A Privacy-Preserving Approach to Effectively Utilize Distributed Data for Malaria Image Detection
Abstract
1. Introduction
1.1. Research Background
1.2. Problem Statement and Rationale of the Research
- Medical images are highly heterogeneous compared to the normal images, therefore it is quite challenging to perform ML modelling on the limited data available [9]. To compensate this issue, we are required to form a collaborative framework that allows multiple hospitals and medical institutions to share data in a privacy-preserving manner.
- Lab-based data or synthesized data are limited to perform effective ML modelling [10], therefore we were required to have vast quantity of data that can be fulfilled by using the live stream of real-time data.
- It can be possible to use the real-time data to fulfil the data availability problem, however due to general data regulation and protection (GDPR), data sharing cannot be possible. Therefore, our research is inspired to use the privacy-preserving framework of federated learning (FL) to allow the data sharing while following GDPR.
- Is it possible to utilise real-time malaria image data by collaboration of different hospitals and medical institutes in a privacy-preserving manner?
- What are state-of-the-art machine learning approaches for effective malaria image detection?
1.3. Significance of the Research
- Improved security and compliance: The proposed research work provides huge potential considering the privacy and security of medical imaging. The research follows the guidelines as per GDPR and DPA for data security that will be a great revolution in the medical industry.
- Enhanced diagnostic capacity: The research framework that constitutes the hybrid model will ensure the security of data and the accuracy efficiency of disease detection that will ultimately result in early diagnosis and treatment.
- Facilitating collaboration: Research will promote an innovative culture that allows the mutual collaboration of hospitals and medical institutes to achieve the improved advancements.
- Benchmark for potential innovation: Based on the study analysis, it will guide the future scope of innovation in medical imaging for researchers. The proposed research can be a benchmark for the future development of this idea that can be mutually beneficial for medical institutes.
- Scalable and flexible framework: The research illustrates the use of a CNN-based pre-trained model on the federated learning framework that is highly scalable towards the multiple types of medical images, and provides a robust and enhanced solution for medical image detection.
- Economic influence: Research will bring important changes to ease the economic impact, such as early detection. It will ultimately bring about early disease detection, saving costs and resources in the medical industry.
- Global extent and convenience: Using the FL framework as in the proposed research will ensure data privacy, allowing data from diverse sources to enhance machine learning models’ learning capability.
1.4. Contribution to Knowledge
1.5. Paper Organisation
2. Literature Review
3. Methodology
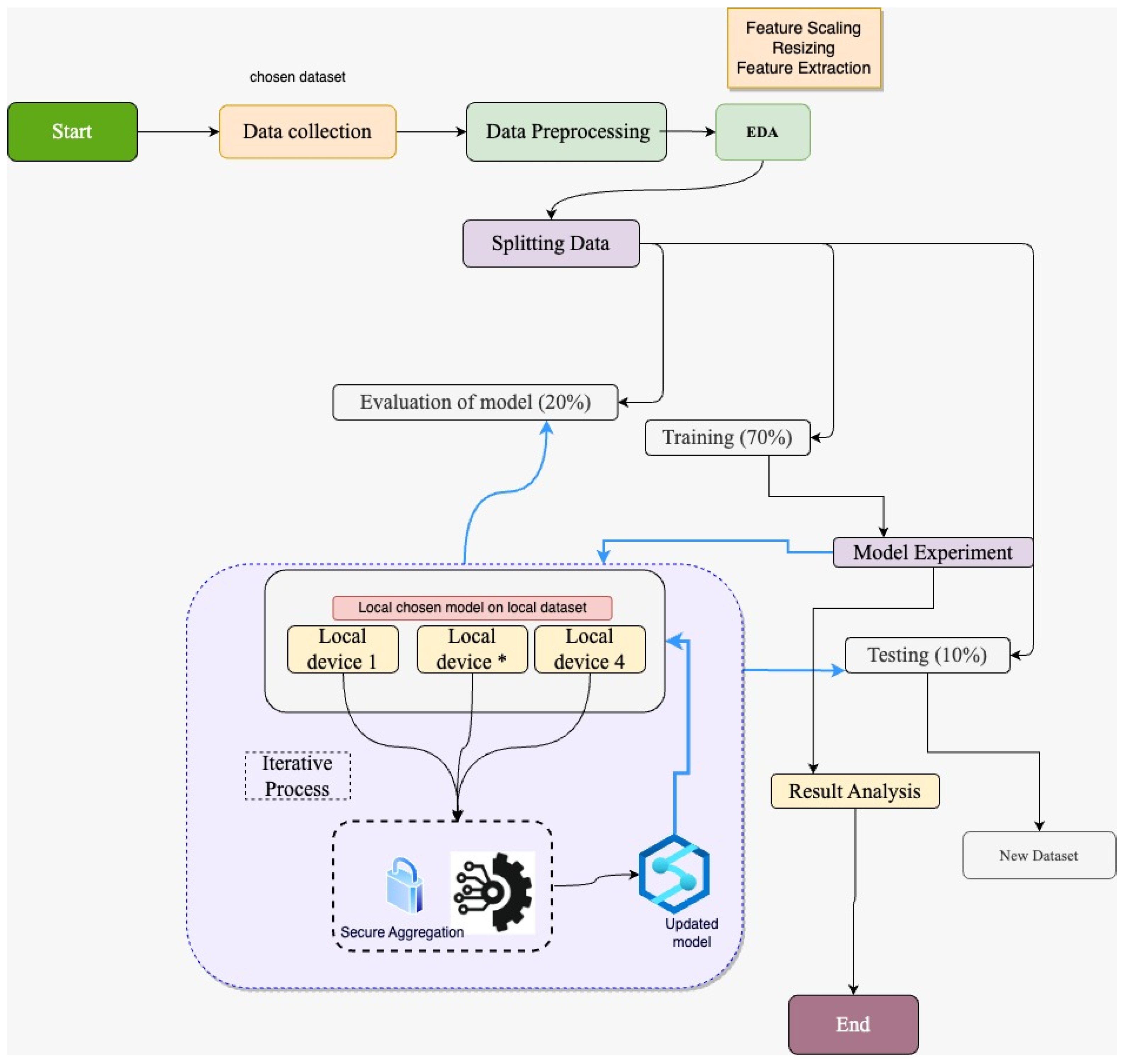
3.1. Research Model Design
3.2. Configuration of Models in Federated Learning
- 8.
