Author Contributions
Conceptualization, A.L.I., T.B. and A.J.T.; methodology, A.L.I., T.B. and A.J.T.; formal analysis, A.L.I., T.A., W.C.M., T.B. and A.J.T.; investigation, A.L.I., J.N.T. and A.J.T.; resources, A.J.T.; data curation, A.L.I., T.B. and A.J.T.; writing—A.L.I., J.N.T., T.B. and A.J.T.; preparation, A.L.I., T.B. and A.J.T.; writing—review and editing, A.L.I., T.A., W.C.M., T.B. and A.J.T.; visualization, A.L.I., T.B. and A.J.T.; supervision, A.J.T.; project administration, A.J.T. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Demonstration of Axolotl DualGraft™ (dHAAM): Sample of dehydrated human amnion–amnion membrane held by sterile tweezers.
Figure 1.
Demonstration of Axolotl DualGraft™ (dHAAM): Sample of dehydrated human amnion–amnion membrane held by sterile tweezers.
Figure 2.
Basic structure of the amniotic membrane: The amniotic membrane is composed of the epithelial layer (epithelium), the basement membrane, and the stroma layer, which is made up of three layers; an inner compact stromal layer, a middle fibroblast layer, and the outermost spongy layer (BioRender.com 2024).
Figure 2.
Basic structure of the amniotic membrane: The amniotic membrane is composed of the epithelial layer (epithelium), the basement membrane, and the stroma layer, which is made up of three layers; an inner compact stromal layer, a middle fibroblast layer, and the outermost spongy layer (BioRender.com 2024).
Figure 3.
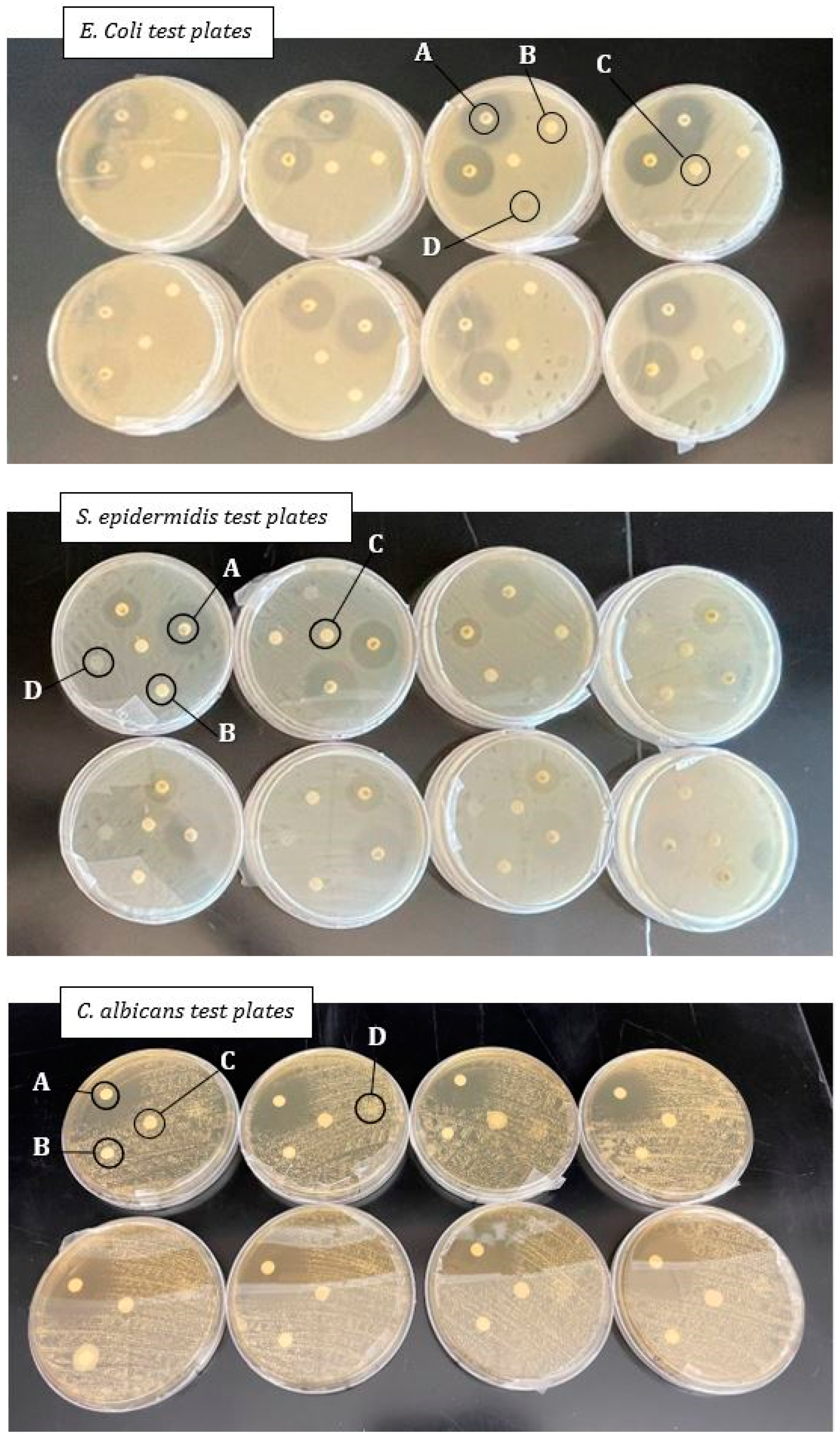
Antimicrobial test plate setup: Disks located in spot A are the antibiotic disks, those in spot B are the dHAAM samples, spot C is the control disk, and spot D is the disk treated with cell conditioned media. E. coli plates are the top collection of plates and the S. epidermidis plates are the middle, both photographed at the 16 h mark. C. albicans is the bottom photo, taken at the 24 h mark. Zones of inhibition, if present, are noted as the clear areas around each disk (Kovacs Z. (2022), A Pilot Study for Antimicrobial Characterization of Two Axolotl Products Using Disk Diffusion. Internal Axolotl Biologix report: unpublished).
Figure 3.
Antimicrobial test plate setup: Disks located in spot A are the antibiotic disks, those in spot B are the dHAAM samples, spot C is the control disk, and spot D is the disk treated with cell conditioned media. E. coli plates are the top collection of plates and the S. epidermidis plates are the middle, both photographed at the 16 h mark. C. albicans is the bottom photo, taken at the 24 h mark. Zones of inhibition, if present, are noted as the clear areas around each disk (Kovacs Z. (2022), A Pilot Study for Antimicrobial Characterization of Two Axolotl Products Using Disk Diffusion. Internal Axolotl Biologix report: unpublished).
Figure 4.
