A Novel Detection and Classification Framework for Diagnosing of Cerebral Microbleeds Using Transformer and Language
Abstract
1. Introduction
- (1)
- We introduce a decoupled two-stage framework named MM-UniCMBs, characterized by the front-light and back-heavy architecture;
- (2)
- We propose a novel lightweight CMBs detection network called CMBs-YOLO, which incorporates the CMBs-MHA module and NWD loss;
- (3)
- An innovative multimodal classification network named CMBsFormer is proposed by integrating text language descriptions with a transformer-based architecture;
- (4)
- The extensive experimental results demonstrate that our proposed framework can achieve satisfactory performance in both the CMB detection and classification tasks.
2. Related Work
3. Materials and Methods
3.1. Data Acquisition
3.2. Methods
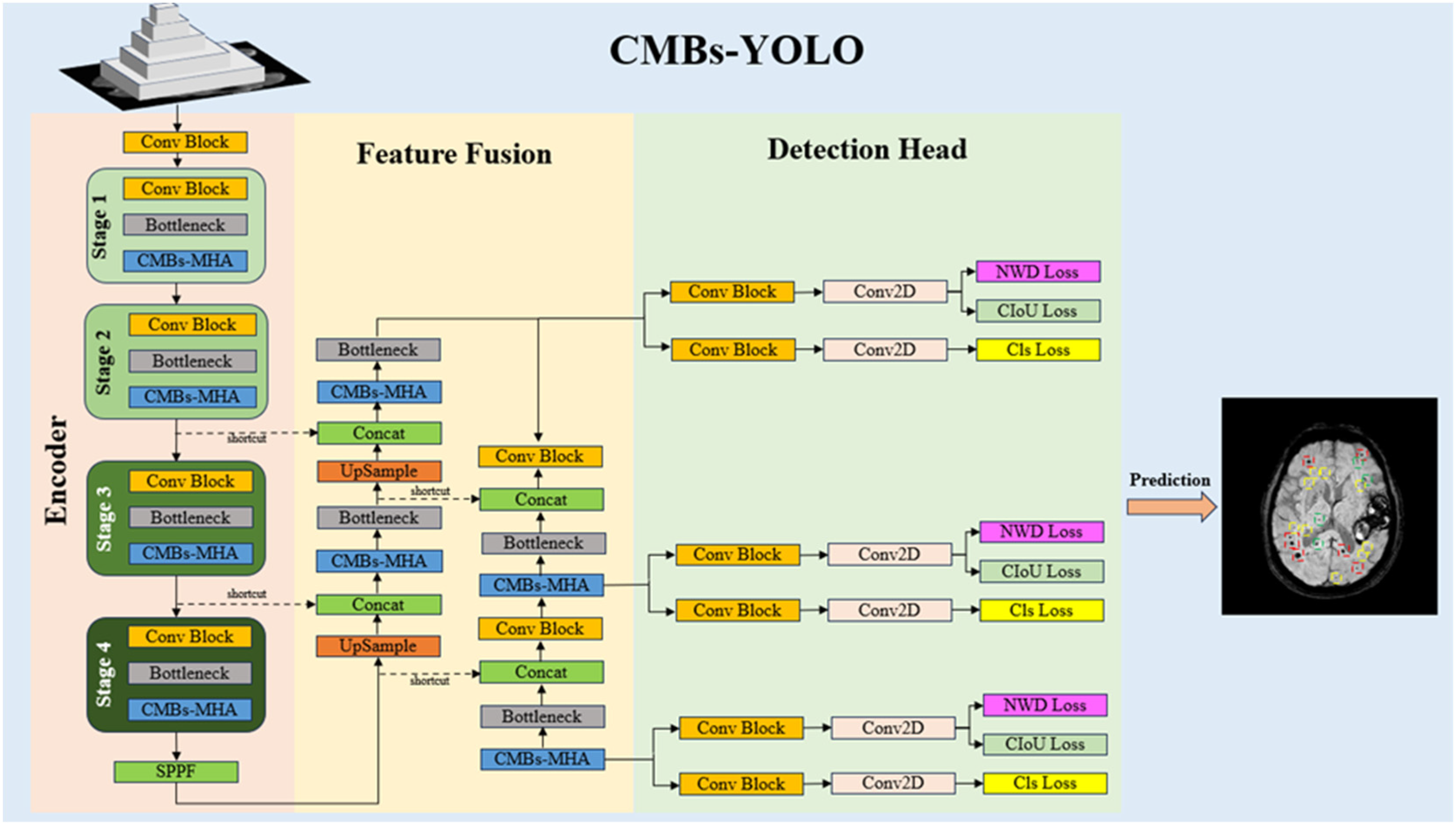
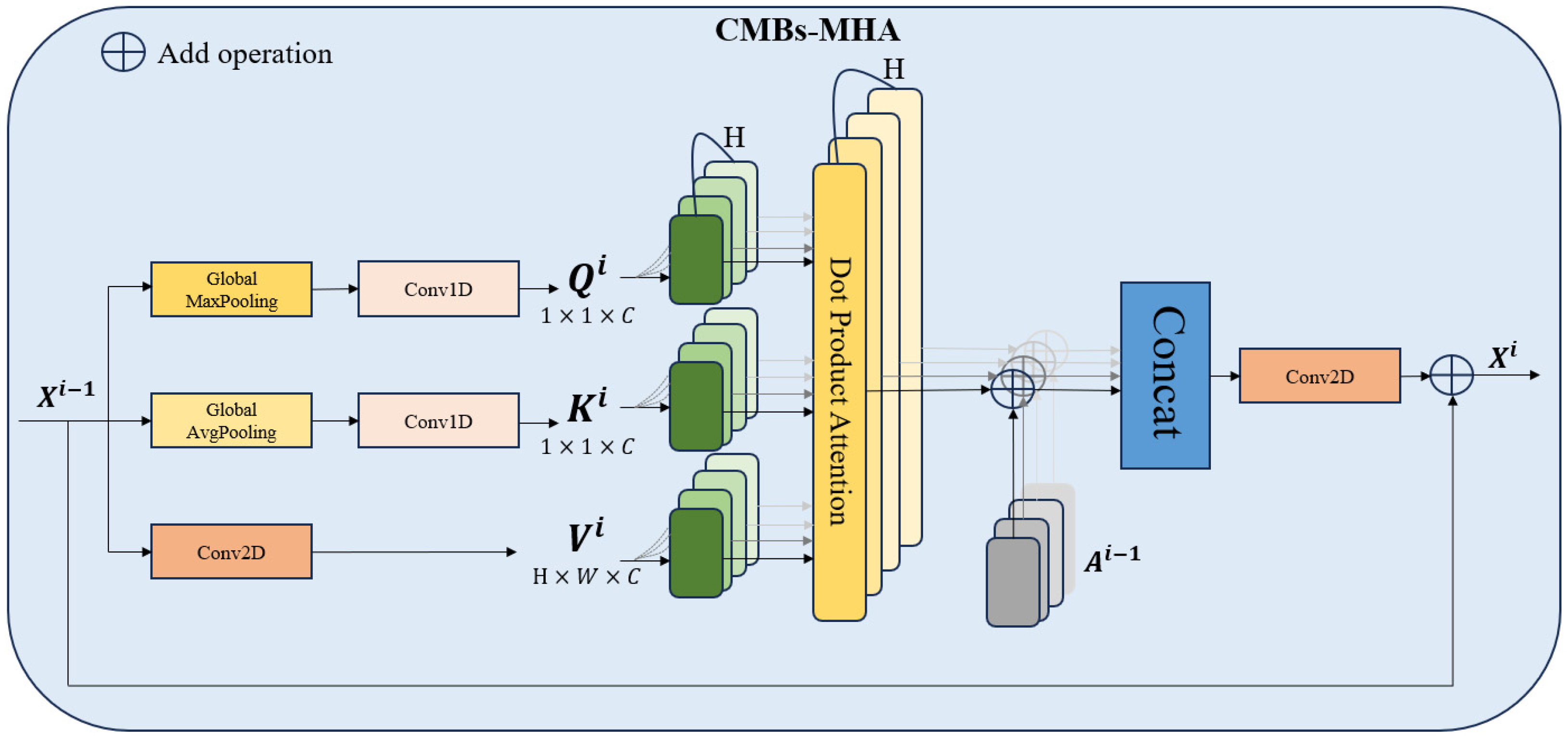
3.2.1. CMBs-YOLO
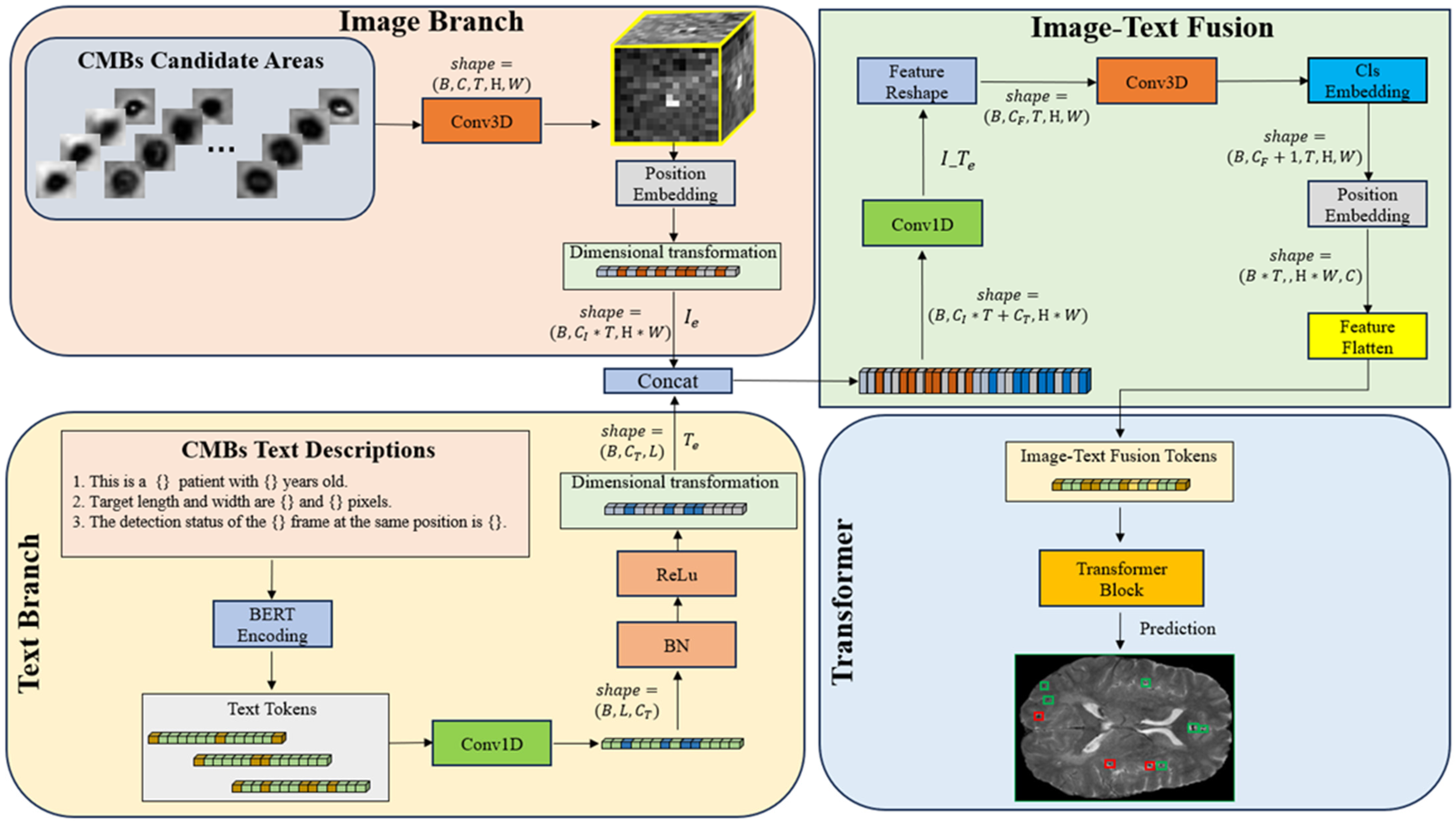
3.2.2. CMBsFormer
4. Results
4.1. Evaluation Metrics
4.2. Experiment Setup
4.3. Evaluation of Model Performance
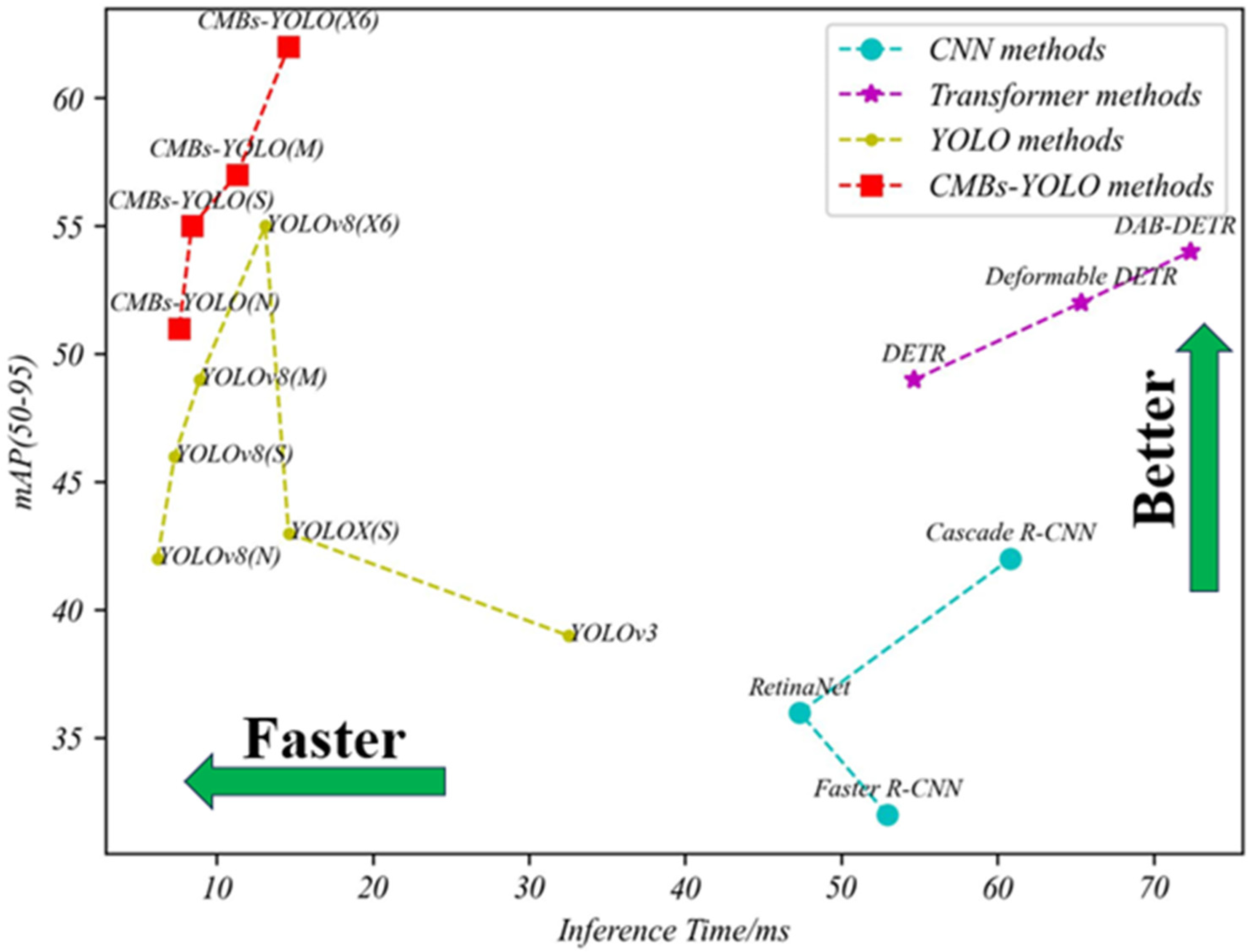
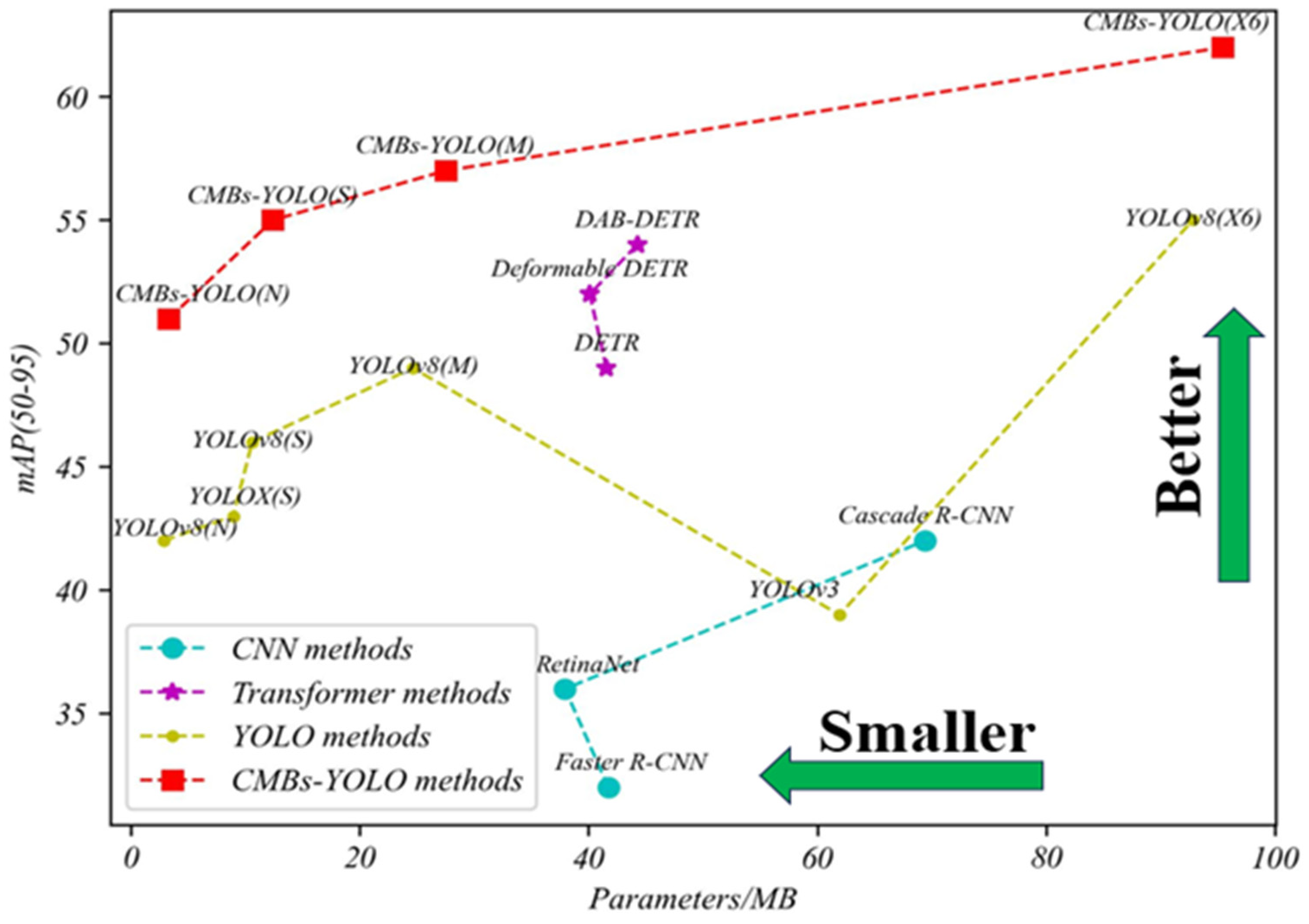
4.3.1. Detection Performance of CMBs
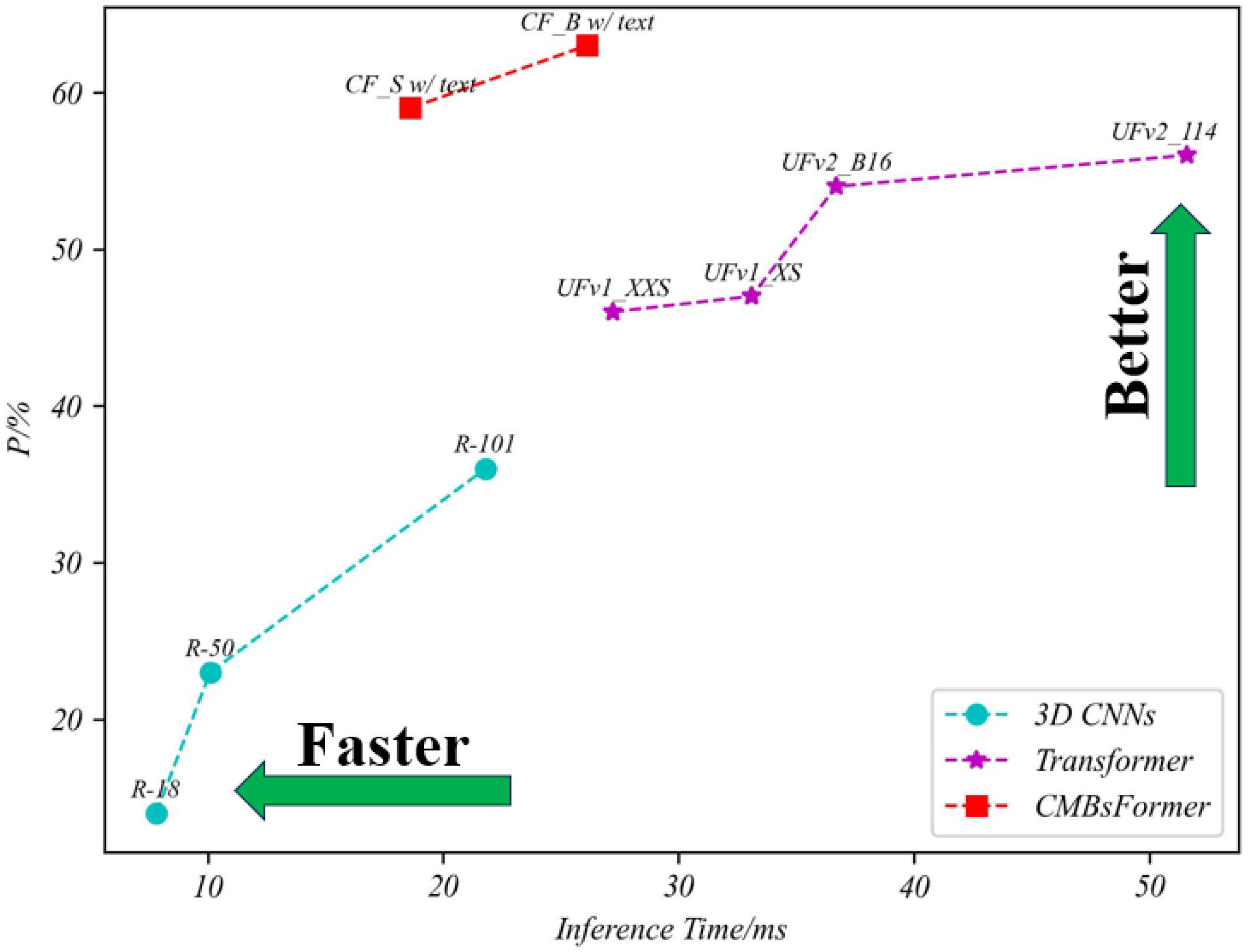
4.3.2. Classification Performance of CMBs
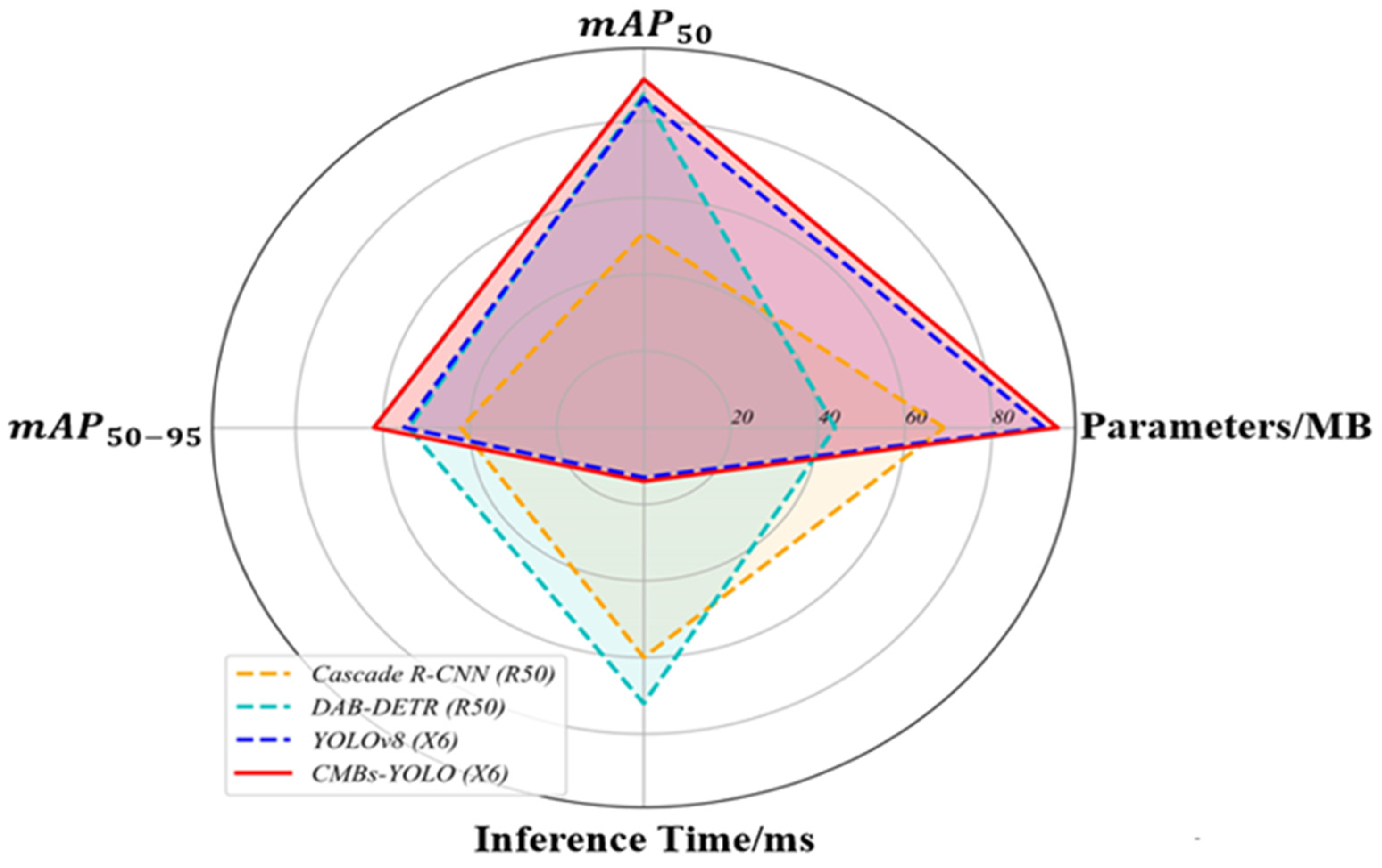
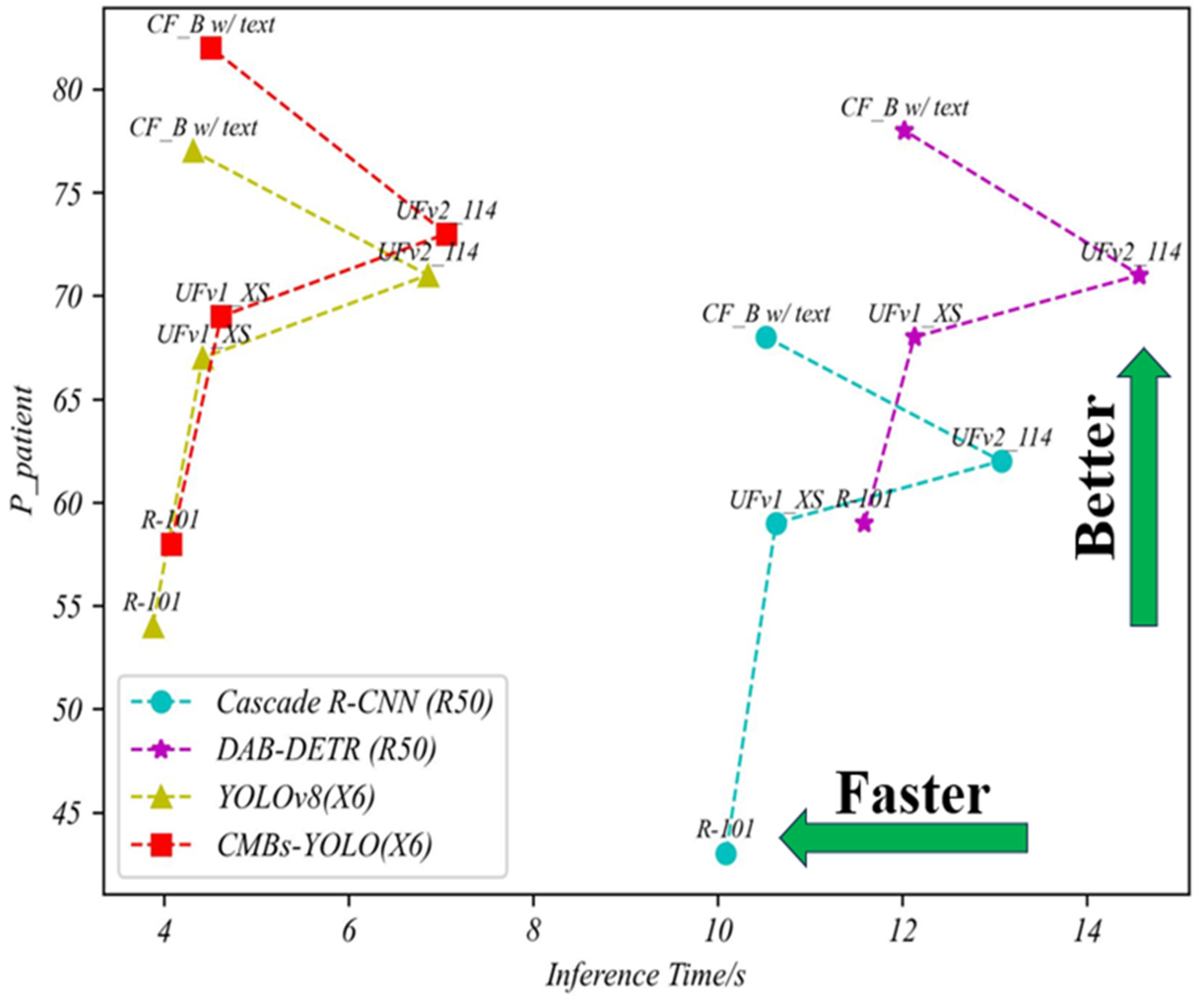
4.3.3. Total Performance of CMBs
5. Ablation Study
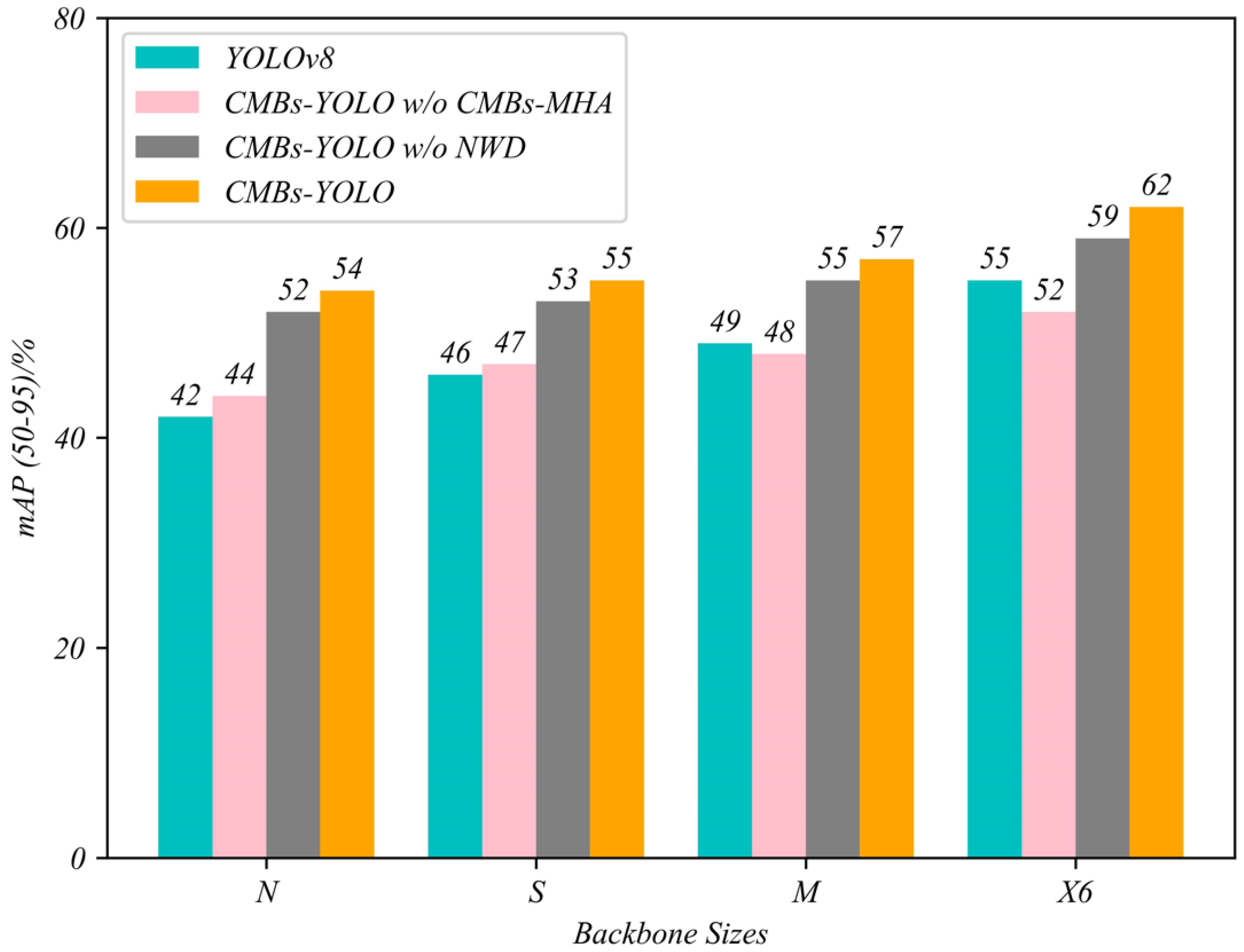
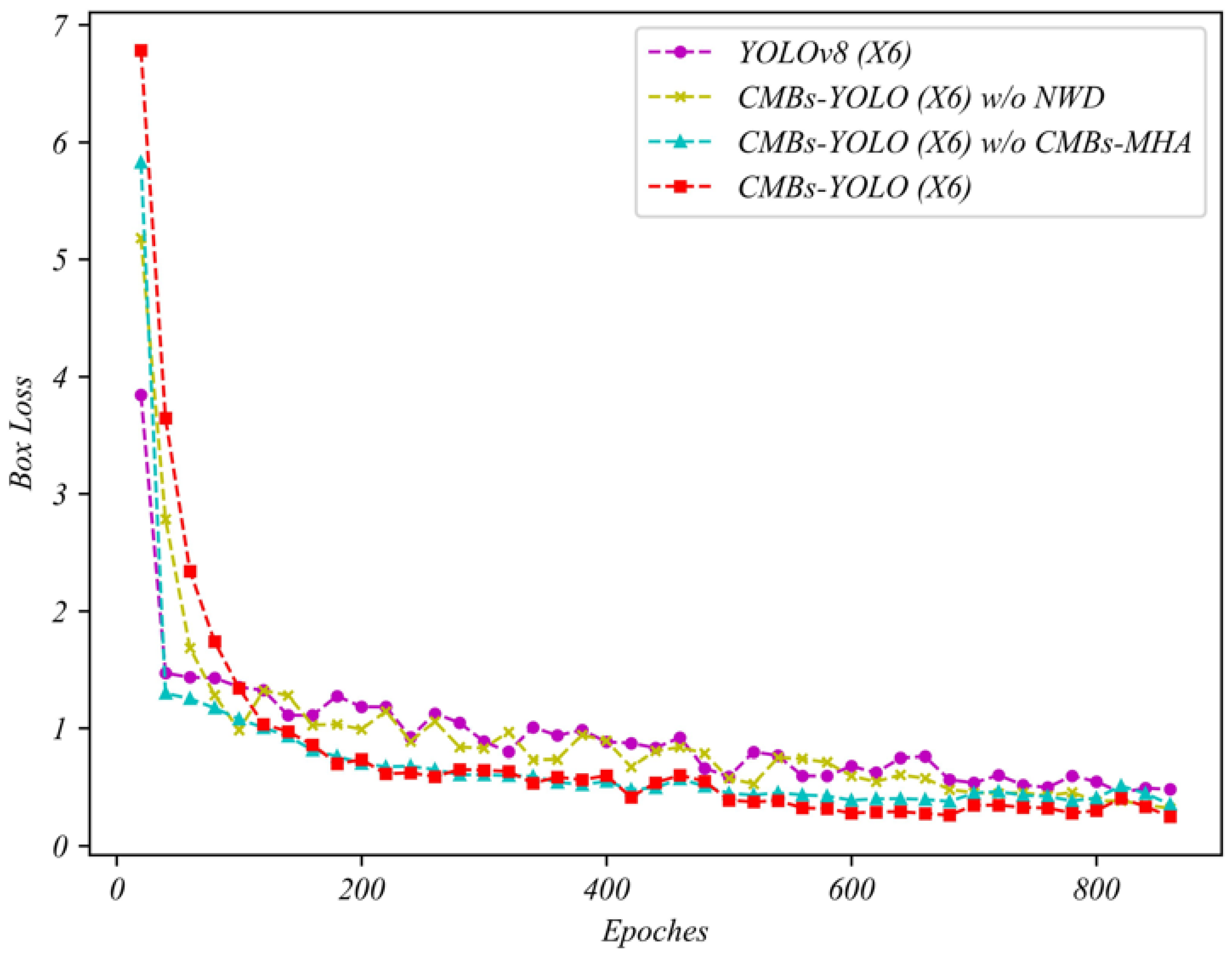
5.1. Ablation Study in Detection Stage
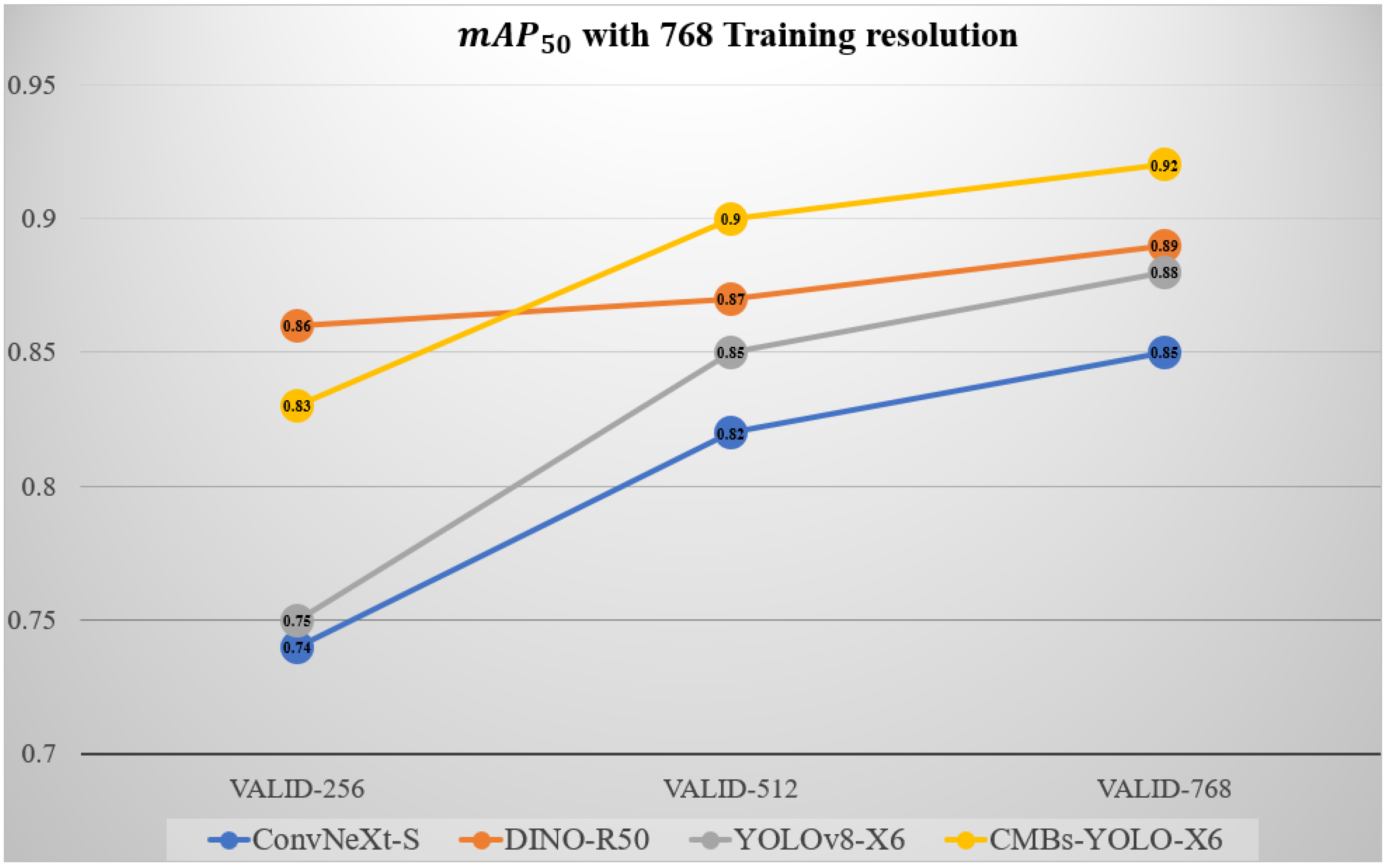
5.2. Ablation Study of Different Input Resolutions in Detection Stage
5.3. Ablation Study of Different Input Resolutions in Classification Stage
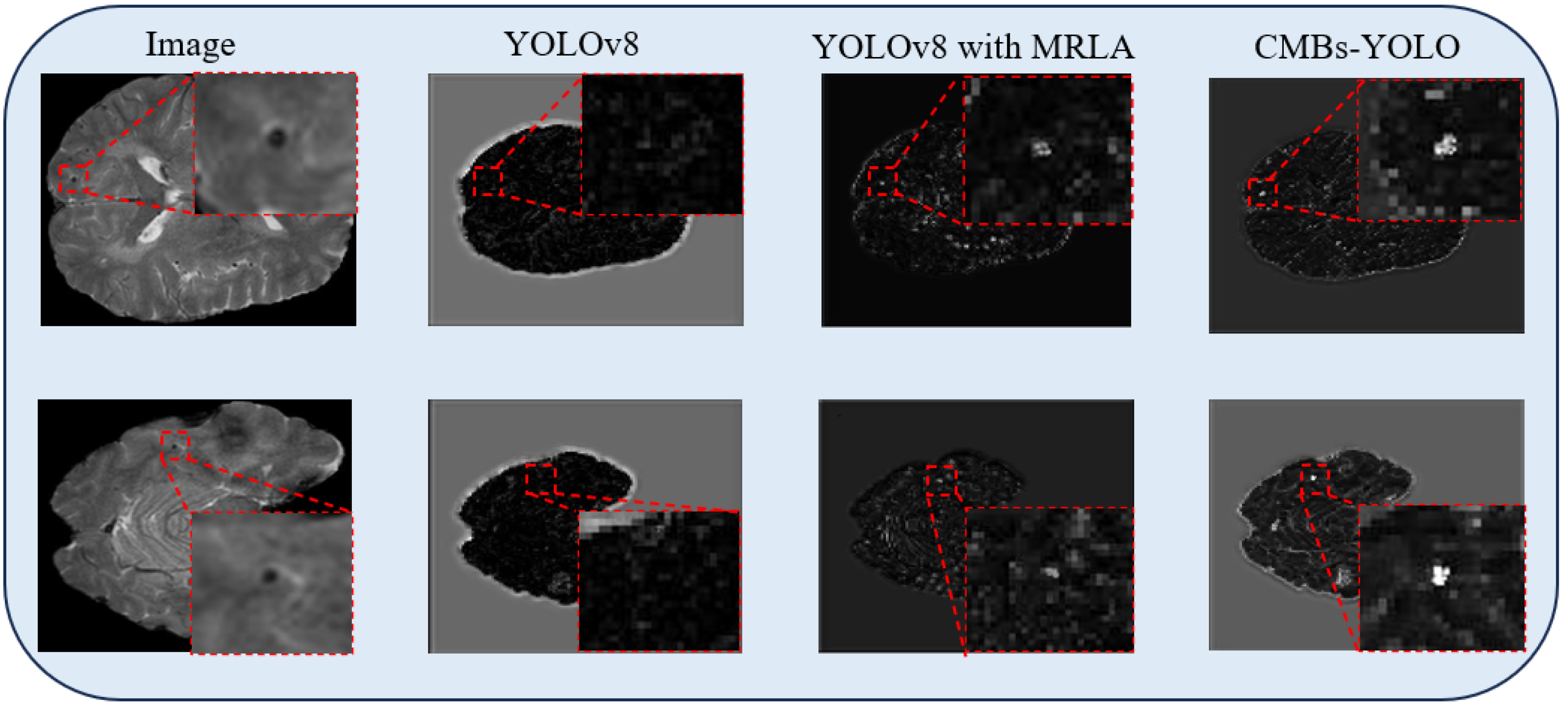
5.4. Ablation Study in Image Branch
5.5. Ablation Study of CLIP Model
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
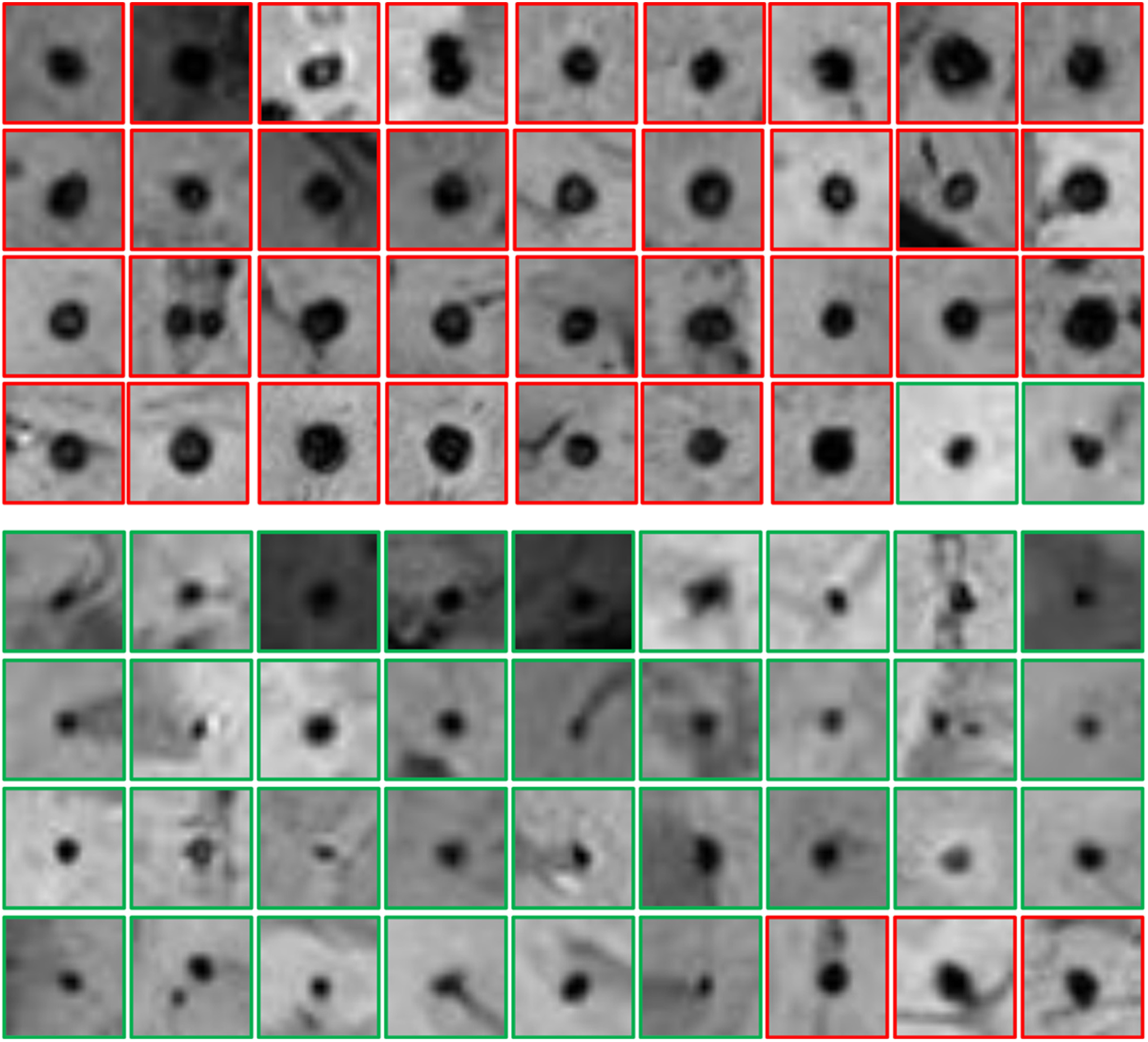
Appendix A
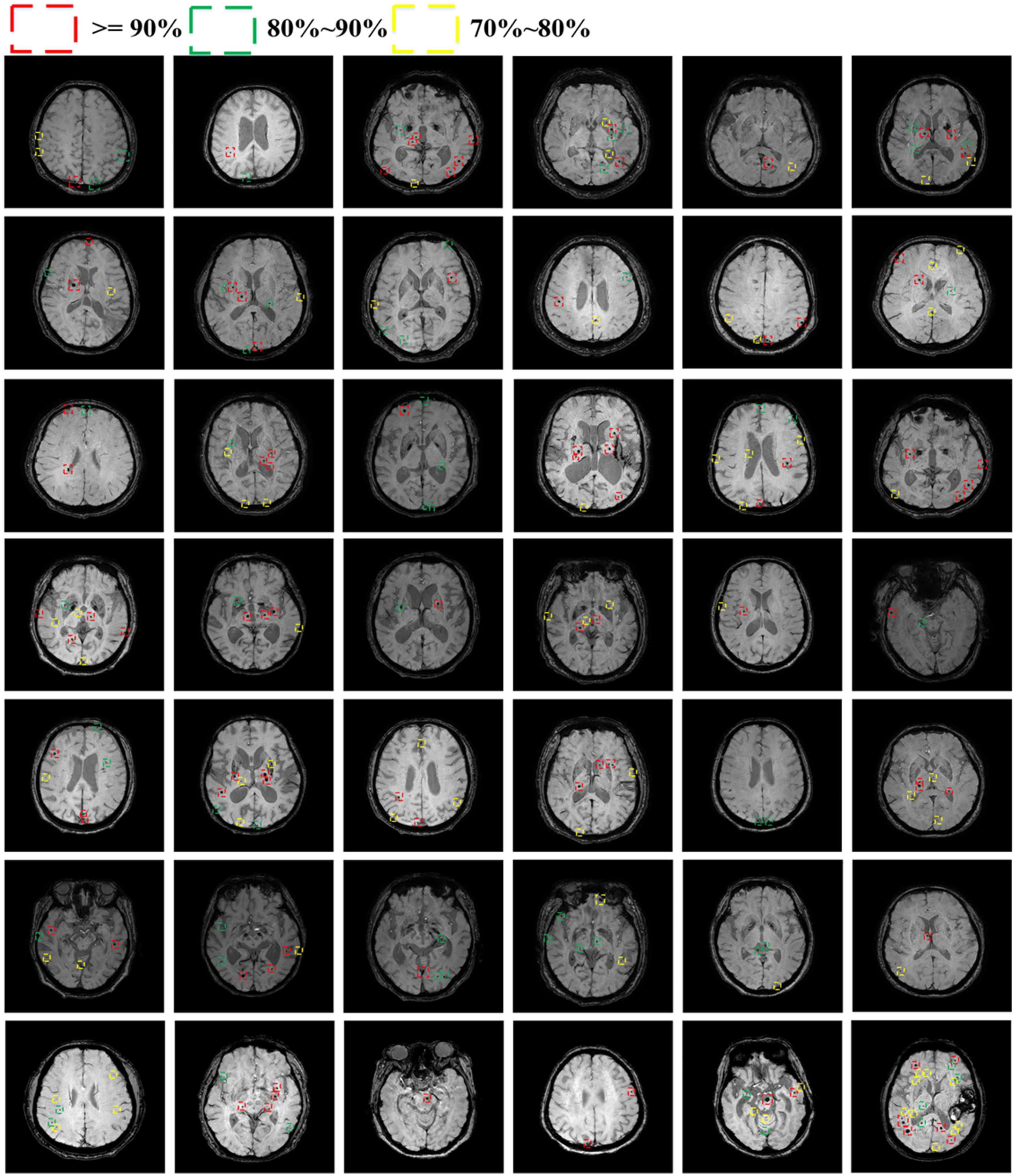
Appendix B
Appendix C
| Abbreviation | Descriptions |
|---|---|
| R-50 [55] | ResNet with 50 convolution depth |
| ConvNeXt-T | ConvNext with tiny backbone size, depth = [3, 3, 9, 3] |
| ConvNeXt-S | ConvNext with small backbone size, depth = [3, 3, 27, 3] |
| YOLO-N/CMBs-YOLO-N | YOLOv8/CMBs-YOLO methods with N backbone size depth_multiple = 0.33, width_multiple = 0.25, max_channels = 1024 |
| YOLO-S/CMBs-YOLO-S | YOLOv8/CMBs-YOLO methods with S backbone size depth_multiple = 0.33, width_multiple = 0.5, max_channels = 1024 |
| YOLO-M/CMBs-YOLO-M | YOLOv8/CMBs-YOLO methods with M backbone size depth_multiple = 0.67, width_multiple = 0.75, max_channels = 768 |
| YOLO-X6/CMBs-YOLO-X6 | YOLOv8/CMBs-YOLO methods with x6 backbone size depth_multiple = 1.0, width_multiple = 1.25, max_channels = 512 |
| Abbreviation | Descriptions |
|---|---|
| R-18 | ResNet with 18 convolution depth |
| R-50 | ResNet with 50 convolution depth |
| R-101 | ResNet with 101 convolution depth |
| UFv1_XXS | UniFormer version 1 with XXS backbone size depth = [2, 5, 8, 2], embed_dim = [56, 112, 224, 448], head_dim = 28 |
| UFv1_XS | UniFormer version 1 with XS backbone size depth = [3, 5, 9, 3], embed_dim = [64, 128, 256, 512], head_dim = 32 |
| UFv2_B16 | UniFormer version 2 with B16 backbone size width = 768, layers = 12, heads = 12, output_dim = 512 |
| UFv2_114 | UniFormer version 2 with 114 backbone size width = 1024, layers = 24, heads = 16, output_dim = 768 |
| CF_S w/o text | CMBsFormer with Small backbone size and text inputs (BERT or CLIP) layers = 6, embed_dims = 512, heads = 4, text_channels = 70 |
| CF_B w/o text | CMBsFormer with Base backbone size and text inputs (BERT or CLIP) layers = 12, embed_dims = 768, heads = 8, text_channels = 70 |
| CF_S w/text | CMBsFormer with Small backbone size layers = 6, embed_dims = 512, heads = 4 |
| CF_B w/text | CMBsFormer with Base backbone size layers = 12, embed_dims = 768, heads = 8 |
References
- Greenberg, S.M.; Vernooij, M.W.; Cordonnier, C.; Viswanathan, A.; Salman, R.A.; Warach, S.; Launer, L.J.; Buchem, M.A.V.B.; Breteler, M.M.B. Cerebral microbleeds: A guide to detection and interpretation. Lancet Neurol. 2009, 8, 165–174. [Google Scholar] [CrossRef]
- Charidimou, A.; Werring, D.J. Cerebral microbleeds: Detection, mechanisms and clinical challenges. Future Neurol. 2011, 6, 587–611. [Google Scholar] [CrossRef]
- Yamazaki, K.; Vo-Ho, V.K.; Bulsara, D.; Le, N. Spiking neural networks and their applications: A review. Brain Sci. 2022, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Dora, S.; Kasabov, N. Spiking neural networks for computational intelligence: An overview. Big Data Cogn. Comput. 2021, 5, 67. [Google Scholar] [CrossRef]