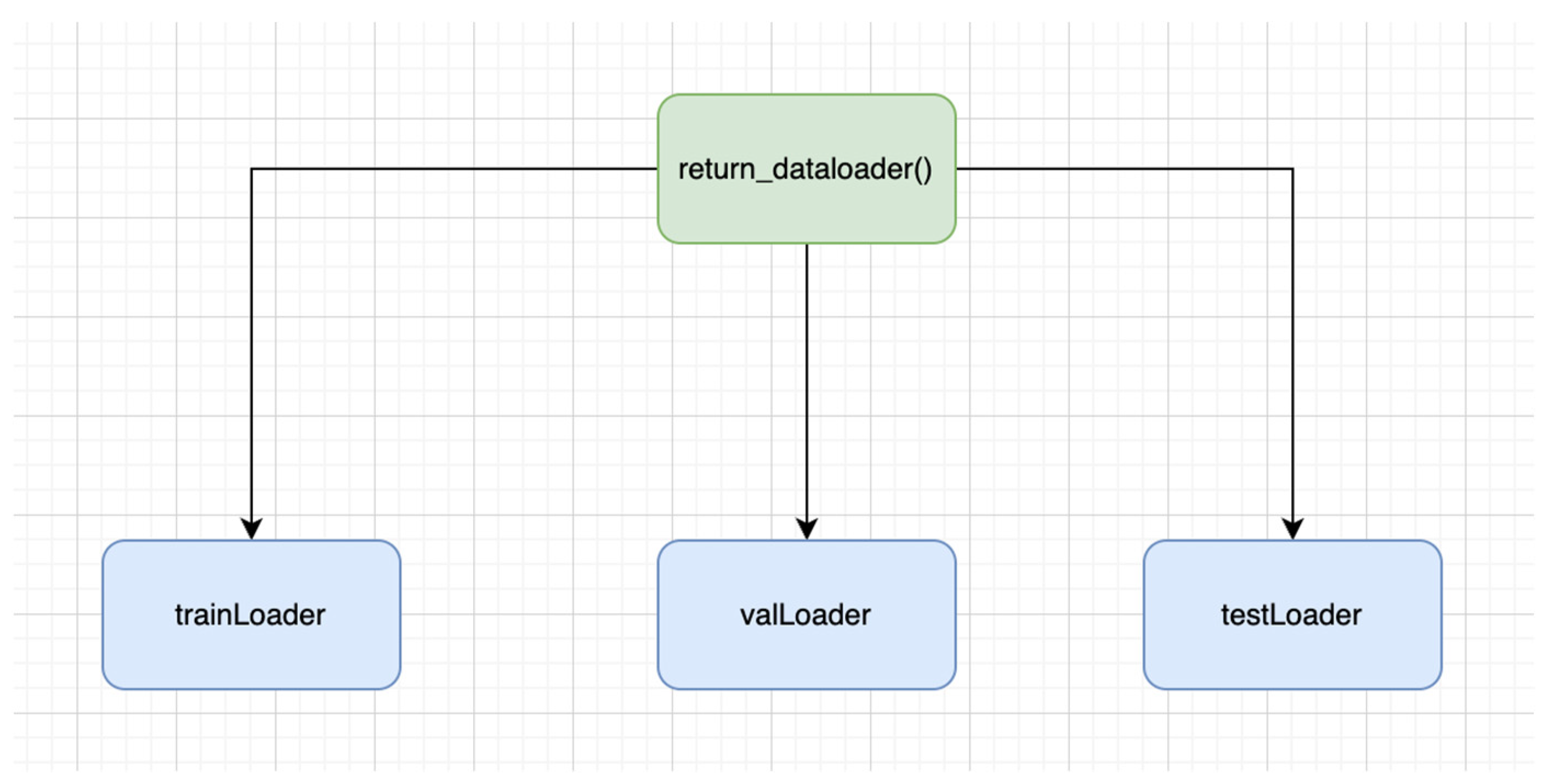
- TrainLoader: It is the type of data loader that determines the training section of the dataset. It is quite useful in iterating the transformed images, as well as in iterating the labels over the batches. Variability is also ensured, as it is involved in the reshuffling of the training data prior to every epoch.
- 9.
- valLoader: It shows the validation section of the dataset. Data iteration takes place over batches on validation images, as well as labels. It is not involved in the data reshuffling; therefore, the order stays intact throughout the epochs.
- 10.
- testLoader: It is the type of data loader that demonstrates the test part of the dataset. The data iteration takes place over the batched over the test images as well as labels. Unlike TrainLoader, it is not involved in data reshuffling.
- 11.
- Initialise the variable and list: The initial step involved in the tracking of the model state, validation loss, and the accuracy for the individual epoch.
- 12.
- Epoch loops: The train model function also loops over a certain number of epochs.
- 13.
- Phase loops: The function also performs the loops over the training as well as the validation stage of individual epochs.
- 14.
- Set Models Mode: The function also keeps the mode to ‘train’ if the model is in training mode. In this way, certain features including dropout as well as batch normalisation are activated. Similarly, during the validation stage, these parameters are disabled.
- 15.
- Batch loops: The function also loops with the data batches.
- 16.
- Forward pass: The function transfers the input label to the corresponding device via model and performs loss calculation.
- 17.
- Backward pass and optimisation: When using the train model function, if the training is zero, then the gradient of the model is calculated by using the optimisers also known as backward pass.
- 18.
- Statistics calculation: In the train_model function, the prediction is calculated, and the model run loss as well as accuracy is updated.
- 19.
- Epoch Statistic Calculation: During the training phase, by the end of every epoch, the loss and accuracy are calculated by the end of every epoch. While in the vase of validation stage, the precision, accuracy, F1-score is calculated while showing the confusion matrix.
- In our case, we have used clients 4, 6 and 8 to train the model individually on the clients while using the ‘train_model’ function, and in this way, model accuracy and loss are given out.
- The weights from the individual clients are kept in ‘w_local’ lists, similarly, the accuracy and loss of the model are kept on their respective lists.
- In the federated learning framework, the ‘fed_avg’ function is used to take the average weight of all models to form the global model.
- The mean loss and accuracy are also calculated on the participants.
- The mean average of all the weights is referred back towards the model on individual devices. It allows every client to receive the similar model update.
- The weight average of the models is stored as the ‘fed_model_client’.
- The output is displayed with the round phase, loss, as well as the accuracy.
- Alterations in ‘validation_loss’ as well as accuracy from the last round are displayed too.
- 20.
- Return best model: Once all corresponding epoch rounds are accomplished, the one with the least validation loss loads up the model.
