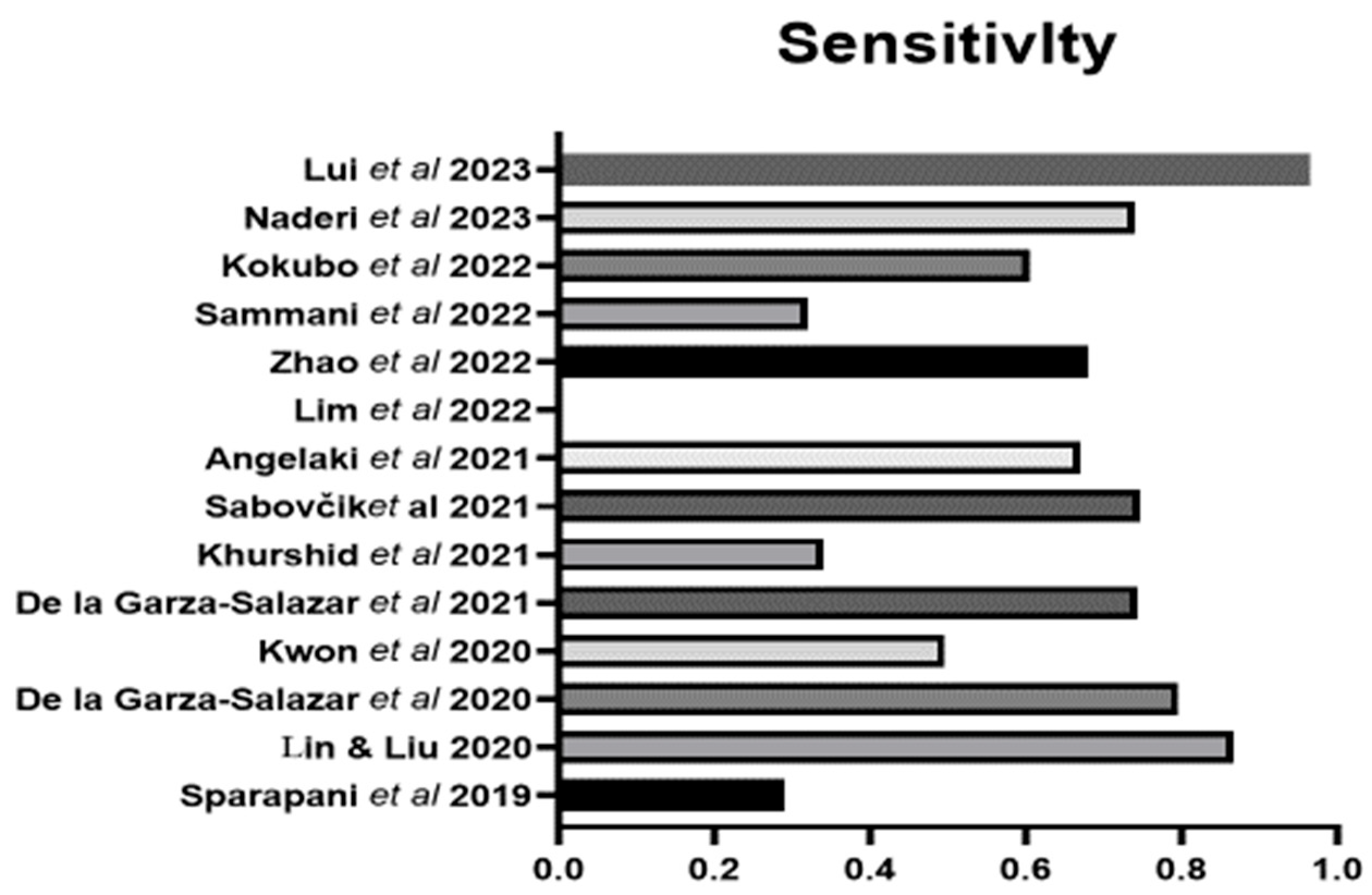
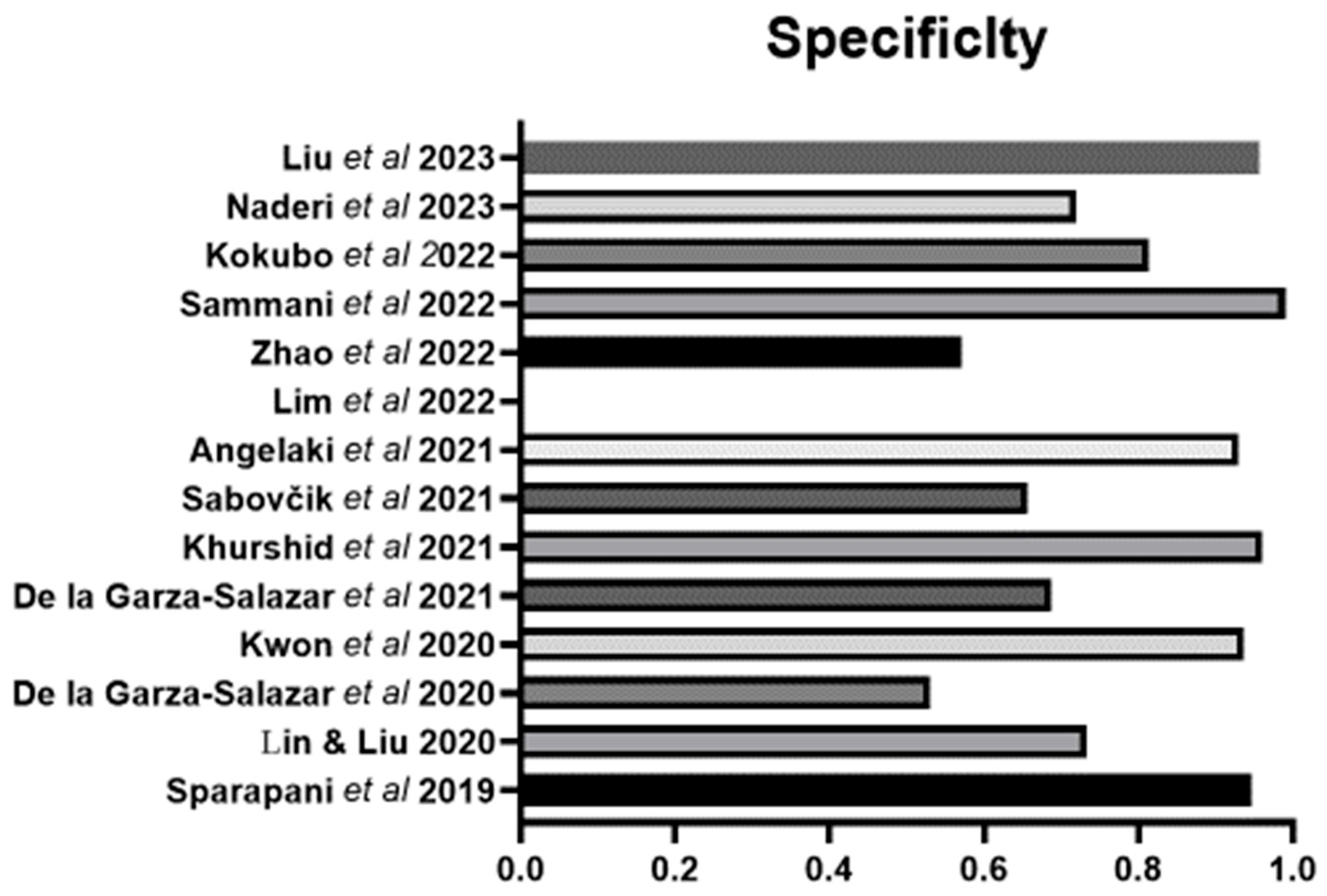


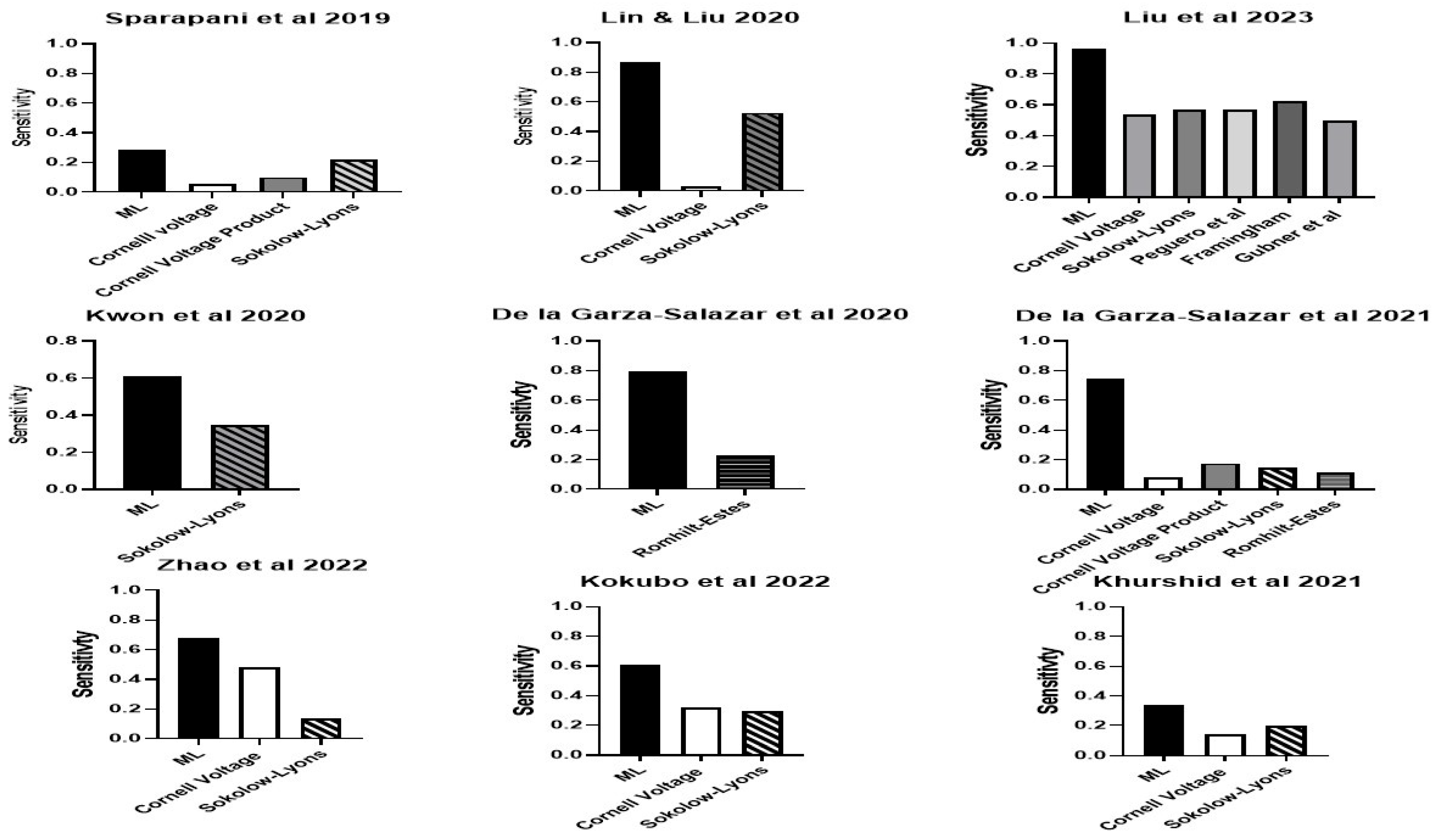
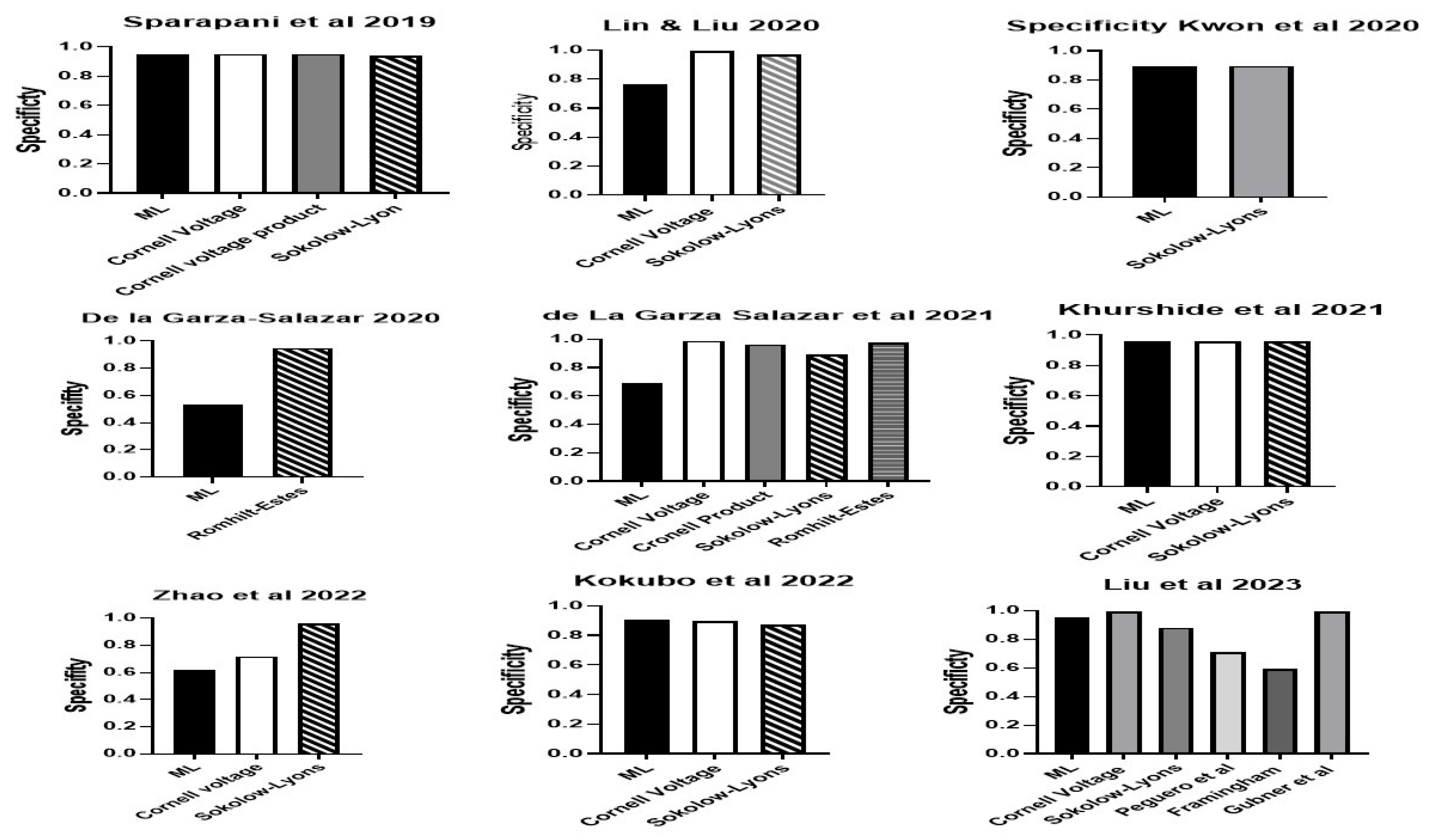
| Authors | Population | Country | Sample Size | Sex (%M) | Age (yrs) | Method LVH | Definition LVH | LVH | Variables | Machine Learning |
|---|---|---|---|---|---|---|---|---|---|---|
| Lin & Lui 2020 [31] | Military | Tawain | 2196 | 100 | 26 | Echocardiogram | ≥116 g/m2 | 6.5% | 31 parameters 3 clinical -age, body height, body weight | Support vector machine classifier (SVM) |
| 28 ECG parameters: duration P, PR, QRS, QT, QTc, P axis QRS axis, T axis plus | ||||||||||
| R amplitude in all 12 leads, S amplitude in avL, V1-6 | ||||||||||
| Sparapani et al., 2019 [32] | Multi-ethnic | USA | 4714 | 46 | MRI | 95th percentile | NA | 556 ECG variables: PR interval, P axis, QRS interval, QRS axis plus 552 amplitudes and durations per ECG | Bayesian additive regression tree | |
| De la Garza-Salazar et al., 2020 [33] | Hospital | Mexico | 432 | 56 | 67 | Echocardiogram | >115 g/m2 (men) | 48% | ECG p wave, QRS complex and ST waves | C5.0 supervised ML algorithm to create a multilevel binary decision tree, |