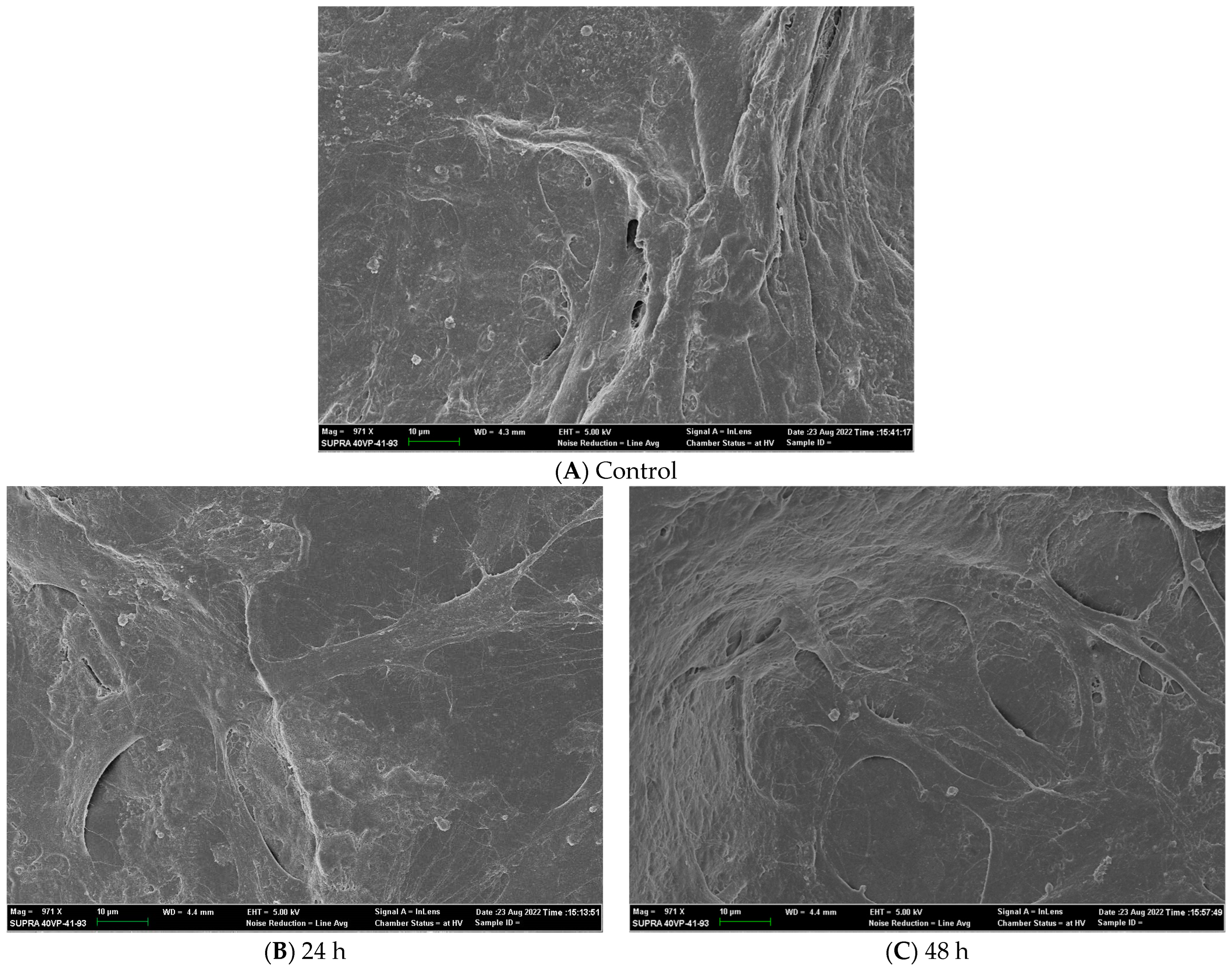
SEM images of adult HDF cells on Axolotl Biologix DualGraftTM: (A) Control SEM image is from a membrane sample fixed after 48 h incubation in DMEM; mag 971×. (B) Adult HDF cells seeded and incubated for 24 h on membrane sample before being fixed; mag 971×. (C) Adult HDF cells seeded and incubated for 48 h on membrane sample before being fixed; mag 971× (we used BioRender.com 2024 to combine images into one unit; images are from Ingraldi, A. (2023) DualGraft with HDF SEM Report. Internal Axolotl Biologix report: unpublished).
Figure 4.
SEM images of adult HDF cells on Axolotl Biologix DualGraftTM: (A) Control SEM image is from a membrane sample fixed after 48 h incubation in DMEM; mag 971×. (B) Adult HDF cells seeded and incubated for 24 h on membrane sample before being fixed; mag 971×. (C) Adult HDF cells seeded and incubated for 48 h on membrane sample before being fixed; mag 971× (we used BioRender.com 2024 to combine images into one unit; images are from Ingraldi, A. (2023) DualGraft with HDF SEM Report. Internal Axolotl Biologix report: unpublished).
Figure 5.
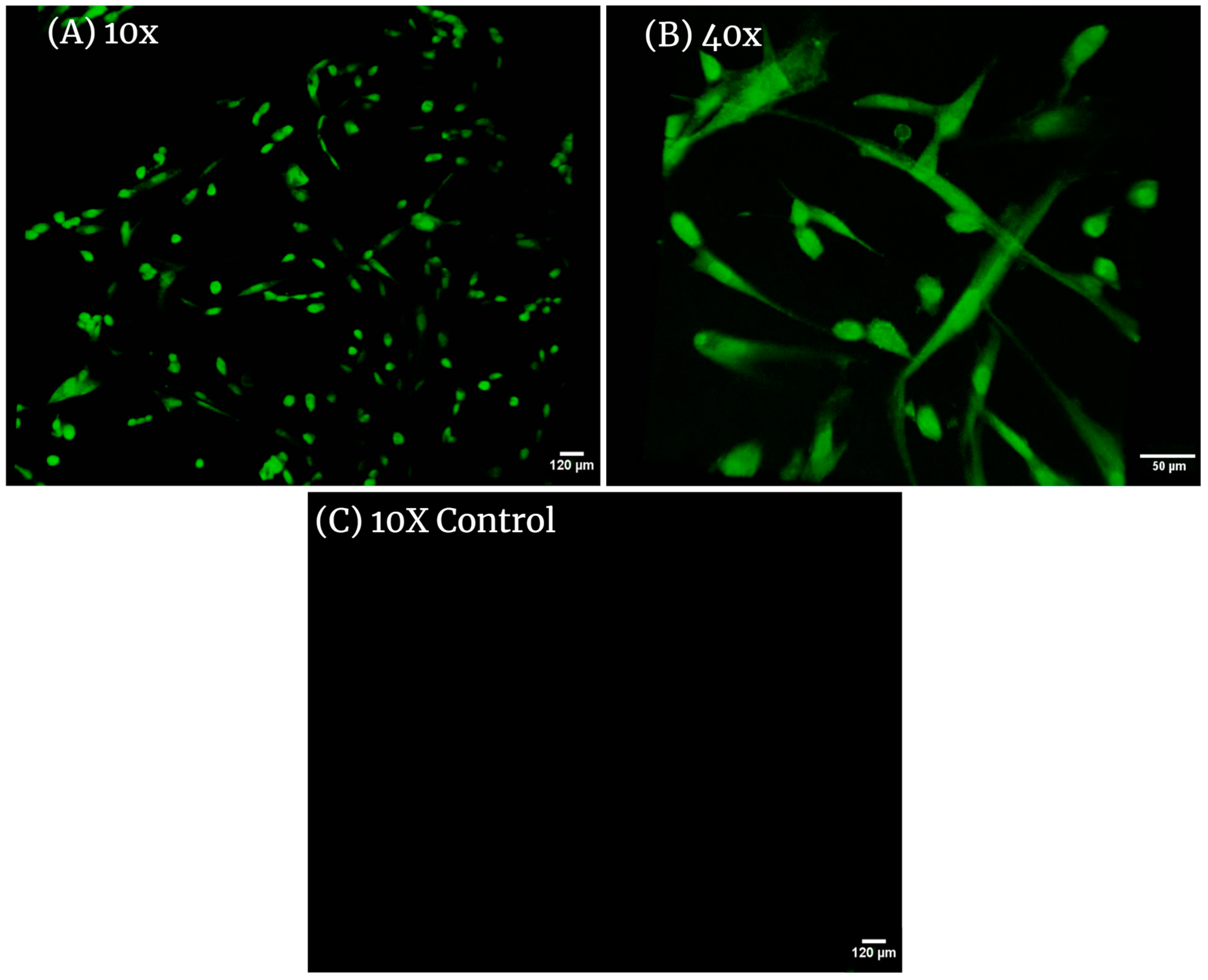
Confocal images of adult HDF cells on Axolotl Biologix DualGraftTM: (A) 10× magnification in confocal microscopy (Leica TCS SPE II) with 488 nm laser shows many cells present within the membrane; this image was taken using the Z-stack feature of the confocal micropscope, outlining the 3D placement of cells within the membrane; (B) 40× magnification presents hDF with normal morphology with elongated arms and rounded cell bodies either recently attached or preparing for division; (C) 10× magnification of unseeded membranes treated with fluorescent probe (control sample); no cells are highlighted, demonstrating that the membrane is decellularized and cells viewed in seeded samples are true positives. We used BioRender.com 2024 to combine images into one unit; images are from Ingraldi, A. (2023) Cell Tracker Green HDF on Membrane Report. Internal Axolotl Biologix report: unpublished.