3.3. Data Gathering
- 21.
- Malaria falciparum blood samples were taken from 150 patients at the Chittaging Medical College Hospital, Bangladesh.
- 22.
- The vivax malaria samples were taken from the same location as above from 150 patients, and also 50 from healthy individuals.
- 23.
- Malaria samples from falciparum were also taken from a similar location in Bangladesh: 148 patients and 45 healthy individuals.
- 24.
- Vibrax samples, which is another form of malaria, were also taken from Bangkok, Thailand, from 171 patients.
- 25.
- In addition, blood cell samples of falciparum malaria were collected from Bangladesh, from 150 patients and others from 50 healthy individuals.
- 26.
- Real-time data, non-synthetic: In the experiments, synthetic data can be quite useful for training the ML models, however, the lab-based model has a limitation to real-time complexity and variance of the disease. The data collected from the real-time provide stronger robust and efficient data for training the models. As in our experiments, we are targeting the real-time data; therefore, the given dataset is collected from the patients in real time, which helps model generalisation.
- 27.
- Geographical relevancy: Another significant impact of using this dataset involves its correspondence with the geographic relevance where malaria is the serious health issue, i.e., in Bangladesh and Thailand. The geographic location of the data helps to make the effective model prediction based on the provided data. The model customisation facility based on geographic location can help achieve higher performance. Similarly, it assists in nonspecific regions for malaria detection as well.
- 28.
- Reliable dataset: Another important aspect of using these data is the reliability, as the data have been collected from the endorsed hospitals that maintain the standards. Therefore, higher reliability is essential for better model performance.
- 29.
- Diverse images: The diverse malaria image collection helps to understand the variations of the causing parasites, which ultimately helps to train the ML model effectively.
- 30.
- Significant global impact: Malaria is one of the serious diseases that affect millions of individuals each year, with major health consequences. Therefore, the reflection of malaria disease constitutes the main global consideration.
3.4. Exploratory Data Analysis (EDA)
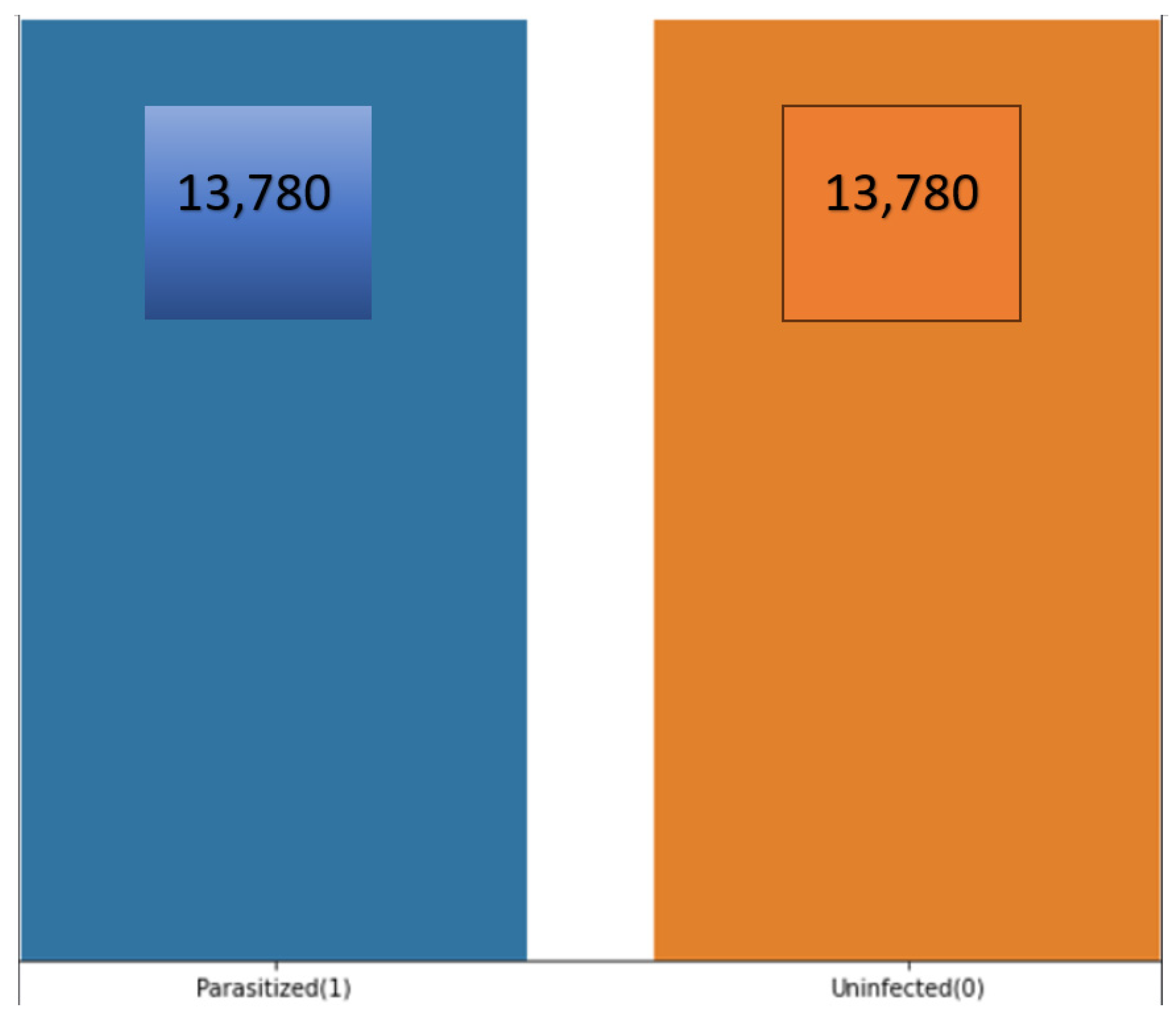
3.4.1. Number of Available Datasets
3.4.2. How Parasitised and Nonparasitised Cells Look
3.5. Data Preparation
- The initial stage involves reading the images from the directory.
- Decoding of the image content that involves converting into grid form as RGB.
- Conversion of images into float point tensor.
- Rescaling the tensors into the form that allows the scale range from 0 and 255 to be 0 s and 1 s as the CNN models takes in the smaller inputs.