- Turkson, R.E.; Qu, H.; Mawuli, C.B.; Eghan, M.J. Classification of Alzheimer’s disease using deep convolutional spiking neural network. Neural Process. Lett. 2021, 53, 2649–2663. [Google Scholar] [CrossRef]
- Ahmadi, M.; Sharifi, A.; Hassantabar, S.; Enayati, S. QAIS-DSNN: Tumor area segmentation of MRI image with optimized quantum matched-filter technique and deep spiking neural network. BioMed Res. Int. 2021, 2021, 6653879. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, R.; Karthick, R.; Meenalochini, P.; Kalaichelvi, T. Deep Convolutional Spiking Neural Network optimized with Arithmetic optimization algorithm for lung disease detection using chest X-ray images. Biomed. Signal Process. Control 2023, 79, 104197. [Google Scholar] [CrossRef]
- Nikseresht, G.; Agam, G.; Arfanakis, K. End-to-end task-guided refinement of synthetic images for data efficient cerebral microbleed detection. In Proceedings of the 26th International Conference on Pattern Recognition (ICPR), Montreal, QC, Canada, 21–25 August 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 2756–2763. [Google Scholar]
- Ferlin, M.A.; Grochowski, M.; Kwasigroch, A.; Mikołajczyk, A.; Szurowska, E.; Grzywińska, M.; Sabisz, A. A comprehensive analysis of deep neural-based cerebral microbleeds detection system. Electronics 2021, 10, 2208. [Google Scholar] [CrossRef]
- Ateeq, T.; Majeed, M.N.; Anwar, S.M.; Maqsood, M.; Rehman, Z.; Lee, J.W.; Muhammad, K.; Wang, S.H.; Baik, S.W.; Mehmood, I. Ensemble-classifiers-assisted detection of cerebral microbleeds in brain MRI. Comput. Electr. Eng. 2018, 69, 768–781. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Yu, L.; Zhao, L.; Qin, J.; Wang, D.; Mok, V.; Shi, L.; Heng, P. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans. Med. Imaging 2016, 35, 1182–1195. [Google Scholar] [CrossRef]
- Xu, C.; Wang, J.; Yang, W.; Yu, H.; Yu, L.; Xia, G. Detecting tiny objects in aerial images: A normalized Wasserstein distance and a new benchmark. ISPRS J. Photogramm. Remote Sens. 2022, 190, 79–93. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU loss: Faster and better learning for bounding box regression. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 12993–13000. [Google Scholar]
- Kaaouana, T.; Bertrand, A.; Ouamer, F.; Ye, B.L.; Pyatigorskaya, N.; Bouyahia, A.; Thiery, N.; Dufouil, C.; Delmaire, C.; Dormont, D.; et al. Improved cerebral microbleeds detection using their magnetic signature on T2*-phase-contrast: A comparison study in a clinical setting. NeuroImage Clin. 2017, 15, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Cheng, H.; Zhang, Y.D.; Liu, J. Detecting cerebral microbleeds with transfer learning. Mach. Vis. Appl. 2019, 30, 1123–1133. [Google Scholar] [CrossRef]
- WANG, H.; Gagnon, B. Cerebral microbleed detection by wavelet entropy and naive Bayes classifier. In Proceedings of the 2nd International Conference on Biomedical and Biological Engineering 2017 (BBE 2017), Guilin, China, 26–28 May 2017; pp. 505–510. [Google Scholar]
- Bao, F.; Shi, M.; Macdonald, F. Voxelwise detection of cerebral microbleed in CADASIL patients by naïve Bayesian classifier. In Proceedings of the 2018 International Conference on Information Technology and Management Engineering (ICITME 2018), Beijing, China, 26–27 August 2018; pp. 176–180. [Google Scholar]
- Sundaresan, V.; Arthofer, C.; Zamboni, G.; Dineen, R.A.; Rothwell, P.M.; Sotiropoulous, S.N.; Auer, D.P.; Tozer, D.J.; Markus, H.S.; Miller, K.L.; et al. Automated detection of candidate subjects with cerebral microbleeds using machine learning. Front. Neuroinform. 2022, 15, 777828. [Google Scholar] [CrossRef] [PubMed]
- Chesebro, A.G.; Amarante, E.; Lao, P.J.; Meier, I.B.; Mayeus, R.; Brickman, A.M. Automated detection of cerebral microbleeds on T2*-weighted MRI. Sci. Rep 2021, 11, 4004. [Google Scholar] [CrossRef] [PubMed]
- Barnes, S.R.S.; Haacke, E.M.; Ayaz, M.; Boikov, A.S.; Kirsch, W.; Kido, D. Semiautomated detection of cerebral microbleeds in magnetic resonance images. Magn. Reson. Imaging 2011, 29, 844–852. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Yu, L.; Dhi, L.; Wang, D.; Mok, V.; Heng, P. Automatic cerebral microbleeds detection from MR images via independent subspace analysis based hierarchical features. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 7933–7936. [Google Scholar]