Figure 5.
Confocal images of adult HDF cells on Axolotl Biologix DualGraftTM: (A) 10× magnification in confocal microscopy (Leica TCS SPE II) with 488 nm laser shows many cells present within the membrane; this image was taken using the Z-stack feature of the confocal micropscope, outlining the 3D placement of cells within the membrane; (B) 40× magnification presents hDF with normal morphology with elongated arms and rounded cell bodies either recently attached or preparing for division; (C) 10× magnification of unseeded membranes treated with fluorescent probe (control sample); no cells are highlighted, demonstrating that the membrane is decellularized and cells viewed in seeded samples are true positives. We used BioRender.com 2024 to combine images into one unit; images are from Ingraldi, A. (2023) Cell Tracker Green HDF on Membrane Report. Internal Axolotl Biologix report: unpublished.
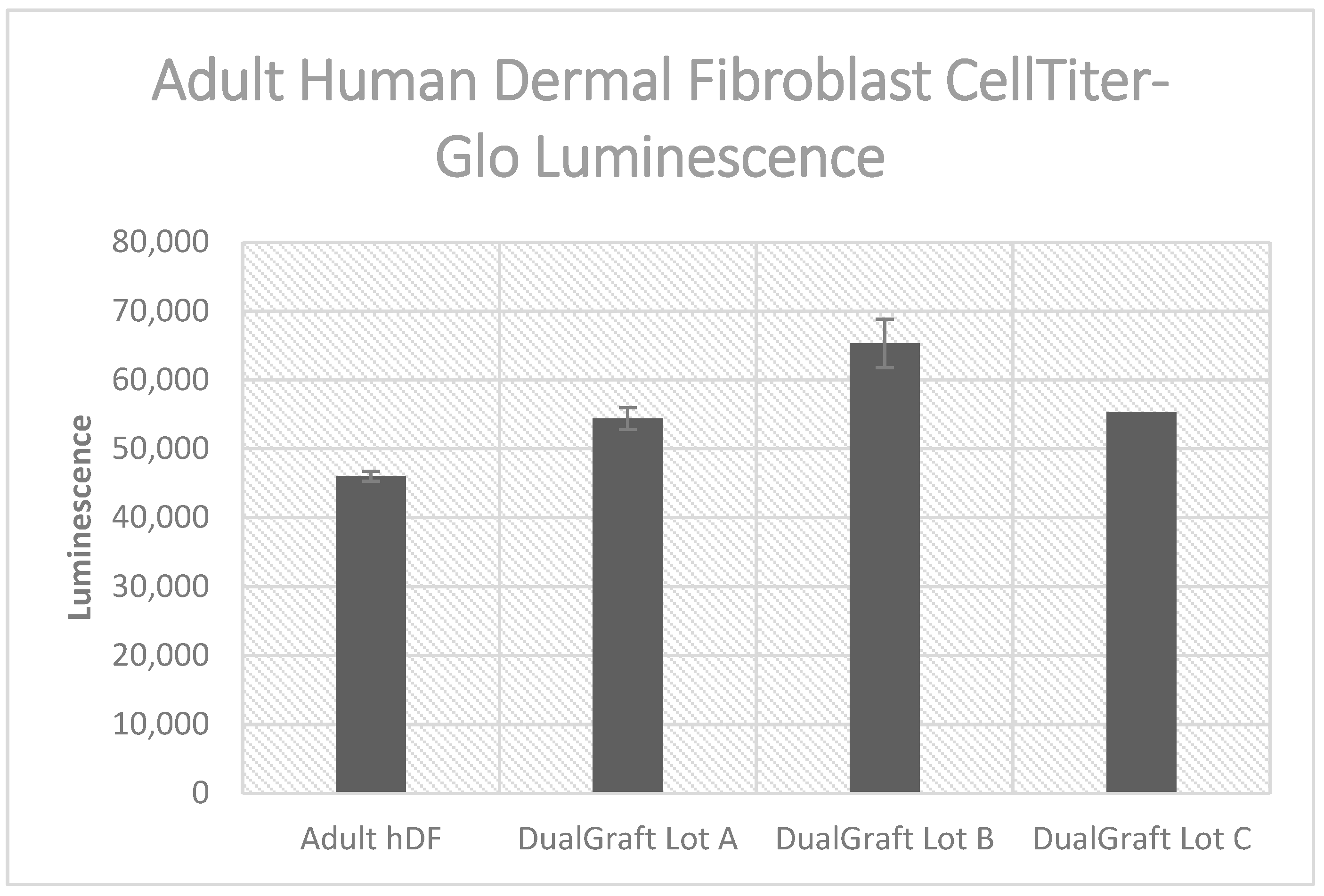
Figure 6.
Luminescent cell viability assay results: Axolotl DualGraft™ samples (Lot A–C, luminescence > 54,300) presented a greater luminescence than adult human dermal fibroblasts cells (luminescence < 46,100) cultured without membrane present (Audet R. and Ingraldi A. (2022), DualGraft Biocompatibility—ATP Assay Report. Internal Axolotl Biologix report: unpublished).
Figure 6.
Luminescent cell viability assay results: Axolotl DualGraft™ samples (Lot A–C, luminescence > 54,300) presented a greater luminescence than adult human dermal fibroblasts cells (luminescence < 46,100) cultured without membrane present (Audet R. and Ingraldi A. (2022), DualGraft Biocompatibility—ATP Assay Report. Internal Axolotl Biologix report: unpublished).
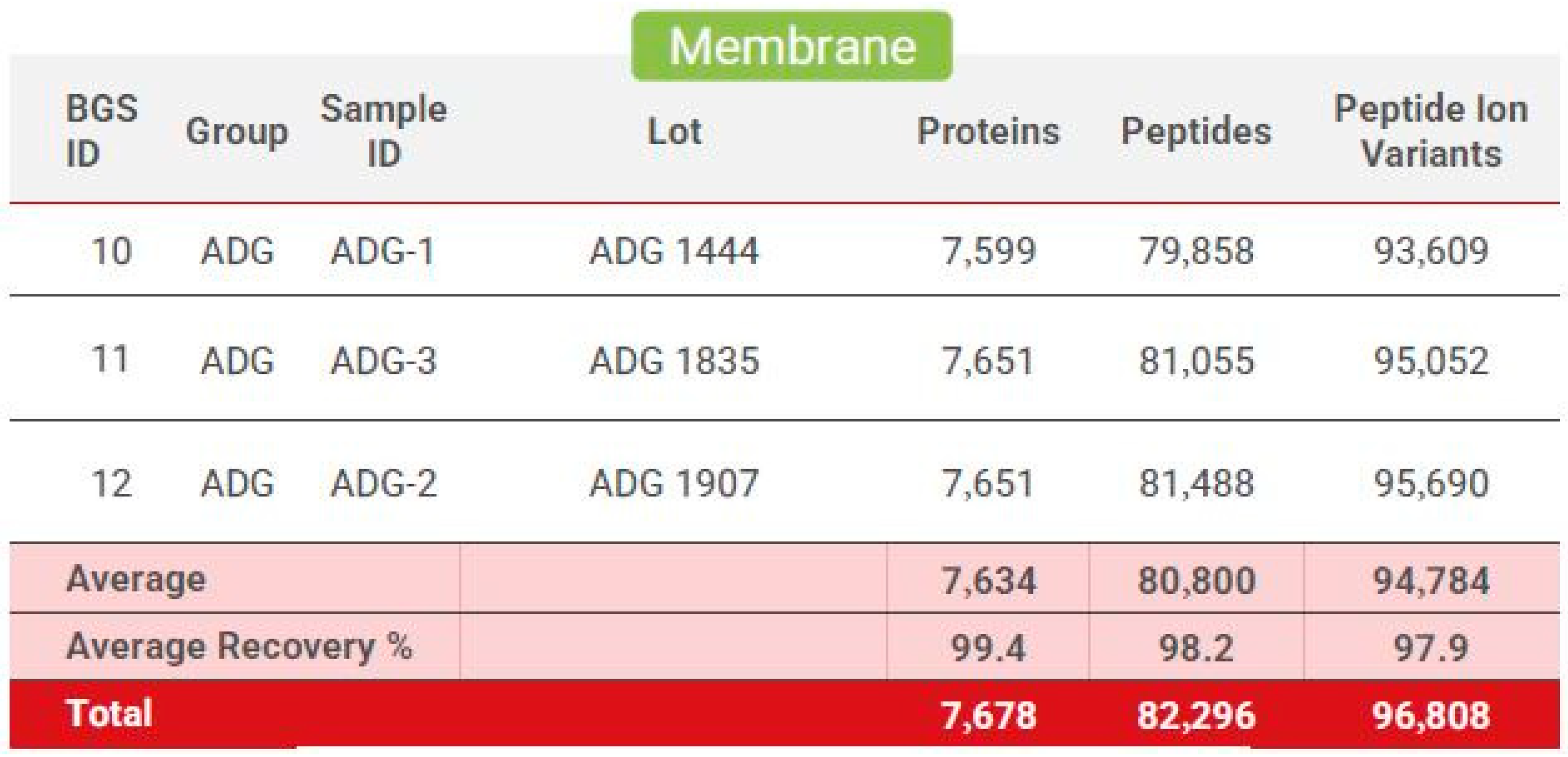
Figure 7.