| >95 g/m2 (women). | ||||||||||
| Kwon et al., 2020 [34] | Hospital based | Korea | 21,286 | 49 | 59 | Echocardiogram | >132 g/m2 in men | 21% | age, sex, weight, height and ECG features, heart rate, presence of atrial fibrillation or flutter, QT, QRS duration, R-wave axis, T-wave | ENN, LR and RF |
| >109 g/m2 in women | ‘Raw’ ECG data with 5000 numbers from each of the 12 leads. | |||||||||
| De la Garza-Salazar et al., 2021 [35] | Hospital | Mexico | 439 | NA | 67 | Echocardiogram | Presumed same as 2020 | 46% | ECG variables including T wave voltage in the lead I, peak-to-peak QRS distance (QRS PPK) in aVF, and peak-to-peak QRS distance in aVL | C5.0 supervised ML algorithm to create a multilevel binary decision tree, |
| Khurshid et al., 2021 [36] | UK data base | UK | 32,239 | 47 | 64 | MRI | 2.6% | |||
| Sabovčik et al., 2021 [37] | General population | Belgium | 1407 | 49 | 51 | Echocardiogram | >115 g/m2 (men) | 19% | 67 variables including clinical, ECG onsets, amplitudes and intervals of P waves, QRS-complexes, and T wave as well as | LR, XGBoost, Random Forest, AdaBoost, Support Vector Machines |