3.6. Visualize the Training Images
4. Malaria Experiments and Results
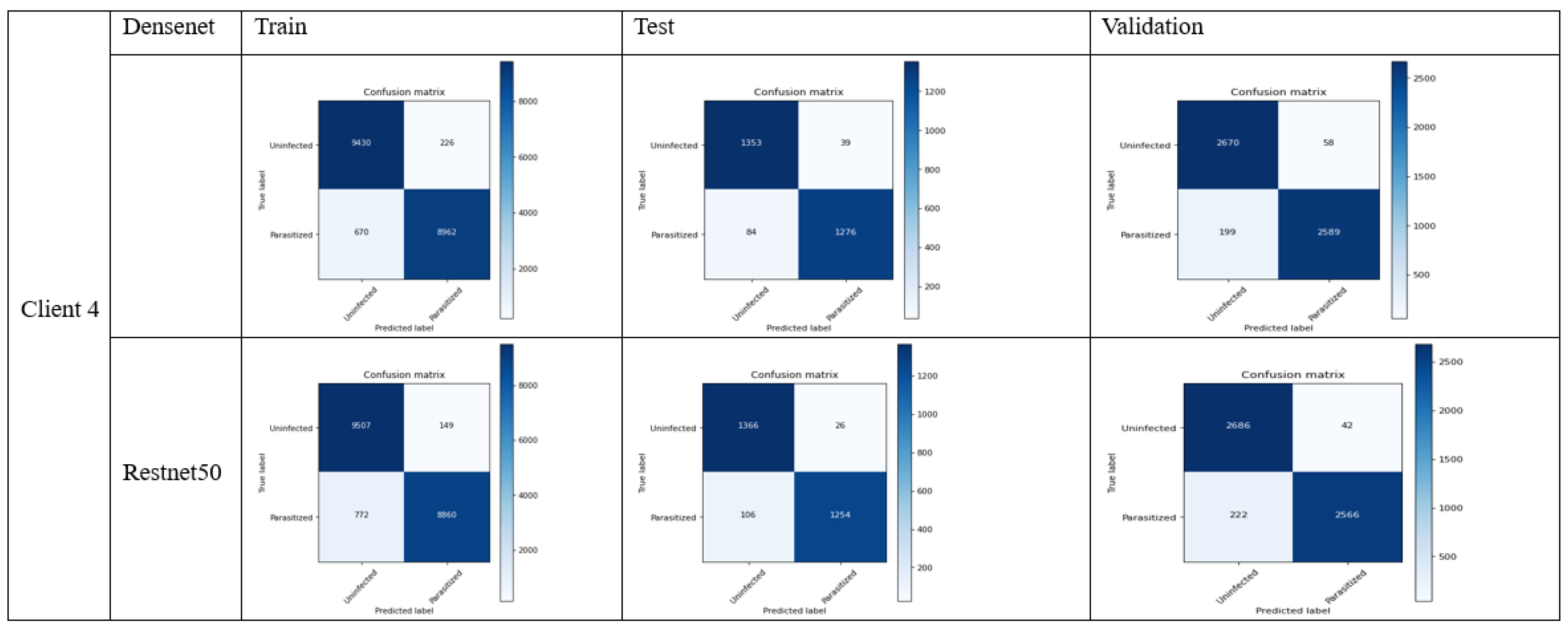
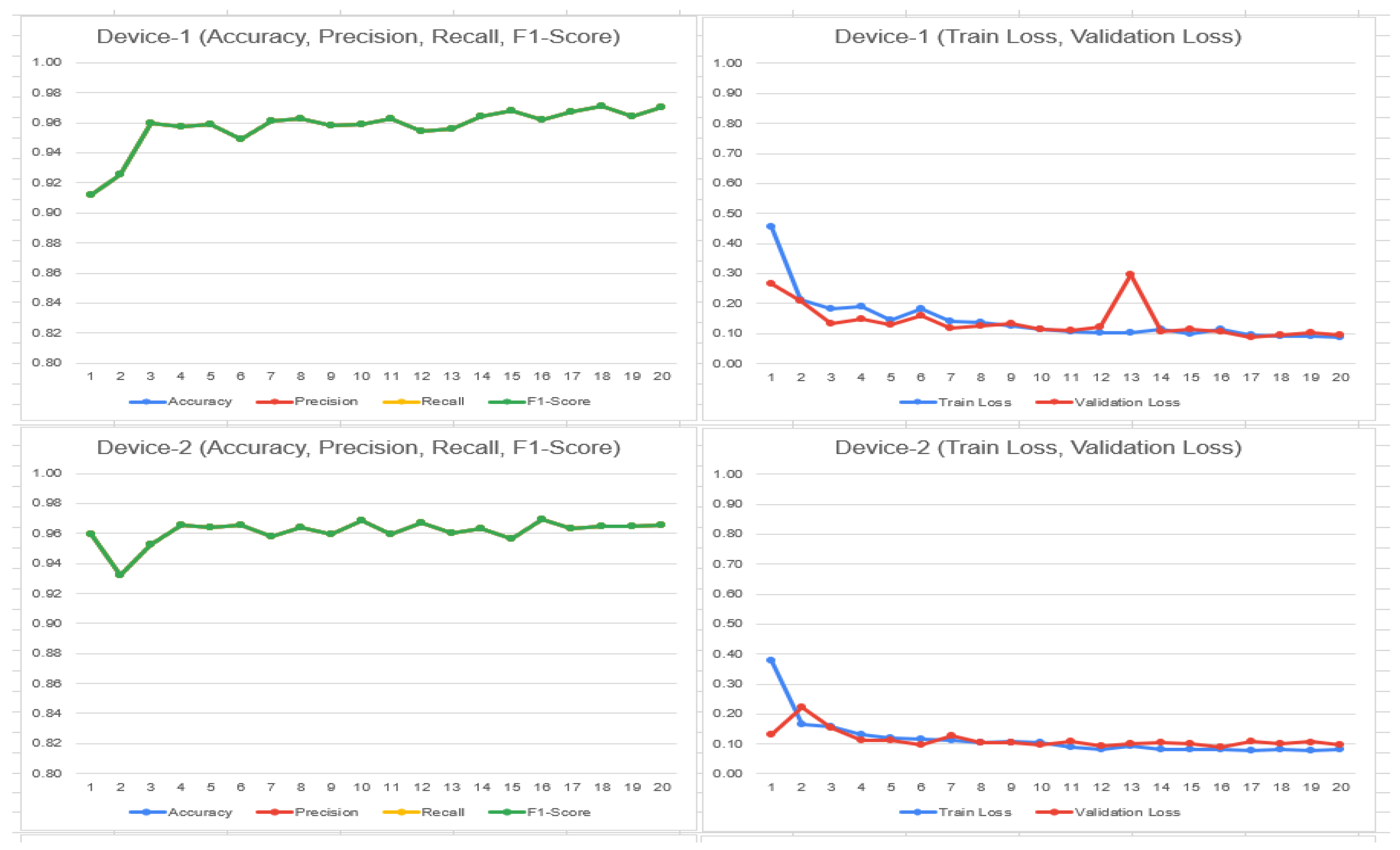
4.1. FL_DenseNet and FL_ResNet-50 (4 Clients)
- 31.