Biognosys proteome protein profile: Generated from three separate dHAAM lots and identifies the total number of proteins, peptides, and peptide ion variants present in each membrane sample (Kamber D. and Soste M. (2022) Proteomic Analysis of Axolotl Ambient and Axolotl DualGraft—Final Report: unpublished).
Figure 7.
Biognosys proteome protein profile: Generated from three separate dHAAM lots and identifies the total number of proteins, peptides, and peptide ion variants present in each membrane sample (Kamber D. and Soste M. (2022) Proteomic Analysis of Axolotl Ambient and Axolotl DualGraft—Final Report: unpublished).
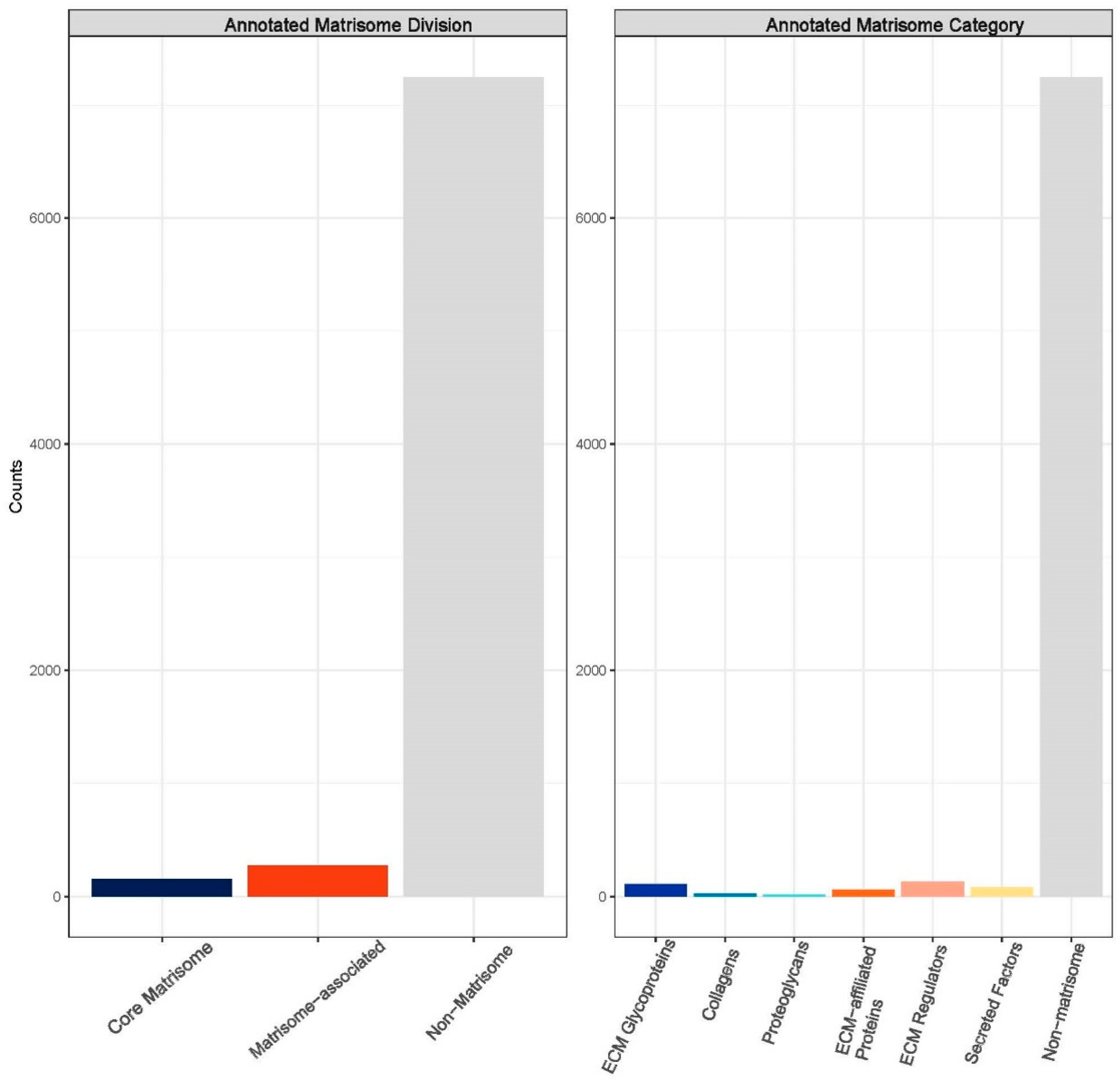
Figure 8.
Annotated matrisome divisions: The matrisome AnalyzeR identified a total of 163 core matrisome proteins, a total of 290 matrisome-associated proteins, and a total of 7228 non-matrisome-associated proteins. (Audet R. and Ingraldi A. (2024) Matrisome AnalyzeR DualGraft Report. Internal Axolotl Biologix report: unpublished).
Figure 8.
Annotated matrisome divisions: The matrisome AnalyzeR identified a total of 163 core matrisome proteins, a total of 290 matrisome-associated proteins, and a total of 7228 non-matrisome-associated proteins. (Audet R. and Ingraldi A. (2024) Matrisome AnalyzeR DualGraft Report. Internal Axolotl Biologix report: unpublished).