| or 95 g/m2(women). | blood count, blood glucose, lipid profile, hormones (plasma renin, leptin, insulin, aldosterone, and cortisol), minerals, | |||||||||
| Angelaki et al. 2021 [38] | NA | Greece | 528 | 44 | 61 | Echocardiogram | >115 g/m2 (men) | 16.8% | clinical variables (sex, age, BMI class, BSA, hypertension, and height | |
| >95 g/m2 (women) | 26 chosen ECG-derived features | Random Forest | ||||||||
| Lim et al., 2021 [39] | Military | Singapore | 17,310 | 100 | 18 | Echocardiogram | >115 g/m2 (men) | 0.8% | clinical variables were: body weight, height, body fat percentage, and systolic blood pressure | Logistic Regression, GLMNet, Random Forests, Gradient Boosting Machines |
| ECG variables included: QT interval, mean QRS duration and R wave in lead I | ||||||||||
| Zhao et al., 2022 [40] | Hospital based | China | 3120 | 42 | 65 | Echocardiogram | >115 g/m2 (men) | 56% | uncertain | CNN |
| >95 g/m2 (women). | Lab: Hgb, PLT, lipids, creatinine, Na, K | |||||||||
| Sammani et al., 2022 [41] | Hospital based | The Netherlands | 2456 | 55 | 61 | Echocardiogram | >115 g/m2 (men) | 0.8% | age, systolic blood pressure and body surface area | XGBoost |
| >95 g/m2 (women). | 20 ECG data: p, QRS and T wave axes, pr, QRS, QT and QTc durations, peak amplitudes of p, Q, R, S and T waves | |||||||||
| Kokubo et al., 2022 [42] | Hospital based | Japan | 12,008 | 64 | 57 | Echocardiogram | >101 g/m2 for men | 16.5% | 19 factors—clinical (age, sex, height and weight) and ECG features (heart rate, rhythm, pr interval, QT interval. QRS axis, p wave axis | ENN |
| >85 g/m2 for women | as well as QRS voltages in leads V1, V2, V5 and V6 | LR, RF | ||||||||
| Naderi et al., 2023 [43] | UK data base | UK | 37,534 | 48 | 64 | MRI | >70 g/m2 (men) | 1.5% | Clinical—blood pressure, diabetes mellitus, lipids, cigarette and alcohol consumption | |
| >55 g/m2 (women) | 23 ECG variables from leads I, II, V1-6 | LR, SVM, RF | ||||||||