- Fazlollahi, A.; Meriaudeau, F.; Giancardo, L.; Villemagne, V.L.; Rowe, C.C.; Yates, P.; Salvado, O.; Bourgeat, P. Computer-aided detection of cerebral microbleeds in susceptibility-weighted imaging. Comput. Med. Imaging Graph 2015, 46, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.; Hess, C.P.; Chang, S.M.; Nelson, S.J.; Lupo, J.M. Computer-aided detection of radiation-induced cerebral microbleeds on susceptibility-weighted MR images. NeuroImage Clin. 2013, 2, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Fazlollahi, A.; Meriaudeau, F.; Villemagne, V.L.; Rowe, C.C.; Yates, P.; Salvado, O.; Bourgeat, P. Efficient machine learning framework for computer-aided detection of cerebral microbleeds using the radon transform. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 113–116. [Google Scholar]
- Morrison, M.A.; Payabvash, S.; Chen, Y.; Avadiappan, S.; Shah, M.; Zou, X.; Hess, C.P.; Lupo, J.M. A user-guided tool for semi-automated cerebral microbleed detection and volume segmentation: Evaluating vascular injury and data labelling for machine learning. NeuroImage Clin. 2018, 20, 498–505. [Google Scholar] [CrossRef]
- Momeni, S.; Fazlollahi, A.; Yates, P.; Rowe, C.; Gao, Y.; Liew, A.; Salvado, O. Synthetic microbleeds generation for classifier training without ground truth. Comput. Methods Programs Biomed. 2021, 207, 106127. [Google Scholar] [CrossRef]
- Liu, S.; Utriainen, D.; Chai, C.; Chen, Y.; Wang, L.; Sethi, S.K.; Xia, S.; Haacke, E.M. Cerebral microbleed detection using susceptibility weighted imaging and deep learning. Neuroimage 2019, 198, 271–282. [Google Scholar] [CrossRef]
- Wu, R.; Liu, H.; Li, H.; Chen, L.; Wei, L.; Huang, X.; Liu, X.; Men, X.; Li, X.; Ham, L.; et al. Deep learning based on susceptibility-weighted MR sequence for detecting cerebral microbleeds and classifying cerebral small vessel disease. BioMedical Eng. Online 2023, 22, 99. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zou, Y.; Bai, P.; Li, S.; Wang, H.; Chen, X.; Meng, Z.; Kang, Z.; Zhou, G. Detecting cerebral microbleeds via deep learning with features enhancement by reusing ground truth. Comput. Methods Programs Biomed. 2021, 204, 106051. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.D.; Hou, X.X.; Chen, Y.; Chen, H.; Yang, M.; Yang, J.Q.; Wang, S.H. Voxelwise detection of cerebral microbleed in CADASIL patients by leaky rectified linear unit and early stopping. Multimed. Tools Appl. 2018, 77, 21825–21845. [Google Scholar] [CrossRef]
- Crouzet, C.; Jeong, G.; Chae, R.H.; Lopresti, K.T.; Dunn, C.E.; Xie, D.F.; Agu, C.; Fang, C.; Nunes, A.C.F.; Lau, W.L.; et al. Spectroscopic and deep learning-based approaches to identify and quantify cerebral microhemorrhages. Sci. Rep. 2021, 11, 10725. [Google Scholar] [CrossRef] [PubMed]
- Rashid, T.; Abdulkadir, A.; Nasrallah, I.M.; Ware, J.B.; Liu, H.F.; Spincemaille, P.; Romero, J.R.; Bryan, R.N.; Heckbert, S.R.; Habes, M. DEEPMIR: A deep neural network for differential detection of cerebral microbleeds and iron deposits in MRI. Sci. Rep. 2021, 11, 14124. [Google Scholar] [CrossRef]
- Lee, H.; Kim, J.H.; Lee, S.; Jung, K.J.; Kim, W.E.; Noh, Y.; Kim, E.Y.; Kang, K.M.; Sohn, C.H.; Lee, D.Y.; et al. Detection of cerebral microbleeds in MR images using a single-stage triplanar ensemble detection network (TPE-Det). J. Magn. Reson. Imaging 2023, 58, 272–283. [Google Scholar] [CrossRef]
- Al-Masni, M.A.; Kim, W.R.; Kim, E.Y.; Noh, Y.; Kim, D.H. Automated detection of cerebral microbleeds in MR images: A two-stage deep learning approach. NeuroImage Clin. 2020, 28, 102464. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, R.; Guo, L.; Xia, T.; Zeng, Y.; Wu, X. Knowledge-guided 2.5 D CNN for cerebral microbleeds detection. Biomed. Signal Process. Control 2023, 86, 105078. [Google Scholar] [CrossRef]
- Lu, S.Y.; Nayak, D.R.; Wang, S.H.; Zhang, Y.D. A cerebral microbleed diagnosis method via featurenet and ensembled randomized neural networks. Appl. Soft Comput. 2021, 109, 107567. [Google Scholar] [CrossRef]
- Koschmieder, K.; Paul, M.M.; Van den Heuvel, T.L.A.; Eerden, A.V.D.; Ginneken, B.V.; Manniesing, R. Automated detection of cerebral microbleeds via segmentation in susceptibility-weighted images of patients with traumatic brain injury. NeuroImage Clin. 2022, 35, 103027. [Google Scholar] [CrossRef] [PubMed]
- Stanley, B.F.; Franklin, S.W. Automated cerebral microbleed detection using selective 3D gradient co-occurance matrix and convolutional neural network. Biomed. Signal Process. Control 2022, 75, 103560. [Google Scholar] [CrossRef]
- Chen, Y.; Villanueva-Meyer, J.E.; Morrison, M.A.; Lupo, J.M. Toward automatic detection of radiation-induced cerebral microbleeds using a 3D deep residual network. J. Digit. Imaging 2019, 32, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, C.; Sun, J.; Zhang, Y. Cerebral micro-bleeding detection based on densely connected neural network. Front. Neurosci. 2019, 13, 422. [Google Scholar] [CrossRef] [PubMed]