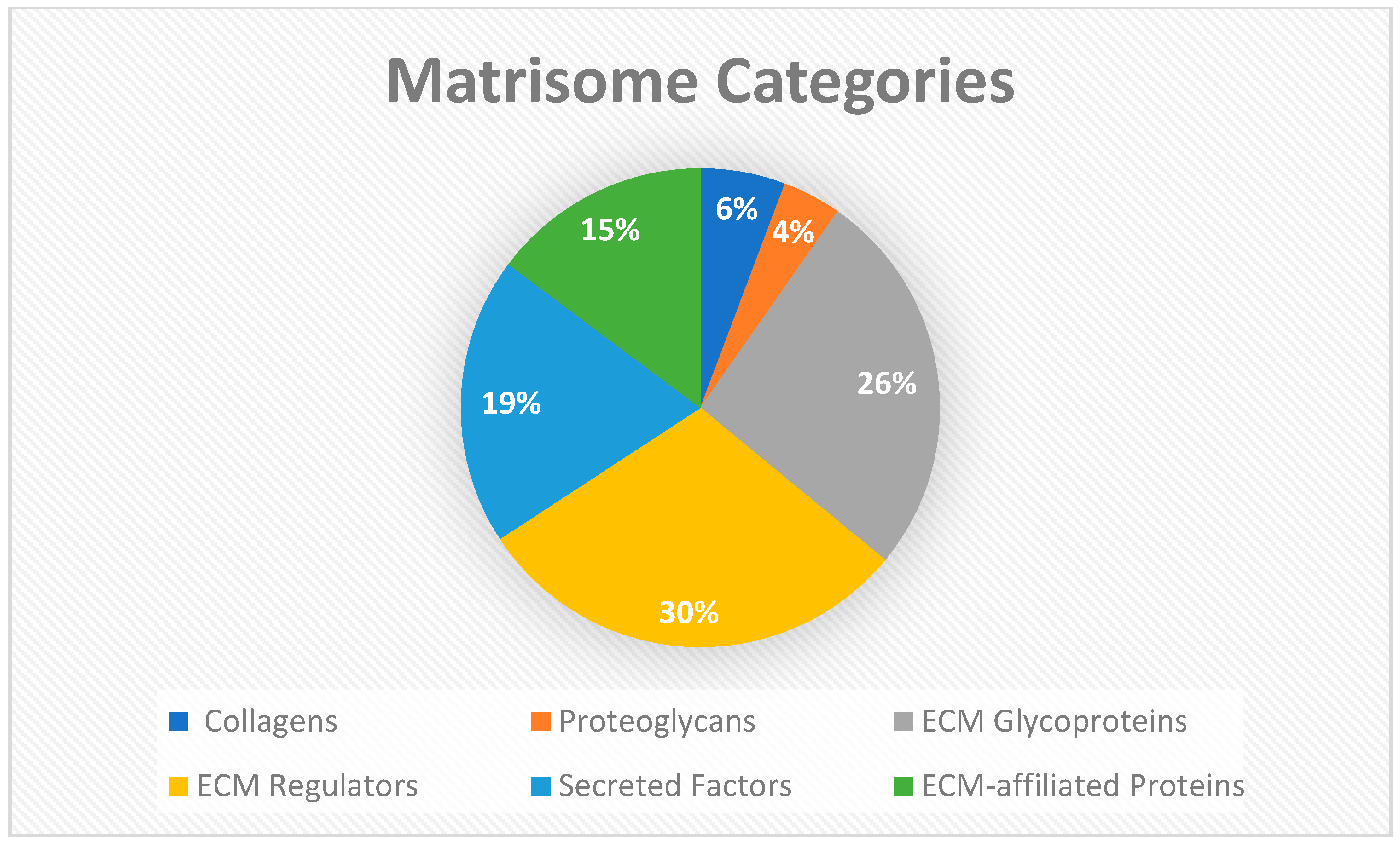
Figure 9.
Matrisome categories: The matrisome AnalyzeR identified a total of 26 collagens, 18 proteoglycans, and 119 ECM glycoprotein genes for total of 163 core matrisome genes. A total of 135 ECM regulators, 88 secreted factors, and 67 ECM-affiliated proteins were identified for a total of 290 matrisome-associated genes. (Audet R. and Ingraldi A. (2024) Matrisome AnalyzeR DualGraft Report. Internal Axolotl Biologix report: unpublished).
Figure 9.
Matrisome categories: The matrisome AnalyzeR identified a total of 26 collagens, 18 proteoglycans, and 119 ECM glycoprotein genes for total of 163 core matrisome genes. A total of 135 ECM regulators, 88 secreted factors, and 67 ECM-affiliated proteins were identified for a total of 290 matrisome-associated genes. (Audet R. and Ingraldi A. (2024) Matrisome AnalyzeR DualGraft Report. Internal Axolotl Biologix report: unpublished).
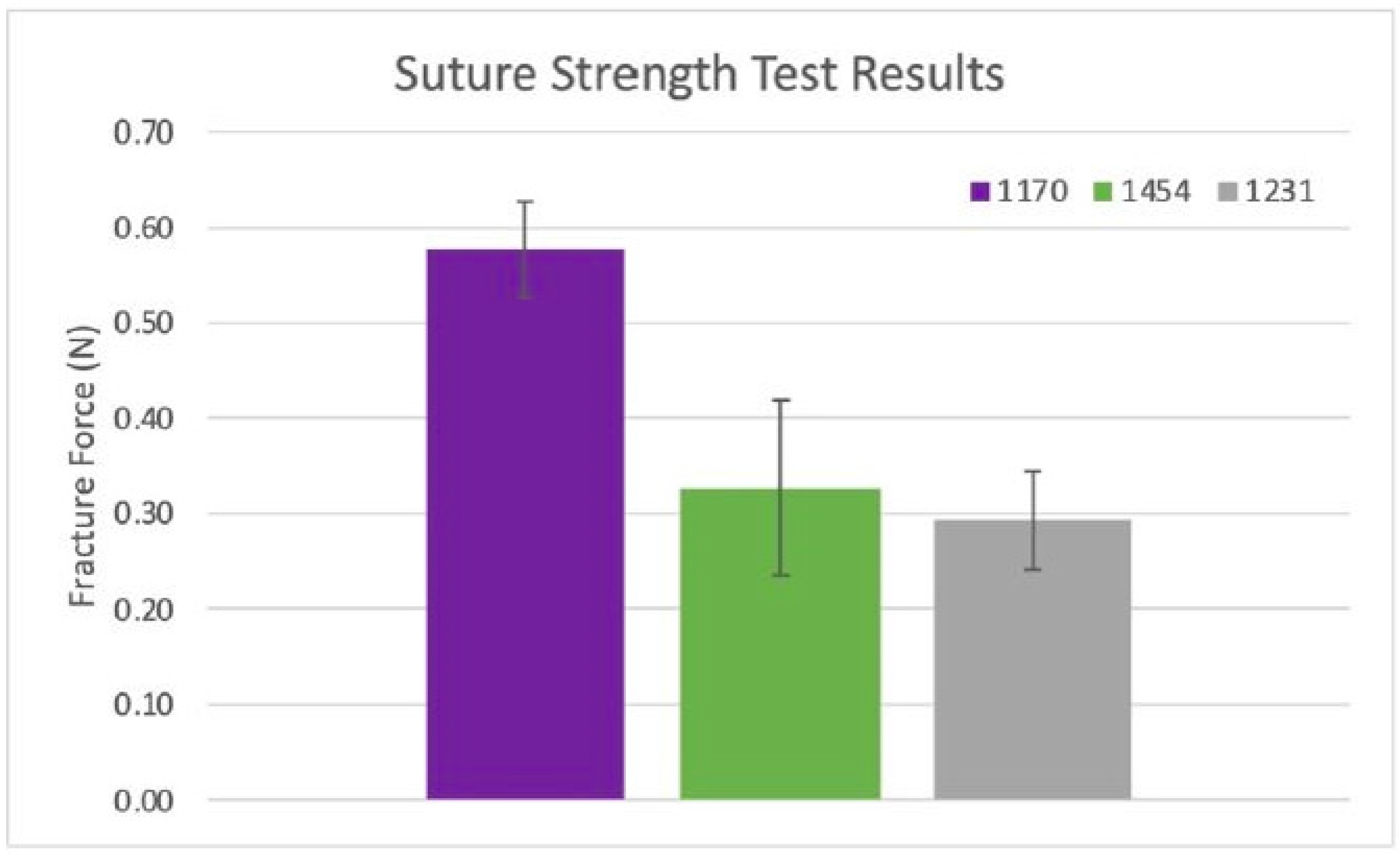
Figure 10.
Suture strength results: Ultimate suture strength based on measured forces from two samples per dHAAM donor lot (1170, 1454, 1231). (Becker T., (2023). Suture Strength Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 10.
Suture strength results: Ultimate suture strength based on measured forces from two samples per dHAAM donor lot (1170, 1454, 1231). (Becker T., (2023). Suture Strength Testing Report. Internal Axolotl Biologix report: unpublished).