| Liu et al., 2023 [44] | Military Hospital | Tawain | 952 | 90 | Echocardiogram | >115 g/m2 (men) | 18% | 24 features which consisted of R peak and S valley amplitudes automatically obtained from the output of ECG signal | Decision tree SVM and Back propagated Neural Network | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabkin, S.W. Searching for the Best Machine Learning Algorithm for the Detection of Left Ventricular Hypertrophy from the ECG: A Review. Bioengineering 2024, 11, 489. https://doi.org/10.3390/bioengineering11050489
Rabkin SW. Searching for the Best Machine Learning Algorithm for the Detection of Left Ventricular Hypertrophy from the ECG: A Review. Bioengineering. 2024; 11(5):489. https://doi.org/10.3390/bioengineering11050489
Chicago/Turabian StyleRabkin, Simon W. 2024. "Searching for the Best Machine Learning Algorithm for the Detection of Left Ventricular Hypertrophy from the ECG: A Review" Bioengineering 11, no. 5: 489. https://doi.org/10.3390/bioengineering11050489
APA StyleRabkin, S. W. (2024). Searching for the Best Machine Learning Algorithm for the Detection of Left Ventricular Hypertrophy from the ECG: A Review. Bioengineering, 11(5), 489. https://doi.org/10.3390/bioengineering11050489