- Accuracy: (TP + TN)/(TP + FP + FN + TN)
- DenseNet: (1276 + 1353)/(1276 + 1353 + 39 + 84) = 0.9463 or 94.63%
- ResNet-50: (1254 + 1366)/(1254 + 1366 + 26 + 106) = 0.9486 or 94.86%
- 32.
- Precision: TP/(TP + FP)
- DenseNet: 1276/(1276 + 39) = 0.9703 or 97.03%
- ResNet-50: 1254/(1254 + 26) = 0.9796 or 97.96%
- 33.
- Recall (sensitivity): TP/(TP + FN)
- DenseNet: 1276/(1276 + 84) = 0.9382 or 93.82%
- ResNet-50: 1254/(1254 + 106) = 0.9220 or 92.20%
- 34.
- Specificity: TN/(TN + FP)
- DenseNet: 1353/(1353 + 39) = 0.9720 or 97.20%
- ResNet-50: 1366/(1366 + 26) = 0.9813 or 98.13%
- 35.
- F1 Score: 2 × (Precision × Recall)/(Precision + Recall)
- DenseNet: 2 × (0.9703 × 0.9382)/(0.9703 + 0.9382) = 0.9540 or 95.40%
- ResNet-50: 2 × (0.9796 × 0.9220)/(0.9796 + 0.9220) = 0.9501 or 95.01%
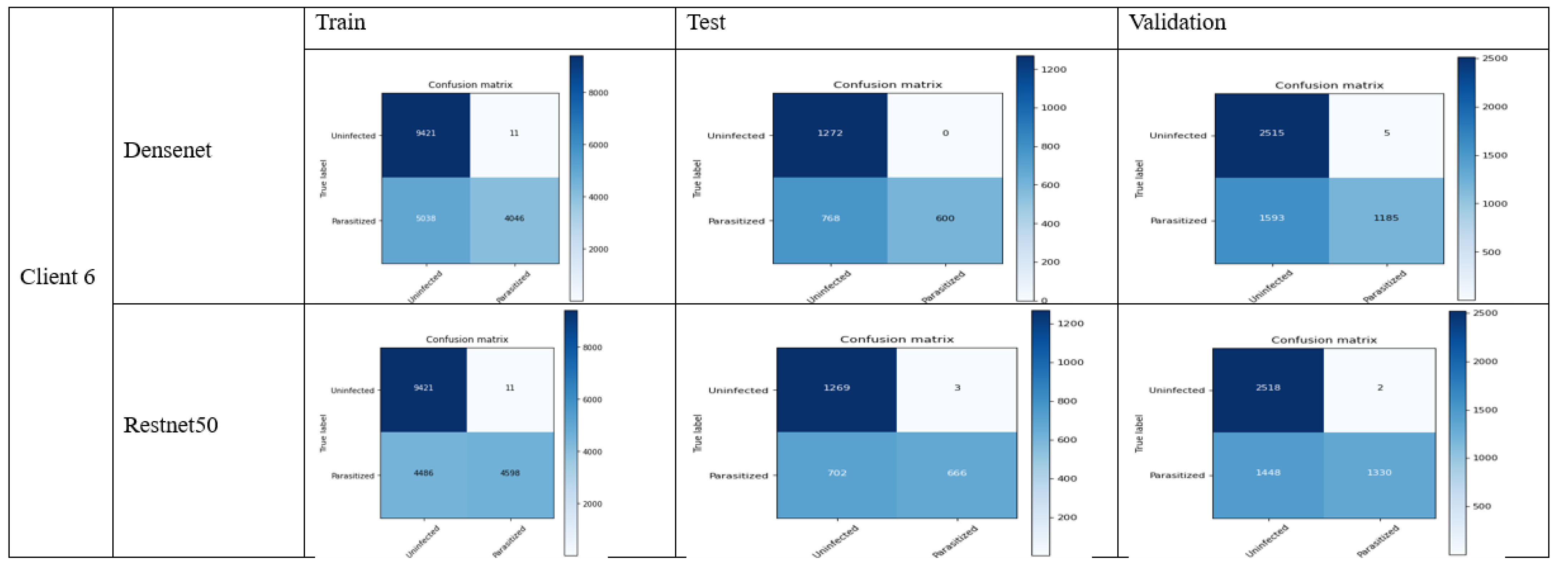
4.2. FL_DenseNet and FL_ResNet-50 (6 Clients)
- 36.
- Accuracy:
- DenseNet: (688 + 1272)/(688 + 0 + 168 + 1272) = 0.9213 (92.13%)
- ResNet-50: (666 + 1269)/(666 + 3 + 703 + 1269) = 0.7250 (72.50%)
- 37.
- Precision:
- DenseNet: 688/(688 + 0) = 1 (100%)
- ResNet-50: 666/(666 + 3) = 0.9955 (99.55%)
- 38.
- Recall:
- DenseNet: 688/(688 + 168) = 0.8037 (80.37%)
- ResNet-50: 666/(666 + 703) = 0.4864 (48.64%)
- 39.
- F1-score:
- DenseNet: 2 × (1 × 0.8037)/(1 + 0.8037) = 0.8911 (89.11%)
- ResNet-50: 2 × (0.9955 × 0.4864)/(0.9955 + 0.4864) = 0.6530 (65.30%)
- 40.