- Boecking, B.; Usuyama, N.; Bannur, S.; Castro, D.C.; Schwaighofer, A.; Hyland, S.; Wetscherek, M.; Naumann, T.; Nori, A.; Alvarez-Valle, J.; et al. Making the most of text semantics to improve biomedical vision–language processing. In European Conference on Computer Vision; Springer Nature: Cham, Switzerland, 2022; pp. 1–21. [Google Scholar]
- Li, Z.; Li, Y.; Li, Q.; Wang, P.; Guo, D.; Lu, L.; Jin, D.; Zhang, Y.; Hong, Q. Lvit: Language meets vision transformer in medical image segmentation. IEEE Trans. Med. Imaging 2023, 43, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Y.; Chen, J.N.; Xiao, J.; Lu, Y.; Landman, B.A.; Yuan, Y.; Yuille, A.; Tang, Y.; Zhou, Z. Clip-driven universal model for organ segmentation and tumor detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–6 October 2023; pp. 21152–21164. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. Int. Conf. Mach. Learn. PMLR 2021, 8748–8763. [Google Scholar]
- Bhalodia, R.; Hatamizadeh, A.; Tam, L.; Xu, Z.; Wang, X.; Turkbey, E.; Xu, D. Improving pneumonia localization via cross-attention on medical images and reports. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 571–581. [Google Scholar]
- Sudre, C.H.; Van Wijnen, K.; Dubost, F.; Adams, H.; Atkinson, D.; Barkhof, F.; Birhanu, M.A.; Bron, E.E.; Camarasa, R.; Chaturvedi, N.; et al. Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021. Med. Image Anal. 2024, 91, 103029. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In European Conference on Computer Vision; Springer International Publishing: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar]
- Ultralytics. YOLOv8—Ultralytics Open-Source Research. Available online: https://github.com/ultralytics/ultralytics (accessed on 26 September 2024).
- Yu, T.; Li, C.; Huang, J.; Xiao, X.; Zhang, X.; Li, Y.; Fu, B. ReF-DDPM: A novel DDPM-based data augmentation method for imbalanced rolling bearing fault diagnosis. Reliab. Eng. Syst. Saf. 2024, 251, 110343. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.Y.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar]
- Suwalska, A.; Wang, Y.; Yuan, Z.; Jiang, Y.; Zhu, D.; Chen, J.; Cui, M.; Chen, X.; Suo, C.; Polanska, J. CMB-HUNT: Automatic detection of cerebral microbleeds using a deep neural network. Comput. Biol. Med. 2022, 151, 106233. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Ferlin, M.; Klawikowska, Z.; Grochowski, M.; Grzywinska, M.; Szurowska, E. Exploring the landscape of automatic cerebral microbleed detection: A comprehensive review of algorithms, current trends, and future challenges. Expert Syst. Appl. 2023, 232, 120655. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lin, T. Focal Loss for Dense Object Detection. arXiv 2017, arXiv:1708.02002. [Google Scholar]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: High quality object detection and instance segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Bai, S.; Xie, L.; Qi, H.; Huang, Q.; Tian, Q. Centernet: Keypoint triplets for object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6569–6578. [Google Scholar]
- Zhang, H.; Chang, H.; Ma, B.; Wang, N.; Chen, X. Dynamic R-CNN: Towards high quality object detection via dynamic training. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part XV 16; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 260–275. [Google Scholar]
- Sun, P.; Zhang, R.; Jiang, Y.; Kong, T.; Xu, C.F.; Zhan, W.; Tomizuka, M.; Li, L.; Yuan, Z.H.; Wang, C.H.; et al. Sparse r-cnn: End-to-end object detection with learnable proposals. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 14454–14463. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrel, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Zhu, X.; Su, W.; Lu, L.; Li, B.; Wang, X.G.; Dai, J. Deformable detr: Deformable transformers for end-to-end object detection. arXiv 2020, arXiv:2010.04159. [Google Scholar]
- Guo, M.H.; Xu, T.X.; Liu, J.J.; Liu, Z.N.; Jiang, P.T.; Mu, T.J.; Zhang, S.H.; Martin, R.R.; Cheng, M.M.; Hu, S.M. Attention mechanisms in computer vision: A survey. Comput. Vis. Media 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Zhang, H.; Li, F.; Liu, S.; Zhang, L.; Su, H.; Zhu, J.; Ni, L.M.; Shum, H.Y. Dino: Detr with improved denoising anchor boxes for end-to-end object detection. arXiv 2022, arXiv:2203.03605. [Google Scholar]
- Redmon, J. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Ge, Z. Yolox: Exceeding yolo series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Sun, Z.; Ke, Q.; Rahmani, H.; Bennamoun, M.; Wang, G.; Liu, J. Human action recognition from various data modalities: A review. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 3200–3225. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, W.; Xie, B.; Sun, Q.; Wu, L.; Wang, X.; Huang, T.; Wang, X.; Cao, Y. Eva: Exploring the limits of masked visual representation learning at scale. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 19358–19369. [Google Scholar]