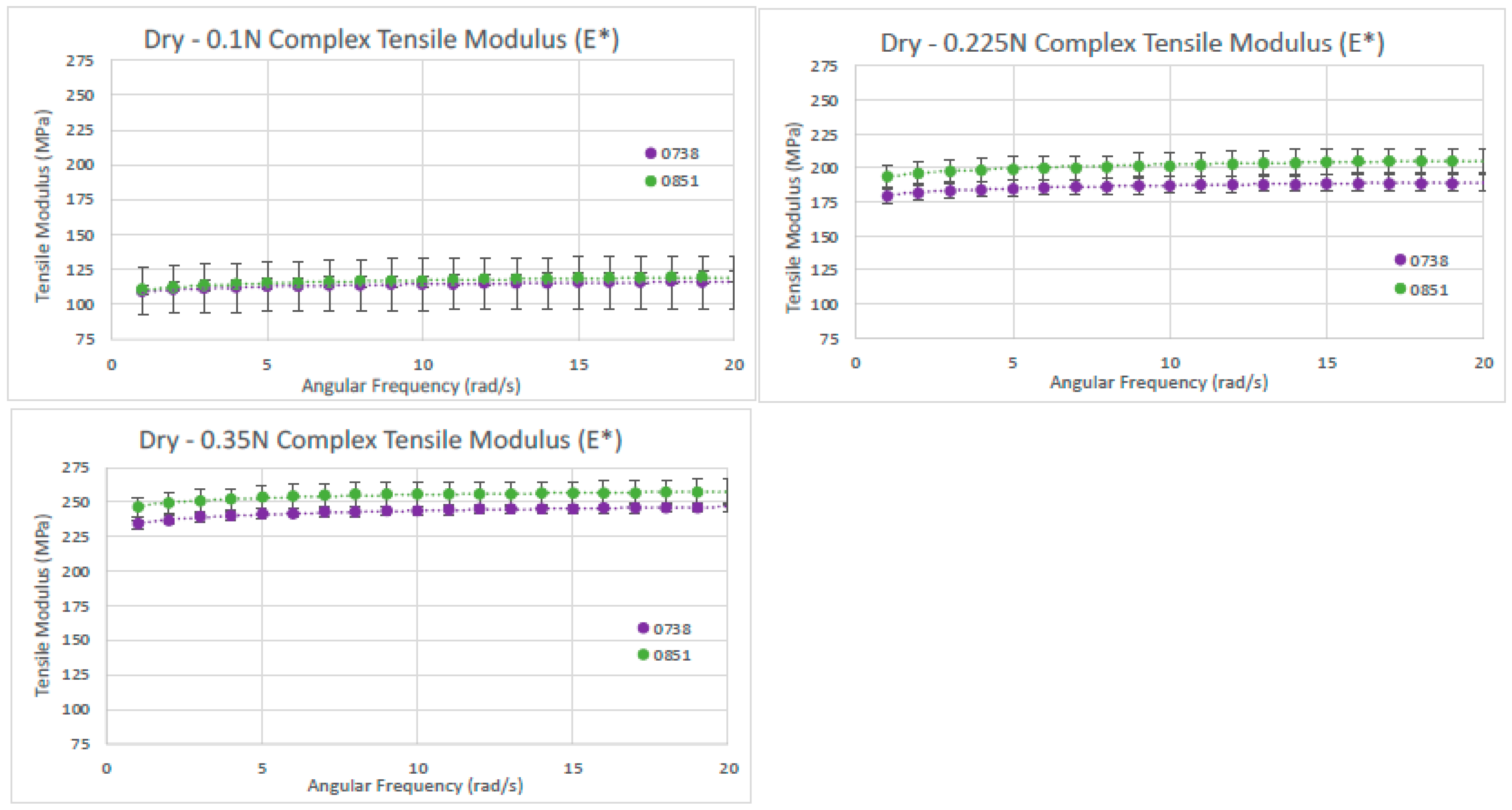
Figure 11.
Dry tensile modulus results. Tensile moduli (E*) for two dry dHAAM samples tested at tensile forces of (left) 0.1 N, (right) 0.225 N, and (lower left) 0.35 N (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report, unpublished).
Figure 11.
Dry tensile modulus results. Tensile moduli (E*) for two dry dHAAM samples tested at tensile forces of (left) 0.1 N, (right) 0.225 N, and (lower left) 0.35 N (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report, unpublished).
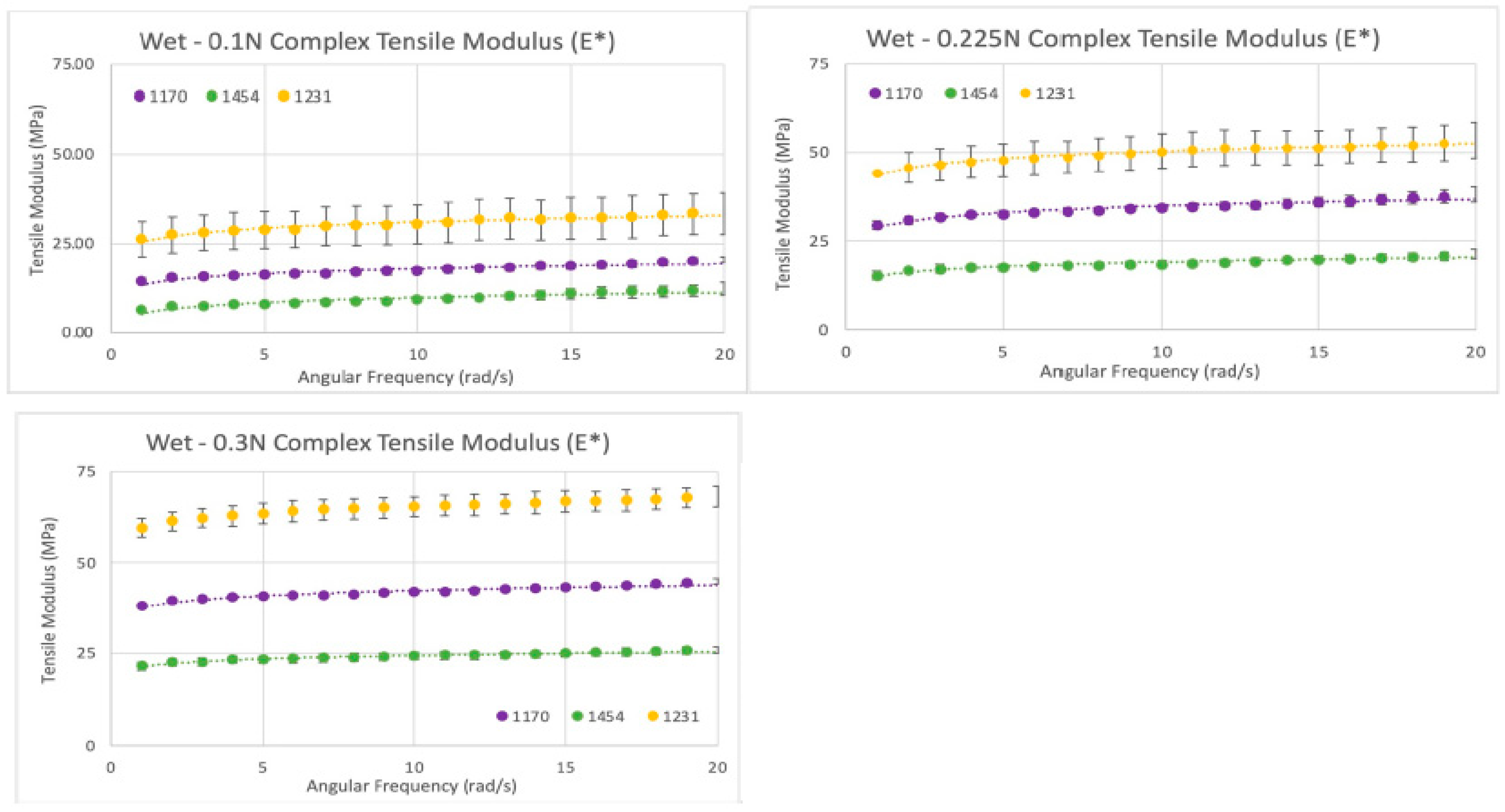
Figure 12.
Wet tensile modulus results: Tensile moduli (E*) for wet samples tested at tensile forces of 0.1 N (top left), 0.225 N (top right), and 0.3 N (bottom left) (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 12.
Wet tensile modulus results: Tensile moduli (E*) for wet samples tested at tensile forces of 0.1 N (top left), 0.225 N (top right), and 0.3 N (bottom left) (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 13.