- Specificity:
- DenseNet: 1272/(1272 + 0) = 1 (100%)
- ResNet-50: 1269/(1269 + 3) = 0.9976 (99.76%)
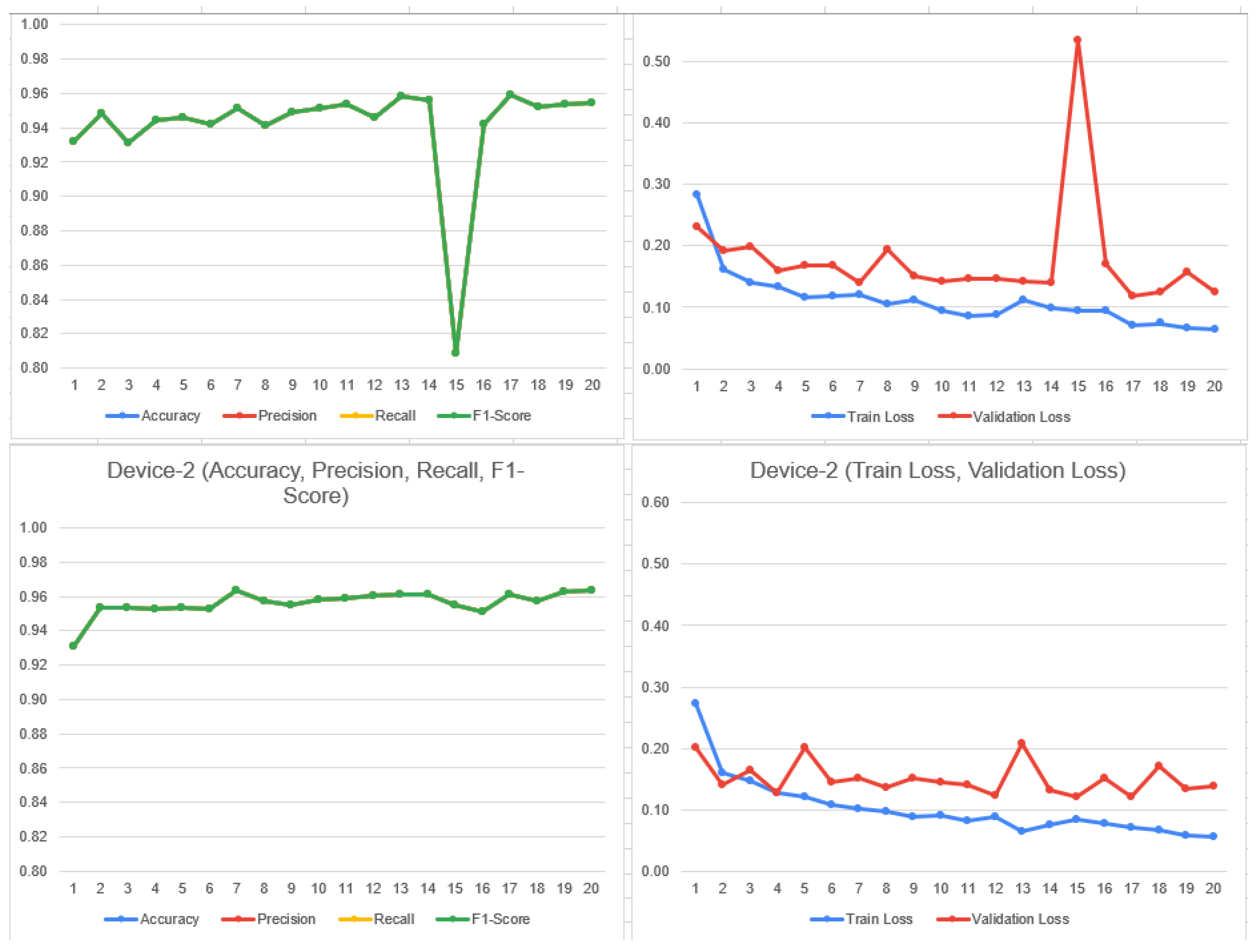
4.3. FL_DenseNet and FL_ResNet-50 (8 Clients)
- 41.
- Accuracy:
- DenseNet: (1308 + 712)/(1308 + 552 + 68 + 712) = 0.7504 (75.04%)
- ResNet-50: (666 + 1269)/(666 + 3 + 703 + 1269) = 0.7250 (72.50%)
- 42.
- Precision:
- DenseNet: 1308/(1308 + 552) = 0.7033 (70.33%)
- ResNet-50: 666/(666 + 3) = 0.9955 (99.55%)
- 43.
- Recall (sensitivity):
- DenseNet: 1308/(1308 + 68) = 0.9506 (95.06%)
- ResNet-50: 666/(666 + 703) = 0.4864 (48.64%)
- 44.
- F1-Score:
- DenseNet: 2 × (0.7033 × 0.9506)/(0.7033 + 0.9506) = 0.8087 (80.87%)
- ResNet-50: 2 × (0.9955 × 0.4864)/(0.9955 + 0.4864) = 0.6530 (65.30%)
- 45.
- Specificity (True Negative Rate):
- DenseNet: 712/(712 + 552) = 0.5635 (56.35%)
- ResNet-50: 1269/(1269 + 3) = 0.9976 (99.76%)
4.4. Significance Test
5. Discussion
- In the case where it is necessary to determine actual malaria disease as much as possible, DenseNet is preferred due to higher recall.
- In the case where it is required to determine actual malaria disease, and in reality, it is malaria, then ResNet-50 is preferred due to its higher precision.
- In the case where it is required to determine the balancing among the false positive as well as false negative, then DenseNet performs well due to higher recall.
- In the case where it is required to correctly determine the negative cases, that is, no-malaria, ResNet-50 performs well due to its effective specificity results.
6. Summary
7. Limitation and Constraints
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jakaite, L.; Schetinin, V.; Maple, C. Bayesian Assessment of Newborn Brain Maturity from Two-Channel Sleep Electroencephalograms. Comput. Math. Methods Med. 2012, 2012, 629654. [Google Scholar] [CrossRef] [PubMed]
- Jakaite, L.; Schetinin, V.; Maple, C.; Schult, J. Bayesian Decision Trees for EEG Assessment of newborn brain maturity. In Proceedings of the 2010 UK Workshop on Computational Intelligence (UKCI), Colchester, UK, 8–10 September 2010; pp. 1–6. [Google Scholar] [CrossRef]
- Balyan, A.K.; Ahuja, S.; Sharma, S.K.; Lilhore, U.K. Machine Learning-Based Intrusion Detection System For Healthcare Data. In Proceedings of the 2022 IEEE VLSI Device Circuit and System (VLSI DCS), Kolkata, India, 26–27 February 2022; pp. 290–294. [Google Scholar] [CrossRef]
- Schetinin, V.; Jakaite, L.; Jakaitis, J.; Krzanowski, W. Bayesian Decision Trees for predicting survival of patients: A study on the US National Trauma Data Bank. Comput. Methods Programs Biomed. 2013, 111, 602–612. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swapna, G.; Vinayakumar, R.; Soman, K.P. Diabetes detection using deep learning algorithms. ICT Express 2018, 4, 243–246. [Google Scholar]
- WHO. Responding to Community Spread of COVID-19. Reference WHO/COVID-19/Community_Transmission/2020.1; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Musleh, A.A.W.A.; Maghari, A.Y. COVID-19 Detection in X-ray Images using CNN Algorithm. In Proceedings of the 2020 International Conference on Promising Electronic Technologies (ICPET), Jerusalem, Palestine, 16–17 December 2020; pp. 5–9. [Google Scholar] [CrossRef]
- Data Protection Act 2018. Available online: https://www.legislation.gov.uk/ukpga/2018/12/part/2/chapter/2/enacted (accessed on 16 March 2024).