- Xia, P.; Hui, E.S.; Chua, B.J.; Huang, F.; Wang, Z.; Zhang, Z.; Yu, H.; Lau, K.K.; Mak, H.K.F.; Cao, P. Deep-learning-based MRI microbleeds detection for cerebral small vessel disease on quantitative susceptibility mapping. J. Magn. Reson. Imaging 2024, 60, 1165–1175. [Google Scholar] [CrossRef]
- Hong, J.; Wang, S.H.; Cheng, H.; Liu, J. Classification of cerebral microbleeds based on fully-optimized convolutional neural network. Multimed. Tools Appl. 2020, 79, 15151–15169. [Google Scholar] [CrossRef]
- Wu, X.; Hong, D.; Chanussot, J. Convolutional neural networks for multimodal remote sensing data classification. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–10. [Google Scholar] [CrossRef]
- Shao, W.; Peng, Y.; Zu, C.; Wang, M.L.; Zhang, D.Q. Hypergraph based multi-task feature selection for multimodal classification of Alzheimer’s disease. Comput. Med. Imaging Graph. 2020, 80, 101663. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Kurz, A.; Hauser, K.; Mehrtens, H.A.; Henning, E.K.; Hekler, A.; Kather, J.N.; Frohling, S.; Kalle, C.V.; Brinker, T.J. Uncertainty estimation in medical image classification: Systematic review. JMIR Med. Inform. 2022, 10, e36427. [Google Scholar] [CrossRef]
- Raj, R.J.S.; Shobana, S.J.; Pustokhina, I.V.; Pustokhin, D.A.; Gupta, D.; Shankar, K. Optimal feature selection-based medical image classification using deep learning model in internet of medical things. IEEE Access 2020, 8, 58006–58017. [Google Scholar] [CrossRef]
- Yang, X.; Yan, J.; Ming, Q.; Wang, W.; Zhang, X.; Tian, Q. Rethinking rotated object detection with gaussian wasserstein distance loss. International conference on machine learning. PMLR 2021, 139, 11830–11841. [Google Scholar]
- Han, Y.; Liu, X.; Sheng, Z.; Ren, Y.; Han, X.; You, J.; Liu, R.; Luo, Z. Wasserstein loss-based deep object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 998–999. [Google Scholar]
- Diwan, T.; Anirudh, G.; Tembhurne, J.V. Object detection using YOLO: Challenges, architectural successors, datasets and applications. Multimed. Tools Appl. 2023, 82, 9243–9275. [Google Scholar] [CrossRef]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamazdeh, M.; Fieguth, P.; Cao, X.C.; Khosravi, A.; Acharya, U.R.; et al. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Faghani, S.; Moassefi, M.; Rouzrokh, P.; Khosravi, B.; Baffour, F.I.; Ringler, M.D.; Erickson, B.J. Quantifying uncertainty in deep learning of radiologic images. Radiology 2023, 308, e222217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wang, K.; Xiao, Y.; Gao, F.; Gao, Z. Leveraging Monte Carlo Dropout for Uncertainty Quantification in Real-Time Object Detection of Autonomous Vehicles. IEEE Access 2024, 12, 33384–33399. [Google Scholar] [CrossRef]
- Abbaszadeh Shahri, A.; Shan, C.; Larsson, S. A novel approach to uncertainty quantification in groundwater table modeling by automated predictive deep learning. Nat. Resour. Res. 2022, 31, 1351–1373. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Abbaszadeh Shahri, A.; Asheghi, R. Prediction of bedload transport rate using a block combined network structure. Hydrol. Sci. J. 2022, 67, 117–128. [Google Scholar] [CrossRef]
- Abbaszadeh Shahri, A.; Maghsoudi Moud, F. Landslide susceptibility mapping using hybridized block modular intelligence model. Bull. Eng. Geol. Environ. 2021, 80, 267–284. [Google Scholar] [CrossRef]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising with block-matching and 3D filtering. Image processing: Algorithms and systems, neural networks, and machine learning. SPIE 2006, 6064, 354–365. [Google Scholar]
- Naveed, K.; Abdullah, F.; Madni, H.A.; Khan, M.A.U.; Khan, T.M.; Naqvi, S.S. Towards automated eye diagnosis: An improved retinal vessel segmentation framework using ensemble block matching 3D filter. Diagnostics 2021, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zeng, L.; Yu, W.; Gong, C. Noise suppression–guided image filtering for low-SNR CT reconstruction. Med. Biol. Eng. Comput. 2020, 58, 2621–2629. [Google Scholar] [CrossRef] [PubMed]





| CMBs-Public | Number of Patients | Number of Patients with CMBs | Average Age of Patients with CMBs | Average Age of Patients without CMBs | Number of Slices per Patient | Slice Image Resolution | Number of CMBs Bounding Boxes | Male Proportion | Female Proportion |
|---|---|---|---|---|---|---|---|---|---|
| Training set | 50 | 37 | × | × | 35 | 512 × 512 | 353 | × | × |
| Validation set | 7 | 4 | × | × | 35 | 512 × 512 | 43 | × | × |
| Testing set | 15 | 9 | × | × | 35 | 512 × 512 | 102 | × | × |