Complex tensile modulus results: (Left Graph) Tensile moduli (E.) for dry samples; moduli vs. applied forces (0.1 N, 0.225 N, and 0.35 N). Blue and orange represent the 2 different dHAAM lots tested. (Right Graph) Tensile moduli (E) for three lots of samples vs. all applied forces (0.1 N, 0.225 N, and 0.3 N) at 6 rad/s (1 Hz) (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 13.
Complex tensile modulus results: (Left Graph) Tensile moduli (E.) for dry samples; moduli vs. applied forces (0.1 N, 0.225 N, and 0.35 N). Blue and orange represent the 2 different dHAAM lots tested. (Right Graph) Tensile moduli (E) for three lots of samples vs. all applied forces (0.1 N, 0.225 N, and 0.3 N) at 6 rad/s (1 Hz) (Becker T., (2023). Complex Tensile Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 14.
Stress versus strain: Example of tensile stress vs. strain for lot 1231 of dHAAM sample, the slope included = tensile modulus. (Becker T., (2023). Tensile Strength Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 14.
Stress versus strain: Example of tensile stress vs. strain for lot 1231 of dHAAM sample, the slope included = tensile modulus. (Becker T., (2023). Tensile Strength Testing Report. Internal Axolotl Biologix report: unpublished).
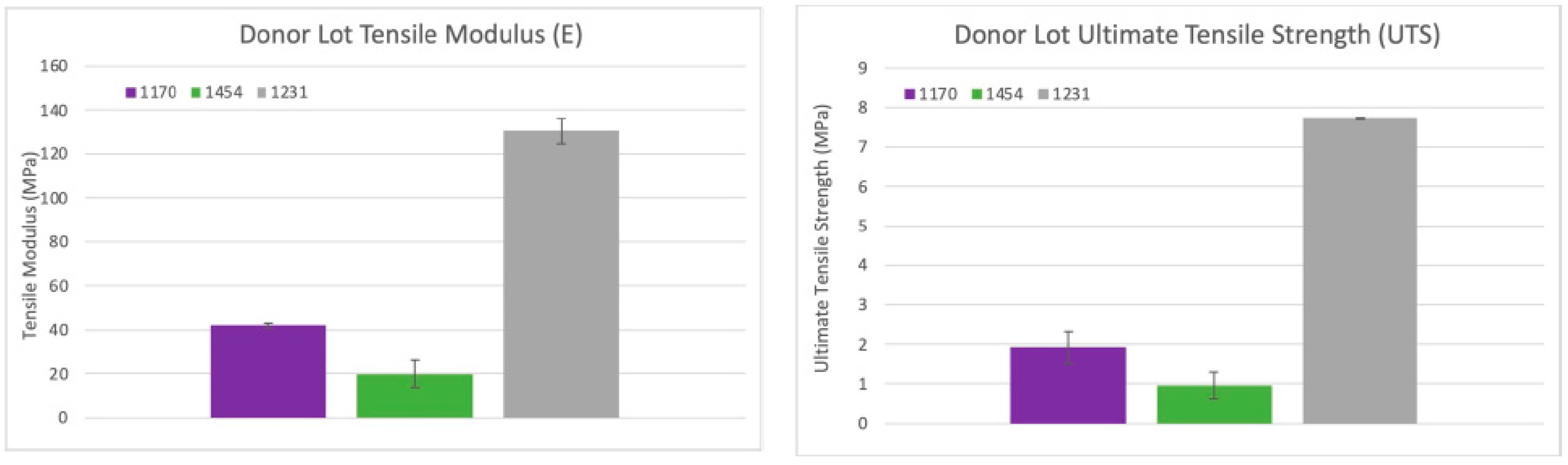
Figure 15.
Tensile strength results: Average tensile moduli for the three dHAAM lots tested; ultimate tensile strength for the three donor lots tested. (Becker T., (2023). Tensile Strength Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 15.
Tensile strength results: Average tensile moduli for the three dHAAM lots tested; ultimate tensile strength for the three donor lots tested. (Becker T., (2023). Tensile Strength Testing Report. Internal Axolotl Biologix report: unpublished).
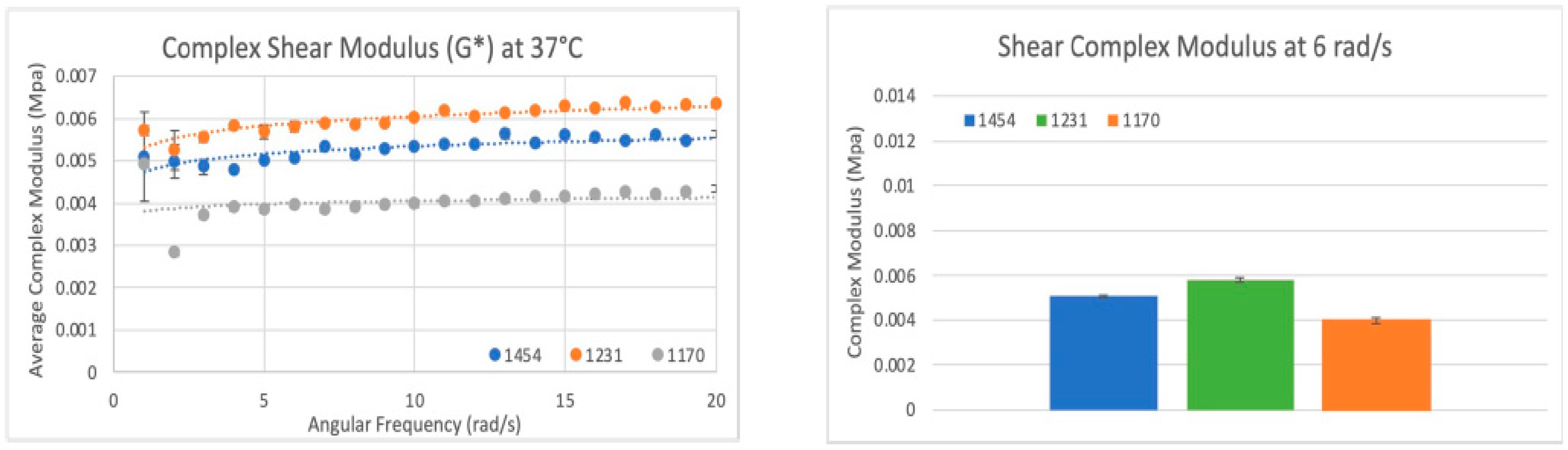
Figure 16.
Shear modulus results: (Left) Scatter plot of the complex shear modulus (G*) for three sample dHAAM lots tested at 37 °C. (Right) Bar graph comparison of shear modulus (G*) for the three samples lots tested at shear rate 6 rad/s (60 BPM); all lot p values < 0.01. (Becker T., (2023). Shear Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
Figure 16.
Shear modulus results: (Left) Scatter plot of the complex shear modulus (G*) for three sample dHAAM lots tested at 37 °C. (Right) Bar graph comparison of shear modulus (G*) for the three samples lots tested at shear rate 6 rad/s (60 BPM); all lot p values < 0.01. (Becker T., (2023). Shear Modulus Testing Report. Internal Axolotl Biologix report: unpublished).