- Zhang, M.; Qu, L.; Singh, P.; Kalpathy-Cramer, J.; Rubin, D.L. Splitavg: A heterogeneity-aware federated deep learning method for medical imaging. IEEE J. Biomed. Health Inform. 2022, 26, 4635–4644. [Google Scholar] [CrossRef] [PubMed]
- Techavipoo, U.; Sinsuebphon, N.; Prompalit, S.; Thongvigitmanee, S.; Narkbuakaew, W.; Kiang-Ia, A.; Srivongsa, T.; Thajchayapong, P.; Chaumrattanakul, U. Image Quality Evaluation of a Digital Radiography System Made in Thailand. BioMed Res. Int. 2021, 2021, e3102673. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Raut, S. Detection of Diabetic Retinopathy from fundus images using Resnet50. In Proceedings of the 2023 2nd International Conference on Paradigm Shifts in Communications Embedded Systems, Machine Learning and Signal Processing (PCEMS), Nagpur, India, 5–6 April 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Sundari, S.M.; Rao, M.D.S.; Rani, M.S.; Durga, K.; Kranthi, A. COVID-19 X-ray Image Detection using ResNet50 and VGG16 in Convolution Neural Network. In Proceedings of the 2022 IEEE Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation (IATMSI), Gwalior, India, 14–16 March 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Schetinin, V.; Jakaite, L.; Nyah, N.; Novakovic, D.; Krzanowski, W. Feature Extraction with GMDH-Type Neural Networks for EEG-Based Person Identification. Int. J. Neural Syst. 2018, 28, 1750064. [Google Scholar] [CrossRef] [PubMed]
- Huy, V.T.Q.; Lin, C.-M. An Improved Densenet Deep Neural Network Model for Tuberculosis Detection Using Chest X-Ray Images. IEEE Access 2023, 11, 42839–42849. [Google Scholar] [CrossRef]
- Anjugam, S.; Arul Leena Rose, P.J. Study of Deep Learning Approaches for Diagnosing COVID-19 Disease using Chest CT Images. In Proceedings of the 2023 7th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 22–23 October 2023; pp. 263–269. [Google Scholar] [CrossRef]
- Chowdhury, D.; Unnikannan, A.; Ghosh, A.; Dutta, A.; Deo, D.S.; Saha, D.; Bhowmick, M.; Majumdar, M.; Bhowmik, S.; De, S.; et al. Detection of SARS-CoV-2 from human chest CT images in Multi-Convolutional Neural Network’s environment. In Proceedings of the 2023 11th International Conference on Internet of Everything, Microwave Engineering, Communication and Networks (IEMECON), Jaipur, India, 10–11 February 2023; pp. 1–7. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Wang, Q.; Toe, T.T. Detection of Pneumonia Based on ResNet Improved by Attention Mechanism. In Proceedings of the 2023 IEEE 3rd International Conference on Power, Electronics and Computer Applications (ICPECA), Shenyang, China, 29–31 January 2023; pp. 859–863. [Google Scholar] [CrossRef]
- Çinar, A.; Yildirim, M. Classification of malaria cell images with deep learning architectures. Ingénierie Des Systèmes D’information 2020, 25, 35. [Google Scholar] [CrossRef]
- Tariq, T.; Suhail, Z.; Nawaz, Z. Knee Osteoarthritis Detection and Classification Using X-rays. IEEE Access 2023, 11, 48292–48303. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Tardy, M.; Ballester, M.A.G.; Mateus, D.; Piella, G. Memory-aware curriculum federated learning for breast cancer classification. Comput. Methods Programs Biomed. 2023, 229, 107318. [Google Scholar] [CrossRef] [PubMed]
- Sohan, M.F.; Basalamah, A. A Systematic Review on Federated Learning in Medical Image Analysis. IEEE Access 2023, 11, 28628–28644. [Google Scholar] [CrossRef]
- Yang, D.; Xu, Z.; Li, W.; Myronenko, A.; Roth, H.R.; Harmon, S.; Xu, S.; Turkbey, B.; Turkbey, E.; Wang, X.; et al. Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from China Italy Japan. Med. Image Anal. 2021, 70, 101992. [Google Scholar] [CrossRef]
- Qayyum, A.; Ahmad, K.; Ahsan, M.A.; Al-Fuqaha, A.; Qadir, J. Collaborative federated learning for healthcare: Multi-modal COVID-19 diagnosis at the edge. IEEE Open J. Comput. Soc. 2022, 3, 172–184. [Google Scholar] [CrossRef]
- Salam, M.A.; Taha, S.; Ramadan, M. COVID-19 detection using federated machine learning. PLoS ONE 2021, 16, e0252573. [Google Scholar]
- Adnan, M.; Kalra, S.; Cresswell, J.C.; Taylor, G.W.; Tizhoosh, H.R. Federated learning and differential privacy for medical image analysis. Sci. Rep. 2022, 12, 1953. [Google Scholar] [CrossRef]
- Deng, Y.; Ren, J.; Tang, C.; Lyu, F.; Liu, Y.; Zhang, Y. A hierarchical knowledge transfer framework for heterogeneous federated learning. In Proceedings of the IEEE INFOCOM 2023-IEEE Conference on Computer Communications, New York, NY, USA, 17–20 May 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–10. [Google Scholar]
- Wang, Z.; Xiao, J.; Wang, L.; Yao, J. A novel federated learning approach with knowledge transfer for credit scoring. Decis. Support Syst. 2024, 177, 114084. [Google Scholar] [CrossRef]
- Albarqouni, S.; Bakas, S.; Bano, S.; Cardoso, M.J.; Khanal, B.; Landman, B.; Li, X.; Qin, C.; Rekik, I.; Rieke, N.; et al. (Eds.) Distributed, Collaborative, and Federated Learning, and Affordable AI and Healthcare for Resource Diverse Global Health: Third MICCAI Workshop, DeCaF 2022, and Second MICCAI Workshop, FAIR 2022, Held in Conjunction with MICCAI 2022, Singapore, September 18 and 22, 2022, Proceedings; Springer Nature: Berlin, Germany, 2022; Volume 13573. [Google Scholar]
- Available online: https://www.kaggle.com/datasets/iarunava/cell-images-for-detecting-malaria (accessed on 7 January 2024).