| CMBs-Private | Number of Patients | Number of Patients with CMBs | Average Age of Patients with CMBs | Average Age of Patients without CMBs | Number of Slices per Patient | Slice Image Resolution | Number of CMBs Bounding Boxes | Male Proportion | Female Proportion |
|---|---|---|---|---|---|---|---|---|---|
| Training set | 400 | 178 | 66 | 57 | 130 | 512 × 512 | 8015 | 61% | 39% |
| Validation set | 50 | 17 | 68 | 61 | 130 | 512 × 512 | 687 | 54% | 46% |
| Testing set | 150 | 56 | 64 | 59 | 130 | 512 × 512 | 2432 | 66% | 34% |
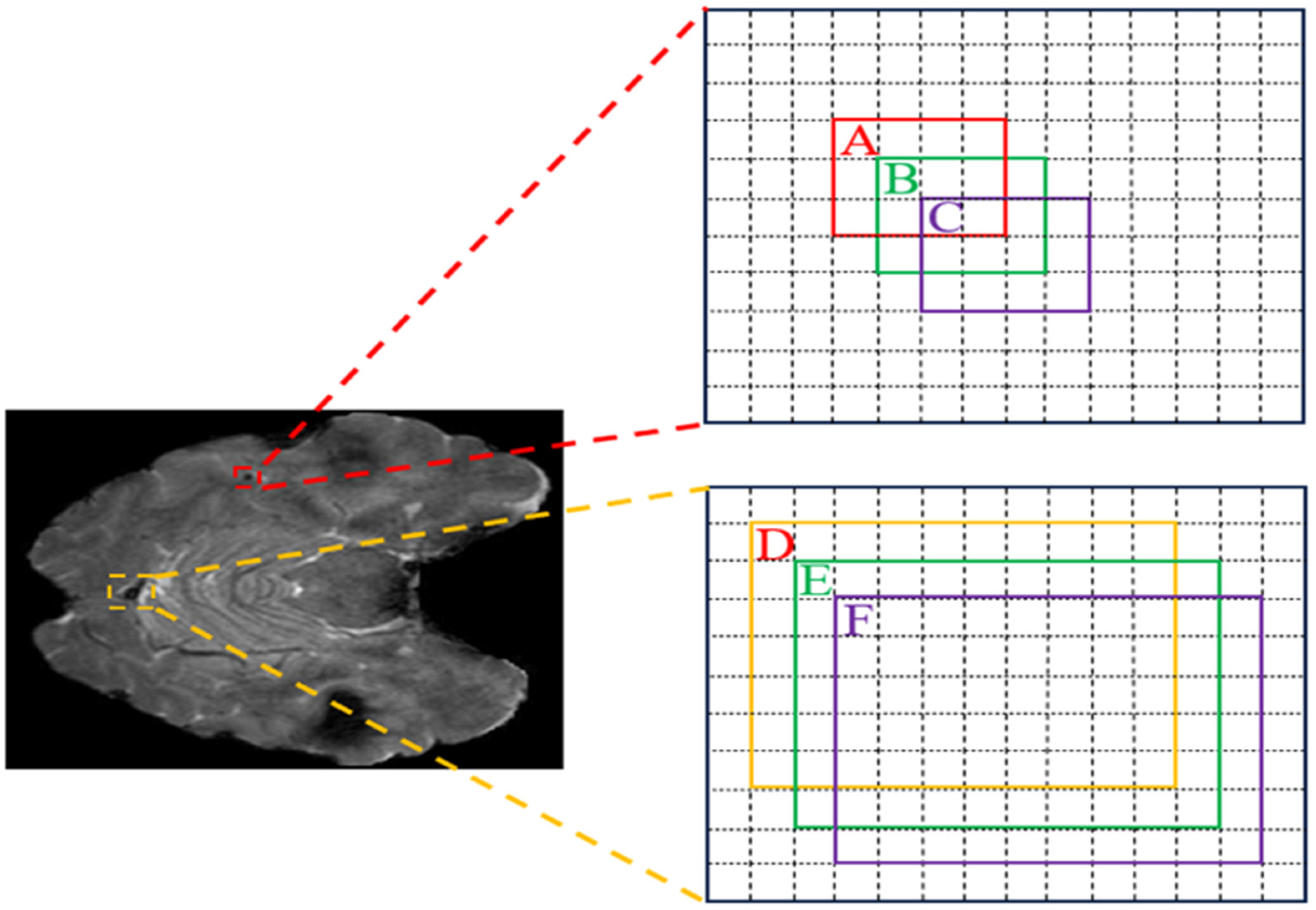
| Loss Name | Value (A, B) | Value (A, C) | Trend | Value (D, E) | Value (D, F) | Trend |
|---|---|---|---|---|---|---|
| IoU [12] | 0.29 | 0.06 | −0.23 | 0.92 | 0.85 | −0.07 |
| GIoU [51] | 0.16 | −0.26 | −0.42 | 0.92 | 0.85 | −0.07 |
| DIoU [12] | 0.29 | 0.06 | −0.23 | 0.92 | 0.85 | −0.07 |
| CIoU [12] | 0.29 | 0.06 | −0.23 | 0.92 | 0.85 | −0.07 |
| NWD | 0.9 | 0.8 | −0.1 | 0.9 | 0.8 | −0.1 |
| Text Prompts | Descriptions |
|---|---|
| This is a {male/female} patient with {} years old. | Describe the patient’s gender and age. |
| Target length and width are {} and {} pixels. | Describe the length and width of the detected CMBs. |
| The detection status of the {previous/next} frame at the same position is {yes/no}. | Describe the detection status of CMBs in the previous and next slice images at the same position. |
| Method Name | Model Name | Proposed Year | Backbone | Dropout Rate | Batch Size | Learning Rate | Epoches |
|---|---|---|---|---|---|---|---|
| CNN | Faster R-CNN [47] | 2015 | R-50 [55] | 0.0 | 8 | 0.001 | 30 |
| RetinaNet [56] | 2017 | R-50 | 0.0 | 8 | 0.001 | 30 | |
| Cascade R-CNN [57] | 2018 | R-50 | 0.0 | 4 | 0.001 | 45 | |
| CenterNet [58] | 2019 | R-50 | 0.0 | 8 | 0.001 | 30 | |
| Dynamic R-CNN [59] | 2020 | R-50 | 0.0 | 8 | 0.001 | 30 | |
| Sparse R-CNN [60] | 2021 | R-50 | 0.0 | 8 | 0.001 | 30 | |
| ConvNeXt [61] | 2022 | T | 0.5 | 8 | 0.001 | 30 | |
| S | 0.5 | 8 | 0.001 | 30 | |||
| Transformer | DETR [48] | 2020 | R-50 | 0.0 | 4 | 0.0001 | 50 |
| Deformable DETR [62] | 2021 | R-50 | 0.0 | 4 | 0.0001 | 50 | |
| DAB-DETR [63] | 2022 | R-50 | 0.0 | 4 | 0.0001 | 50 | |
| DINO [64] | 2023 | R-50 | 0.0 | 4 | 0.0001 | 50 | |
| YOLO | YOLOv3 [65] | 2018 | DarkNet-53 | 0.0 | 8 | 0.001 | 30 |
| YOLOX [66] | 2021 | S | 0.0 | 8 | 0.001 | 30 | |
| YOLOv8 | 2023 | N | 0.3 | 32 | 0.001 | 30 | |
| S | 0.3 | 32 | 0.001 | 30 | |||
| M | 0.2 | 16 | 0.001 | 30 | |||
| X6 | 0.2 | 8 | 0.001 | 30 | |||
| CMBs-YOLO | N | 0.3 | 32 | 0.001 | 30 | ||
| S | 0.3 | 32 | 0.001 | 30 | |||
| M | 0.2 | 16 | 0.001 | 30 | |||
| X6 | 0.2 | 8 | 0.001 | 30 |
| Method Name | Model Name | Backbone | Parameters (MB) | P | R | Inference Time/ms | ||
|---|---|---|---|---|---|---|---|---|
| CNN | Faster R-CNN | R-50 | 41.75 | 0.47 | 0.38 | 0.43 | 0.32 | 52.96 |
| RetinaNet | R-50 | 37.96 | 0.49 | 0.43 | 0.49 | 0.36 | 47.34 | |
| Cascade R-CNN | R-50 | 69.39 | 0.56 | 0.49 | 0.51 | 0.42 | 60.83 | |
| CenterNet | R-50 | 32.29 | 0.47 | 0.53 | 0.55 | 0.36 | 25.32 | |
| Dynamic R-CNN | R-50 | 41.75 | 0.68 | 0.57 | 0.61 | 0.38 | 34.17 | |
| Sparse R-CNN | R-50 | 106.29 | 0.75 | 0.63 | 0.68 | 0.41 | 57.42 | |
| ConvNeXt | T | 48.09 | 0.79 | 0.66 | 0.73 | 0.43 | 32.95 | |
| S | 106.35 | 0.88 | 0.75 | 0.81 | 0.52 | 65.13 | ||
| Transformer | DETR | R-50 | 41.58 | 0.84 | 0.77 | 0.84 | 0.49 | 54.67 |
| Deformable DETR | R-50 | 40.12 | 0.86 | 0.81 | 0.86 | 0.52 | 65.36 | |
| DAB-DETR | R-50 | 44.27 | 0.89 | 0.83 | 0.87 | 0.54 | 72.39 | |
| DINO | R-50 | 47.55 | 0.92 | 0.91 | 0.88 | 0.64 | 47.69 | |
| YOLO | YOLOv3 | DarkNet-53 | 61.95 | 0.57 | 0.51 | 0.48 | 0.39 | 32.54 |
| YOLOX | S | 8.97 | 0.81 | 0.72 | 0.75 | 0.43 | 14.61 | |
| YOLOv8 | N | 2.87 | 0.72 | 0.63 | 0.66 | 0.42 | 6.2 | |
| S | 10.62 | 0.79 | 0.68 | 0.74 | 0.46 | 7.3 | ||
| M | 24.66 | 0.88 | 0.71 | 0.77 | 0.49 | 8.9 | ||
| X6 | 92.81 | 0.91 | 0.78 | 0.86 | 0.55 | 13.1 | ||
| CMBs-YOLO | N | 3.33 | 0.78 | 0.69 | 0.75 | 0.51 | 7.6 | |
| S | 12.44 | 0.87 | 0.79 | 0.83 | 0.55 | 8.4 | ||
| M | 27.53 | 0.91 | 0.82 | 0.87 | 0.57 | 11.3 | ||
| X6 | 95.43 | 0.94 | 0.86 | 0.91 | 0.62 | 14.6 |
| Method Name | Model Name | Backbone | P | R | ||
|---|---|---|---|---|---|---|
| CNN | Faster R-CNN | R-50 | 0.41 | 0.22 | 0.35 | 0.26 |
| RetinaNet | R-50 | 0.54 | 0.29 | 0.47 | 0.31 | |
| Cascade R-CNN | R-50 | 0.58 | 0.51 | 0.46 | 0.32 | |
| CenterNet | R-50 | 0.64 | 0.43 | 0.57 | 0.34 | |
| Dynamic R-CNN | R-50 | 0.68 | 0.57 | 0.61 | 0.43 | |
| Sparse R-CNN | R-50 | 0.68 | 0.39 | 0.65 | 0.52 | |
| ConvNeXt | T | 0.77 | 0.47 | 0.65 | 0.53 | |
| S | 0.82 | 0.53 | 0.78 | 0.59 | ||
| Transformer | DETR | R-50 | 0.64 | 0.41 | 0.57 | 0.52 |
| Deformable DETR | R-50 | 0.66 | 0.39 | 0.63 | 0.61 | |
| DAB-DETR | R-50 | 0.68 | 0.43 | 0.64 | 0.61 | |
| DINO | R-50 | 0.81 | 0.48 | 0.77 | 0.65 | |
| YOLO | YOLOv3 | DarkNet-53 | 0.43 | 0.31 | 0.38 | 0.32 |
| YOLOX | S | 0.72 | 0.49 | 0.63 | 0.54 | |
| YOLOv8 | N | 0.61 | 0.37 | 0.58 | 0.49 | |
| S | 0.68 | 0.48 | 0.64 | 0.52 | ||
| M | 0.75 | 0.52 | 0.71 | 0.57 | ||
| X6 | 0.79 | 0.55 | 0.74 | 0.63 | ||
| CMBs-YOLO | N | 0.66 | 0.43 | 0.61 | 0.55 | |
| S | 0.71 | 0.51 | 0.68 | 0.59 | ||
| M | 0.78 | 0.57 | 0.73 | 0.61 | ||
| X6 | 0.83 | 0.62 | 0.79 | 0.68 |
| Method Name | Model Name | TPR | TNR | P | Parameter (MB) | Inference Time/ms |
|---|---|---|---|---|---|---|
| 3D-CNN | R-18 | 0.64 | 0.69 | 0.14 | 31.6 | 7.8 |
| R-50 | 0.72 | 0.75 | 0.23 | 60.5 | 10.1 | |
| R-101 | 0.85 | 0.87 | 0.36 | 117.9 | 21.8 | |
| Transformer | UFv1_XXS | 0.83 | 0.84 | 0.46 | 10.1 | 27.2 |
| UFv1_XS | 0.84 | 0.85 | 0.47 | 16.3 | 33.1 | |
| UFv2_B16 | 0.88 | 0.86 | 0.54 | 108.5 | 36.7 | |
| UFv2_114 | 0.89 | 0.88 | 0.56 | 336.9 | 51.6 | |
| CF | CF_S w/o text | 0.86 | 0.85 | 0.49 | 26.68 | 12.7 |
| CF_B w/o text | 0.89 | 0.86 | 0.51 | 107.15 | 18.86 | |
| CF_S w/text | 0.92 | 0.91 | 0.59 | 30.1 | 18.6 | |
| CF_B w/text | 0.94 | 0.93 | 0.63 | 114.5 | 26.12 |
| Input Images | Gender | Age | Ground Truth | R101 | UFv1_XS | UFv2_114 | CF_B w/o text | CF_B w/text | |
|---|---|---|---|---|---|---|---|---|---|
| Patient 1 |  | Female | 55 | Not CMBs | CMBs | CMBs | CMBs | CMBs | Not CMBs |
| Patient 2 | 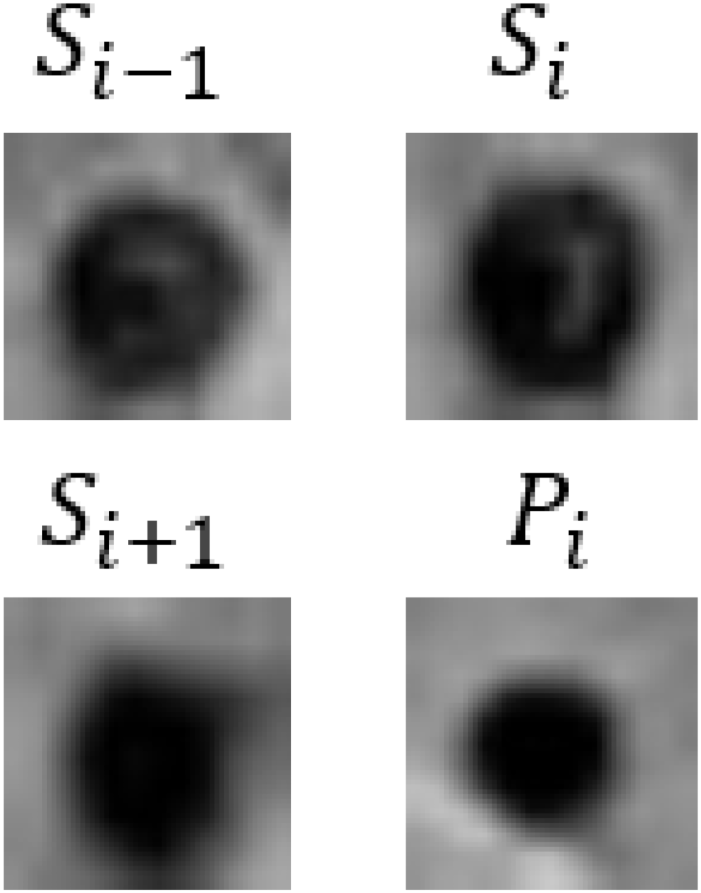 | Male | 68 | CMBs | CMBs | CMBs | CMBs | CMBs | CMBs |
| Patient 3 | 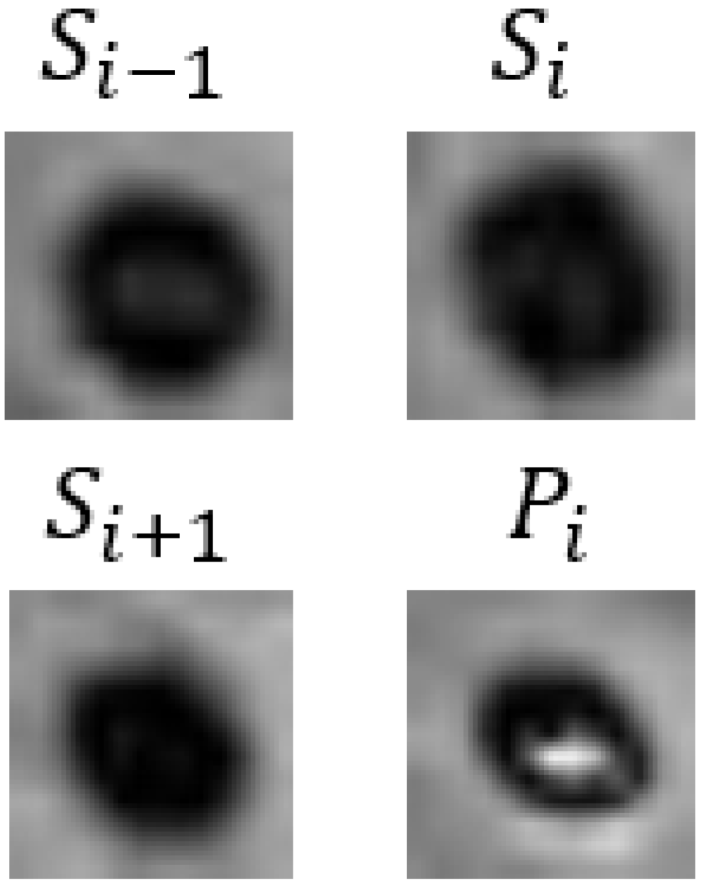 | Female | 57 | Not CMBs | CMBs | CMBs | CMBs | CMBs | Not CMBs |
| Detection Model | Classification Model | FPavg | Total Parameters/MB | Total Inference Time/s | |
|---|---|---|---|---|---|
| Cascade R-CNN (R50) | R-101 | 10.87 | 0.43 | 187.29 | 10.09 |
| UFv1_XS | 7.98 | 0.59 | 85.69 | 10.63 | |
| UFv2_114 | 7.06 | 0.62 | 406.29 | 13.07 | |
| CF_B w/text | 5.49 | 0.68 | 183.86 | 10.52 | |
| DAB-DETR (R50) | R-101 | 9.63 | 0.51 | 162.17 | 11.59 |
| UFv1_XS | 7.14 | 0.68 | 60.57 | 12.13 | |
| UFv2_114 | 5.67 | 0.69 | 381.17 | 14.57 | |
| CF_B w/text | 3.98 | 0.78 | 158.77 | 12.02 | |
| YOLOv8 (X6) | R-101 | 8.19 | 0.54 | 210.71 | 3.88 |
| UFv1_XS | 6.33 | 0.67 | 109.11 | 4.42 | |
| UFv2_114 | 5.72 | 0.71 | 429.71 | 6.86 | |
| CF_B w/text | 3.66 | 0.77 | 207.31 | 4.32 | |
| CMBs-YOLO (X6) | R-101 | 8.42 | 0.58 | 213.03 | 4.08 |
| UFv1_XS | 6.97 | 0.69 | 111.73 | 4.62 | |
| UFv2_114 | 6.21 | 0.73 | 432.33 | 7.06 | |
| CF_B w/text | 3.37 | 0.82 | 209.93 | 4.51 |
| Image | Enlarged View | Age | Gender | Lifestyle Habits | Medical History | Clinical Diagnostic Results | Our Results | |
|---|---|---|---|---|---|---|---|---|
| Patient 1 | 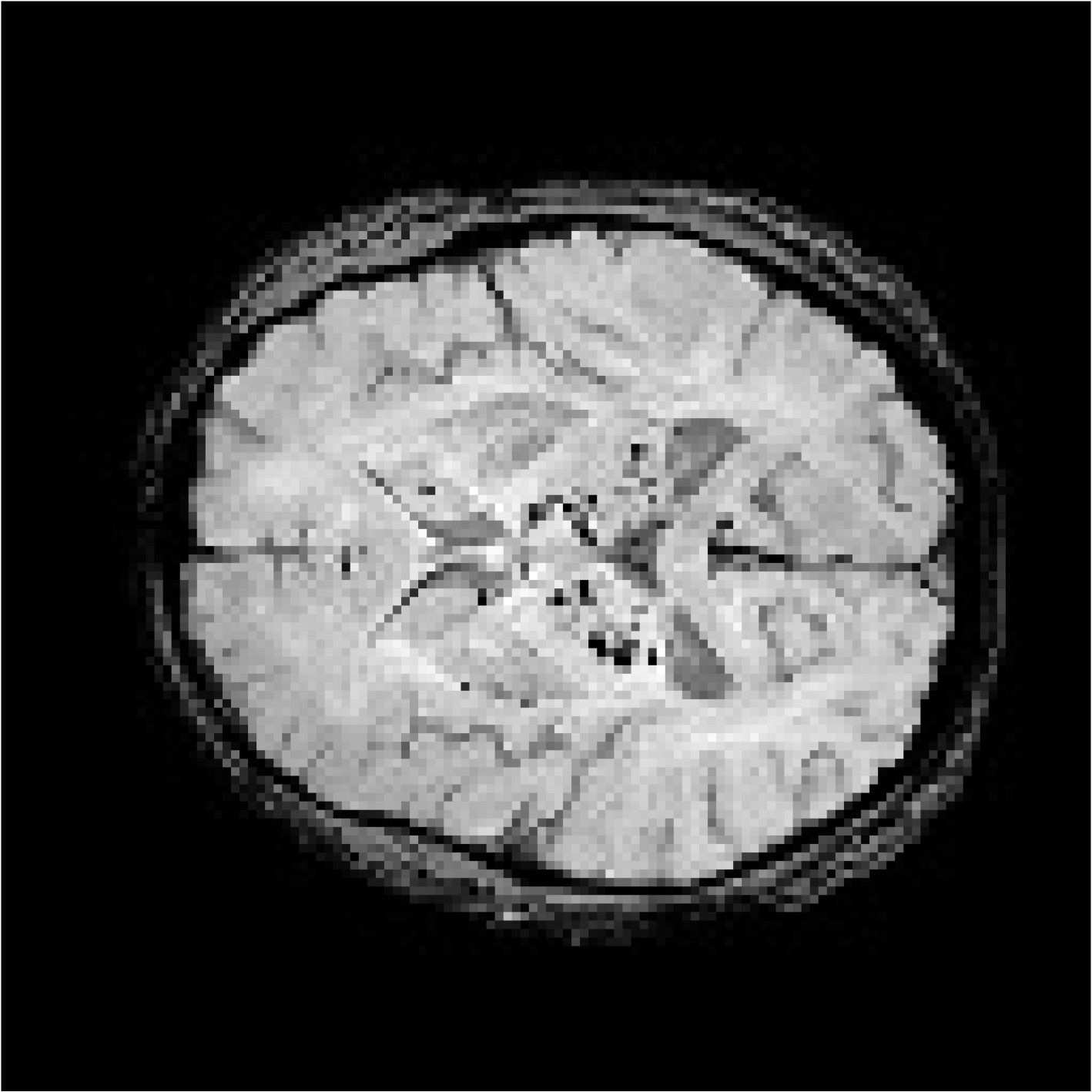 | 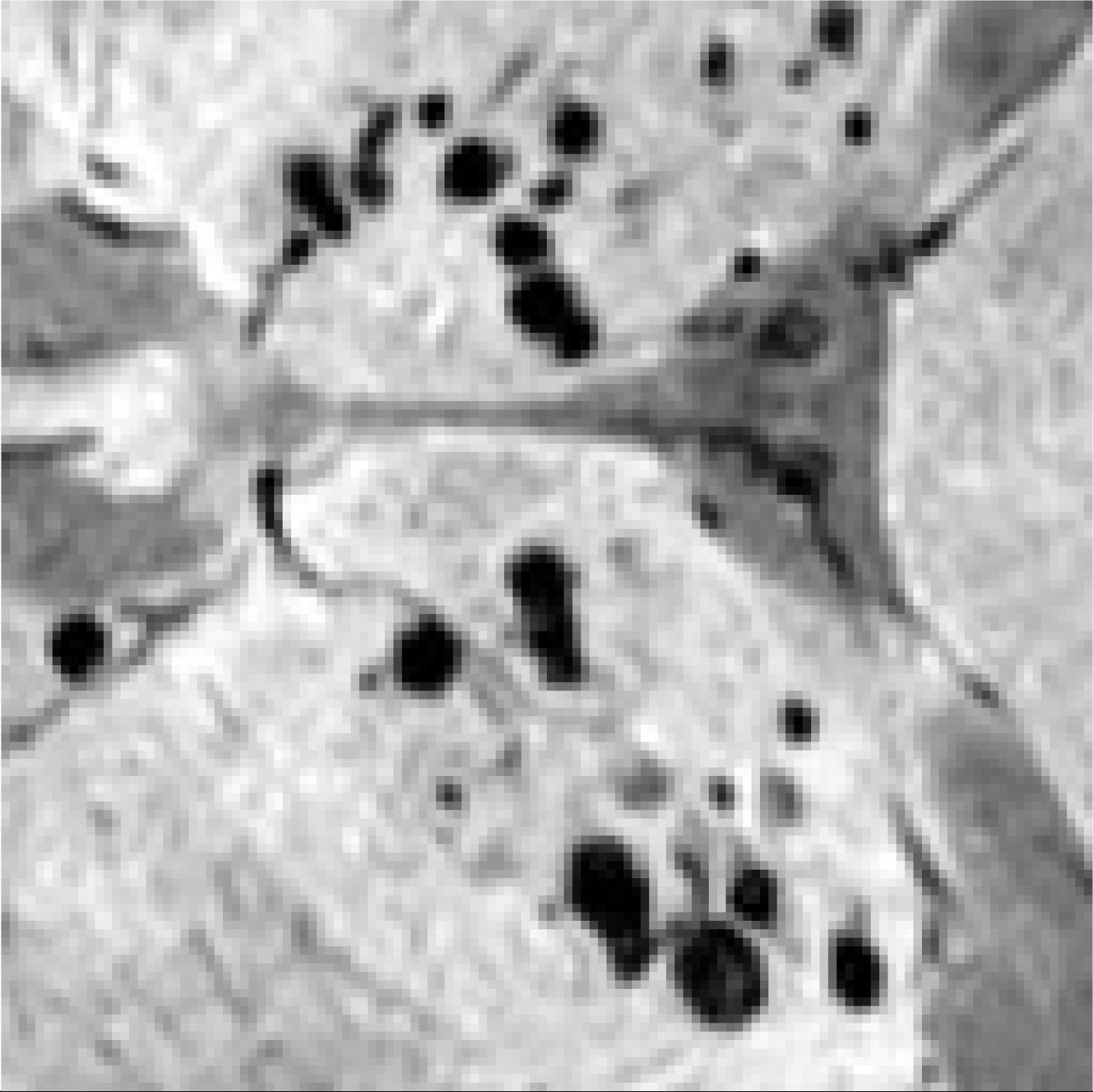 | 53 | Female | CADASIL | CMBs | Not CMBs | |
| Patient 2 |  | 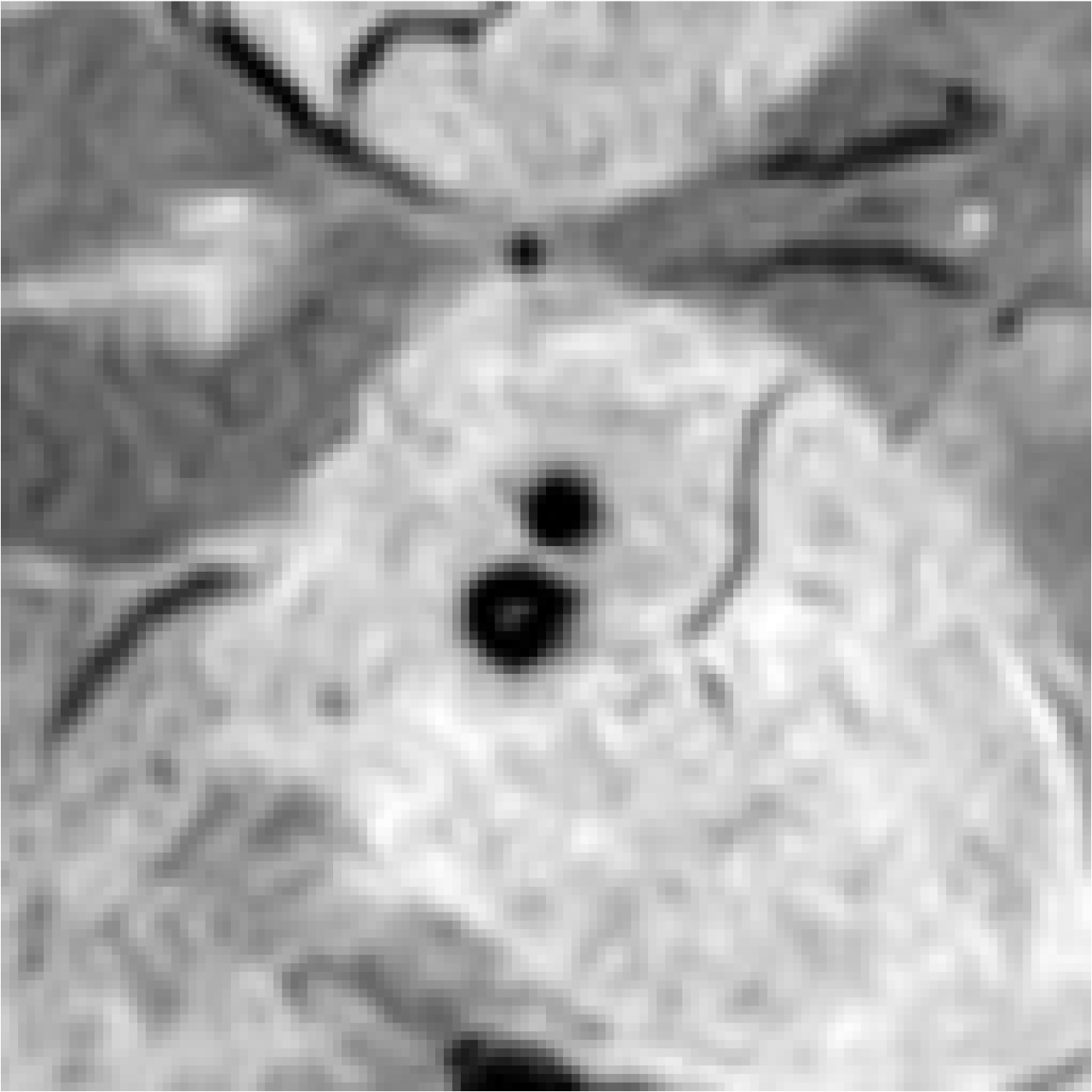 | 55 | Male | Drinking and Smoking | Hypertension | CMBs | Not CMBs |
| Patient 3 |  |  | 78 | Male | Cerebral Calcification (Not CMBs) | CMBs |
| Model Name | Backbone | Box Loss | P | R | ||
|---|---|---|---|---|---|---|
| YOLOv8 | N | 0.49 | 0.72 | 0.63 | 0.66 | 0.42 |
| S | 0.47 | 0.79 | 0.68 | 0.74 | 0.46 | |
| M | 0.43 | 0.88 | 0.71 | 0.77 | 0.49 | |
| X6 | 0.40 | 0.91 | 0.78 | 0.86 | 0.55 | |
| CMBs-YOLO w/o NWD | N | 0.43 | 0.76 | 0.68 | 0.72 | 0.52 |
| S | 0.41 | 0.86 | 0.78 | 0.81 | 0.53 | |
| M | 0.36 | 0.90 | 0.79 | 0.85 | 0.55 | |
| X6 | 0.36 | 0.92 | 0.87 | 0.89 | 0.59 | |
| CMBs-YOLO w/o CMBs-MHA | N | 0.43 | 0.73 | 0.64 | 0.67 | 0.44 |
| S | 0.42 | 0.81 | 0.67 | 0.75 | 0.47 | |
| M | 0.37 | 0.86 | 0.73 | 0.78 | 0.48 | |
| X6 | 0.33 | 0.88 | 0.79 | 0.85 | 0.52 | |
| CMBs-YOLO | N | 0.39 | 0.78 | 0.69 | 0.75 | 0.54 |
| S | 0.37 | 0.87 | 0.79 | 0.83 | 0.55 | |
| M | 0.32 | 0.91 | 0.82 | 0.87 | 0.57 | |
| X6 | 0.31 | 0.94 | 0.86 | 0.91 | 0.62 |
| Model Name | Backbone | Train Resolution | Validation Resolution | P | R | ||
|---|---|---|---|---|---|---|---|
| ConvNeXt | S | 0.80 | 0.66 | 0.76 | 0.49 | ||
| 0.83 | 0.75 | 0.81 | 0.52 | ||||
| 0.88 | 0.77 | 0.82 | 0.54 | ||||
| 0.82 | 0.57 | 0.74 | 0.43 | ||||
| 0.86 | 0.74 | 0.82 | 0.52 | ||||
| 0.89 | 0.81 | 0.85 | 0.57 | ||||
| DINO | R50 | 0.91 | 0.85 | 0.86 | 0.61 | ||
| 0.92 | 0.89 | 0.88 | 0.64 | ||||
| 0.92 | 0.88 | 0.87 | 0.64 | ||||
| 0.89 | 0.88 | 0.86 | 0.63 | ||||
| 0.92 | 0.91 | 0.87 | 0.64 | ||||
| 0.93 | 0.91 | 0.89 | 0.66 | ||||
| YOLOv8 | X6 | 0.83 | 0.74 | 0.76 | 0.52 | ||
| 0.91 | 0.78 | 0.86 | 0.55 | ||||
| 0.92 | 0.78 | 0.85 | 0.54 | ||||
| 0.81 | 0.72 | 0.75 | 0.49 | ||||
| 0.88 | 0.79 | 0.85 | 0.56 | ||||