- Grabchak, M. How Do We Perform a Paired t-Test When We Don’t Know How to Pair? Am. Stat. 2023, 77, 127–133. [Google Scholar] [CrossRef]





| References | Disease | Algorithm | Accuracy | Dataset Size | Pros | Cons |
|---|---|---|---|---|---|---|
| [11] | Diabetic Retinopathy | ResNet-50 | 98% | 3762 images | ResNet-50 is effective when the image pre-processing is enhanced as the model performance can vary depending on the input images. | The performance of this algorithm is not further explored in larger datasets. |
| [12] | COVID-19 | ResNet-50 and VGG16 | ResNet50 was 88% VGG16 was 85% | 10 k | Effective classification considering both algorithms. | Data is imbalanced. In case of ResNet-50 precision is 100% while for VGG16 its 85% |
| [13] | COVID-19 | ResNet-50 | 88% to 94% | 5 K CT scan | Authors have clearly distinguish the use of different hyperparameter tuning to effectively increase the model accuracy for the disease prediction | Limited dataset size. Hyper tuning could be enhanced with use of some optimizer. |
| [14] | Tuberculosis (TB) | DenseNet | 98.8% | 5 K | This research has highlighted the use of different epochs to understand the best possible outcome of the experiment for medical image detection. | Dataset is highly imbalance between the normal and infected images. |
| [16] | SARS-CoV-2 | customised CNN model | 92% | 2481 CT scan | The customised CNN approach has produced in the classification of the SARS-CoV-2 virus | Limited dataset size. Higher number can adversely impact on the performance of the model. |
| [17] | Pneumonia | ResNet-50 | 90% | 5800 images | The performance of the ResNet-50 with the attention mechanism performs well in terms of accuracy | Higher time consumption in training dataset |
| [18] | Malaria | CNN models | 97.83% for DenseNet-201 | 6730 images | The use of gauss filter has increased the overall accuracy | Processor intensive. Gaussian filtering can blur the images which ultimately results in the loss of essential image details including edges which causes ineffective classification. |
| [19] | Knee Osteoarthritis | CNN models | 98% | 9786 images | Effective approach of ensemble for the detection of knee osteoarthritis. | Model overfitting issues where the different models have their own capabilities to input and process the data. |
| Our contribution | Malaria (4 clients) | ResNet-50 DenseNet | ResNet-50 = 94.86% DenseNet = 94.63% | 27 k images | Privacy preserving approach of using dataset from different clients while following GDPR with enhanced accuracy in detection of malaria disease. | Legal regulation of deploying this architecture in the real-time could be challenging as it requires standard operating procedures. The reliance on image annotation from the medical experts can make this process slower. |
| Our contribution | Malaria (6 clients) | ResNet-50 DenseNet | ResNet-50 = 72.50% DenseNet = 92.13% | 27 k images | Privacy preserving approach of using dataset from different clients while following GDPR with enhanced accuracy in detection of malaria disease. | Legal regulation of deploying this architecture in the real-time could be challenging as it requires standard operating procedures. The reliance on image annotation from the medical experts can make this process slower. |
| Our contribution | Malaria (8 clients) | ResNet-50 DenseNet | ResNet-50 = 72.50% DenseNet = 75.04% | 27 k images | Privacy preserving approach of using dataset from different clients while following GDPR with enhanced accuracy in detection of malaria disease. | Legal regulation of deploying this architecture in the real-time could be challenging as it requires standard operating procedures. The reliance on image annotation from the medical experts can make this process slower. |
| Accuracy | Precision | Recall | F1 Score | Specificity | |
|---|---|---|---|---|---|
| ResNet-50 | 96.63% | 97.96% | 92.20% | 95.01% | 98.13% |
| DenseNet | 94.86% | 97.03% | 93.82% | 95.40% | 97.20% |
| Accuracy | Precision | Recall | F1 Score | Specificity | |
|---|---|---|---|---|---|
| ResNet-50 | 72.50% | 99.55% | 48.64% | 65.30% | 100.00% |
| DenseNet | 92.13% | 100.00% | 80.37% | 89.11% | 97.20% |
| Accuracy | Precision | Recall | F1 Score | Specificity | |
|---|---|---|---|---|---|
| ResNet-50 | 72.50% | 99.55% | 48.64% | 65.30% | 99.76% |
| DenseNet | 75.04% | 70.33% | 95.06% | 80.87% | 56.35% |
| Metrics | Model Name | T-Stats | p-Value |
|---|---|---|---|
| Accuracy | FL_DENSENET And FL RESNET-50 | 11.45726 | 0 |
| Precision | FL_DENSENET And FL RESNET-50 | −0.09118 | 0.92746 |
| Re-call | FL_DENSENET And FL RESNET-50 | 0.09637 | 0.92335 |
| F1-Score | FL_DENSENET And FL RESNET-50 | 0.25665 | 0.79778 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kareem, A.; Liu, H.; Velisavljevic, V. A Privacy-Preserving Approach to Effectively Utilize Distributed Data for Malaria Image Detection. Bioengineering 2024, 11, 340. https://doi.org/10.3390/bioengineering11040340
Kareem A, Liu H, Velisavljevic V. A Privacy-Preserving Approach to Effectively Utilize Distributed Data for Malaria Image Detection. Bioengineering. 2024; 11(4):340. https://doi.org/10.3390/bioengineering11040340
Chicago/Turabian StyleKareem, Amer, Haiming Liu, and Vladan Velisavljevic. 2024. "A Privacy-Preserving Approach to Effectively Utilize Distributed Data for Malaria Image Detection" Bioengineering 11, no. 4: 340. https://doi.org/10.3390/bioengineering11040340
APA StyleKareem, A., Liu, H., & Velisavljevic, V. (2024). A Privacy-Preserving Approach to Effectively Utilize Distributed Data for Malaria Image Detection. Bioengineering, 11(4), 340. https://doi.org/10.3390/bioengineering11040340