| 0.92 | 0.81 | 0.88 | 0.57 | ||||
| CMBs-YOLO | X6 | 0.88 | 0.81 | 0.85 | 0.57 | ||
| 0.93 | 0.86 | 0.91 | 0.62 | ||||
| 0.94 | 0.87 | 0.90 | 0.61 | ||||
| 0.86 | 0.83 | 0.83 | 0.55 | ||||
| 0.93 | 0.88 | 0.90 | 0.62 | ||||
| 0.95 | 0.89 | 0.92 | 0.62 |
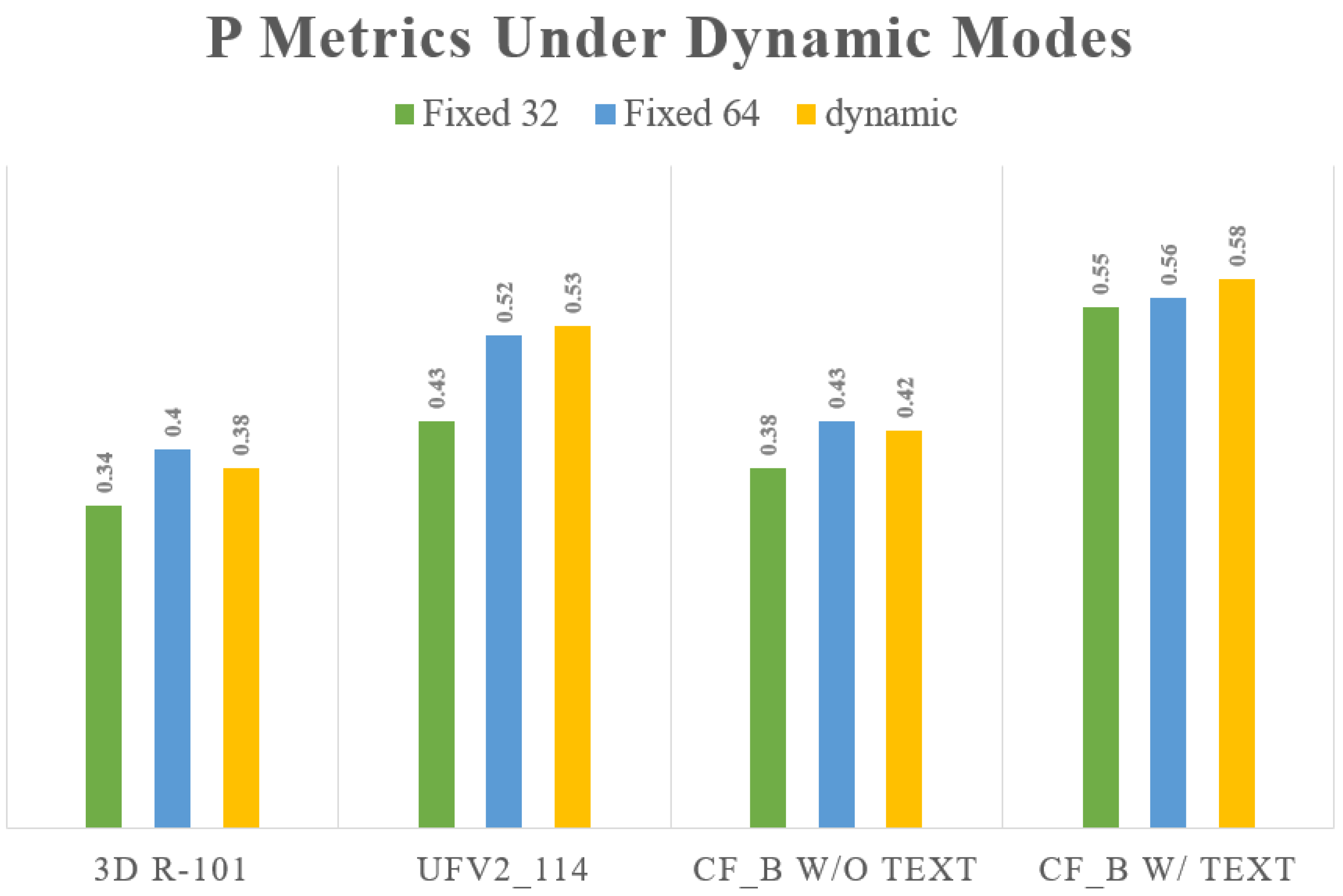
| Method Name | Model Name | Training Mode/Resolution | Testing Mode/Resolution | TPR | TNR | P |
|---|---|---|---|---|---|---|
| 3D-CNN | R-101 | Fixed | Fixed | 0.69 | 0.71 | 0.31 |
| Fixed | 0.83 | 0.84 | 0.34 | |||
| Dynamic | 0.82 | 0.86 | 0.33 | |||
| Fixed | Fixed | 0.79 | 0.82 | 0.32 | ||
| Fixed | 0.85 | 0.87 | 0.36 | |||
| Dynamic | 0.84 | 0.83 | 0.35 | |||
| Dynamic | Fixed | 0.72 | 0.69 | 0.34 | ||
| Fixed | 0.78 | 0.76 | 0.40 | |||
| Dynamic | 0.76 | 0.74 | 0.38 | |||
| Transformer | UFv2_114 | Fixed | Fixed | 0.77 | 0.79 | 0.39 |
| Fixed | 0.81 | 0.83 | 0.42 | |||
| Dynamic | 0.80 | 0.84 | 0.41 | |||
| Fixed | Fixed | 0.84 | 0.83 | 0.49 | ||
| Fixed | 0.89 | 0.88 | 0.56 | |||
| Dynamic | 0.85 | 0.81 | 0.51 | |||
| Dynamic | Fixed | 0.78 | 0.79 | 0.43 | ||
| Fixed | 0.86 | 0.83 | 0.52 | |||
| Dynamic | 0.85 | 0.86 | 0.53 | |||
| CF | CF_B w/o text | Fixed | Fixed | 0.82 | 0.81 | 0.41 |
| Fixed | 0.83 | 0.82 | 0.41 | |||
| Dynamic | 0.81 | 0.80 | 0.39 | |||
| Fixed | Fixed | 0.83 | 0.82 | 0.45 | ||
| Fixed | 0.89 | 0.86 | 0.51 | |||
| Dynamic | 0.84 | 0.81 | 0.41 | |||
| Dynamic | Fixed | 0.79 | 0.75 | 0.38 | ||
| Fixed | 0.83 | 0.79 | 0.43 | |||
| Dynamic | 0.82 | 0.80 | 0.42 | |||
| CF_B w/text | Fixed | Fixed | 0.88 | 0.87 | 0.56 | |
| Fixed | 0.89 | 0.88 | 0.58 | |||
| Dynamic | 0.85 | 0.87 | 0.55 | |||
| Fixed | Fixed | 0.90 | 0.89 | 0.57 | ||
| Fixed | 0.94 | 0.93 | 0.63 | |||
| Dynamic | 0.92 | 0.91 | 0.59 | |||
| Dynamic | Fixed | 0.87 | 0.85 | 0.55 | ||
| Fixed | 0.91 | 0.88 | 0.56 | |||
| Dynamic | 0.91 | 0.87 | 0.58 |
| Method Name | Classification Model Name | TPR | TNR | P |
|---|---|---|---|---|
| 2D-CNN | R-18 | 0.52 | 0.49 | 0.07 |
| R-50 | 0.61 | 0.62 | 0.15 | |
| R-101 | 0.73 | 0.76 | 0.23 | |
| 3D-CNN | R-18 | 0.64 | 0.69 | 0.14 |
| R-50 | 0.72 | 0.75 | 0.23 | |
| R-101 | 0.85 | 0.87 | 0.36 | |
| 2D-CF | CF_S w/text | 0.87 | 0.84 | 0.55 |
| CF_B w/text | 0.89 | 0.87 | 0.57 | |
| 3D-CF | CF_S w/text | 0.92 | 0.91 | 0.59 |
| CF_B w/text | 0.94 | 0.93 | 0.63 |
| 2D Input Image (Averaging) | 3D Input Images | Ground Truth | 2D-R101 | 3D-R101 | 2D-CF_B | 3D-CF_B | |
|---|---|---|---|---|---|---|---|
| Case 1 |  |  | CMBs | Not CMBs | CMBs | CMBs | CMBs |
| Case 2 |  |  | Cerebral Calcification (Not CMBs) | CMBs | Not CMBs | CMBs | Not CMBs |
| Case 3 |  | 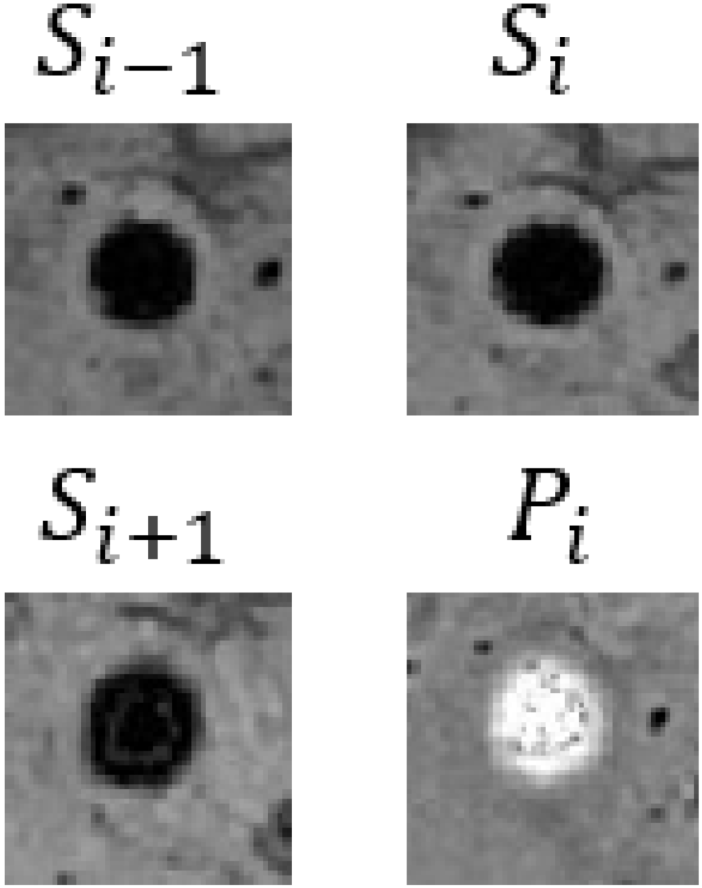 | Cerebral Calcification (Not CMBs) | CMBs | Not CMBs | CMBs | Not CMBs |
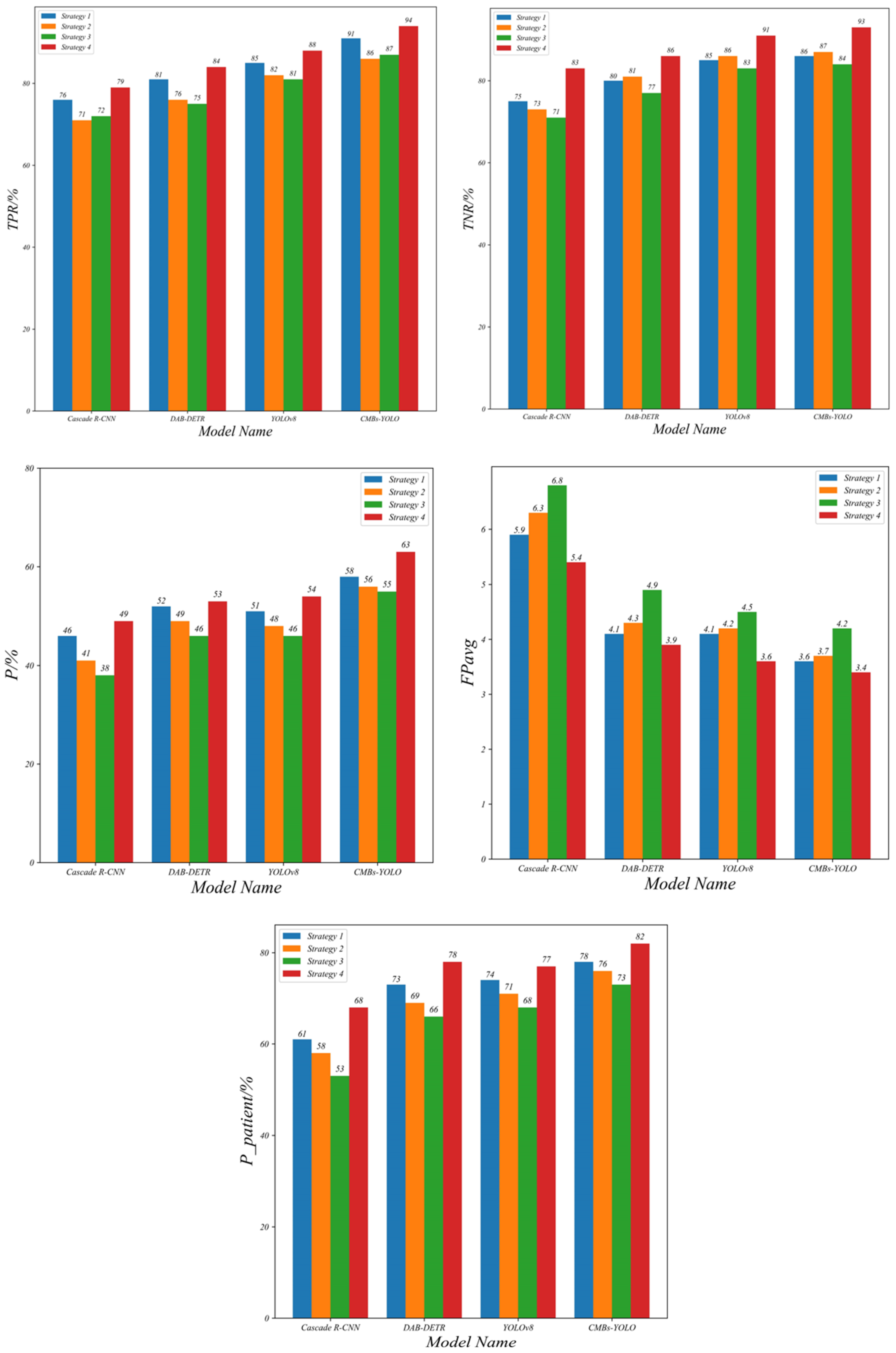
| Detection Model | Classification Model | Comparison Strategy | TPR | TNR | P | FPavg | |
|---|---|---|---|---|---|---|---|
| Cascade R-CNN (R50) | CF_B | 1 | 0.76 | 0.75 | 0.46 | 5.92 | 0.61 |
| 2 | 0.71 | 0.73 | 0.41 | 6.31 | 0.58 | ||
| 3 | 0.72 | 0.71 | 0.38 | 6.87 | 0.53 | ||
| 4 | 0.79 | 0.83 | 0.49 | 5.49 | 0.68 | ||
| DAB-DETR (R50) | CF_B | 1 | 0.81 | 0.80 | 0.52 | 4.12 | 0.73 |
| 2 | 0.76 | 0.81 | 0.49 | 4.37 | 0.69 | ||
| 3 | 0.75 | 0.77 | 0.46 | 4.92 | 0.66 | ||
| 4 | 0.84 | 0.86 | 0.53 | 3.98 | 0.78 | ||
| YOLOv8 (X6) | CF_B | 1 | 0.85 | 0.85 | 0.51 | 4.07 | 0.74 |
| 2 | 0.82 | 0.86 | 0.48 | 4.22 | 0.71 | ||
| 3 | 0.81 | 0.83 | 0.46 | 4.56 | 0.68 | ||
| 4 | 0.88 | 0.91 | 0.54 | 3.66 | 0.77 | ||
| CMBs-YOLO (X6) | CF_B | 1 | 0.91 | 0.86 | 0.58 | 3.61 | 0.78 |
| 2 | 0.86 | 0.87 | 0.56 | 3.77 | 0.76 | ||
| 3 | 0.87 | 0.84 | 0.55 | 4.19 | 0.73 | ||
| 4 | 0.94 | 0.93 | 0.63 | 3.37 | 0.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Zhao, L.-L.; Lang, Q.; Xu, Y. A Novel Detection and Classification Framework for Diagnosing of Cerebral Microbleeds Using Transformer and Language. Bioengineering 2024, 11, 993. https://doi.org/10.3390/bioengineering11100993
Chen C, Zhao L-L, Lang Q, Xu Y. A Novel Detection and Classification Framework for Diagnosing of Cerebral Microbleeds Using Transformer and Language. Bioengineering. 2024; 11(10):993. https://doi.org/10.3390/bioengineering11100993
Chicago/Turabian StyleChen, Cong, Lin-Lin Zhao, Qin Lang, and Yun Xu. 2024. "A Novel Detection and Classification Framework for Diagnosing of Cerebral Microbleeds Using Transformer and Language" Bioengineering 11, no. 10: 993. https://doi.org/10.3390/bioengineering11100993
APA StyleChen, C., Zhao, L.-L., Lang, Q., & Xu, Y. (2024). A Novel Detection and Classification Framework for Diagnosing of Cerebral Microbleeds Using Transformer and Language. Bioengineering, 11(10), 993. https://doi.org/10.3390/bioengineering11100993