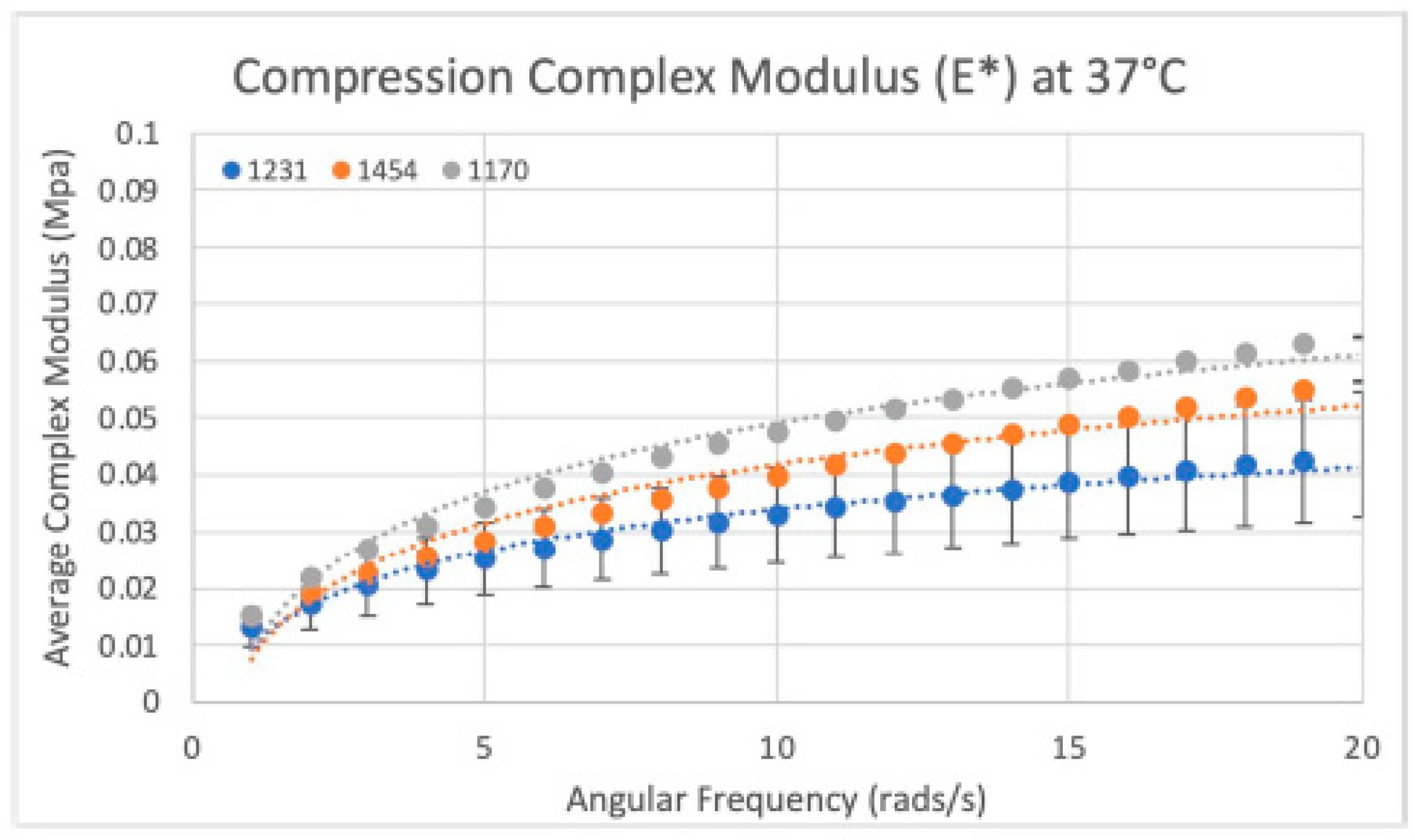
Figure 17.
Compressive modulus results: Compressive modulus (E*) for the three dHAAM lots tested at 37 °C. (Becker T., (2023). Compressive Modulus Report. Internal Axolotl Biologix report: unpublished).
Figure 17.
Compressive modulus results: Compressive modulus (E*) for the three dHAAM lots tested at 37 °C. (Becker T., (2023). Compressive Modulus Report. Internal Axolotl Biologix report: unpublished).
Table 1.
Mean Inhibitory Zone: The average zone of inhibition for each antibiotic treatment per organism in mm (n = 8). The dHAAM and conditioned media samples performed similarly to the control group, overgrown with the organisms on the plate, resulting in no inhibition.
Table 1.
Mean Inhibitory Zone: The average zone of inhibition for each antibiotic treatment per organism in mm (n = 8). The dHAAM and conditioned media samples performed similarly to the control group, overgrown with the organisms on the plate, resulting in no inhibition.
| Mean of Inhibitory Zone (n = 8) |
|---|
| Organism | Tetracycline | Chloramphenicol | Gentamycin | Fluconazole | DualGraft | Cond. Media | Control |
|---|
| E. coli | 26 | 27 | - | - | 0 | 0 | 0 |
| S. epidermidis | 18 | - | 26 | - | 0 | 0 | 0 |
| C. albicans | - | - | - | 30 | 0 | 0 | 0 |
Table 2.
Ultimate suture strength: Ultimate suture strength (USS) determination and suture force based on suture size. (Becker T., (2023). Suture Strength Testing Report. Internal Axolotl Biologix report: unpublished).
Table 2.
Ultimate suture strength: Ultimate suture strength (USS) determination and suture force based on suture size. (Becker T., (2023). Suture Strength Testing Report. Internal Axolotl Biologix report: unpublished).
| Ultimate Suture Strength (USS) Table |
|---|
| Suture Size (USP) | Suture Diameter (mm) | Suture Area (mm2) | USS (MPa) | Suture Force (N) |
|---|
| 6-0 | 0.07 | 0.005 | 8.5 | 0.05 |
| 5-0 | 0.10 | 0.008 | 8.5 | 0.07 |
| 4-0 | 0.15 | 0.012 | 8.5 | 0.10 |
| 3-0 | 0.20 | 0.016 | 8.5 | 0.13 |
| 2-0 | 0.30 | 0.024 | 8.5 | 0.20 |
| 0 | 0.35 | 0.027 | 8.5 | 0.23 |
Table 3.
Category—core matrisome (total identified = 163).
Table 3.
Category—core matrisome (total identified = 163).
| Annotated Gene | Protein Name | Protein Description | Protein Function |
|---|
| DCN | PGS2_HUMAN | Decorin | A small leucine-rich proteoglycan involved in the regulation of collagen fibrillogenesis and matrix organization. |
| COL1A2 | CO1A2_HUMAN | Collagen alpha 2(I) chain | Part of type I collagen, which is a major structural protein in connective tissues. |
| COL1A1 | CO1A1_HUMAN | Collagen alpha 1(I) chain | Also a component of type I collagen, working together with the alpha 2 chain to form the collagen fibrils that provide structural support and strength to various tissues. |
| LUM | LUM_HUMAN | Lumican | A member of the small leucine-rich proteoglycan family, lumican plays a role in collagen fibril organization. |
| OGN | MIME_HUMAN | Mimecan | Mimecan is a small leucine-rich proteoglycan involved in regulating collagen fibrillogenesis and contributing to bone and cartilage matrix organization. |
| PRELP | PRELP_HUMAN | Prolargin | A small leucine-rich proteoglycan that influences collagen fibril formation and tissue repair, playing a role in the structural integrity of connective tissues. |
| DSP | DESP_HUMAN | Desmoplakin | A key component of desmosomes, which are cell structures involved in maintaining cell–cell adhesion and structural integrity in tissues such as the skin and heart. |
| FN1 | FINC_HUMAN | Fibronectin | A glycoprotein which plays a crucial role in cell adhesion, growth, migration, and wound healing. It helps to organize the extracellular matrix and facilitate cellular interactions with the matrix. |
| COL3A1 | CO3A1_HUMAN | Collagen alpha 1(III) chain | Part of type III collagen, which is found in many tissues including skin, blood vessels, and internal organs. Type III collagen provides structural support and flexibility. |
| LTBP4 | LTBP4_HUMAN | Latent-transforming growth factor beta-binding protein 4 | A protein that binds to and regulates the activation of transforming growth factor beta (TGF-β), which is involved in cell growth, differentiation, and extracellular matrix production. |
Table 4.
Category—ECM glycoproteins (total identified = 119).
Table 4.
Category—ECM glycoproteins (total identified = 119).
| Annotated Gene | Protein Name | Protein Description | Protein Function |
|---|
| DSP | DESP_HUMAN | Desmoplakin | Component of desmosomes, which are adhesive junctions that provide mechanical strength to tissues by linking intermediate filaments of the cytoskeleton to the cell membrane, thereby maintaining cell–cell adhesion. |
| FN1 | FINC_HUMAN | Fibronectin | Fibronectin is a multifunctional glycoprotein involved in cell adhesion, migration, and matrix organization. It plays a key role in wound healing and tissue repair by facilitating interactions between cells and the extracellular matrix. |
| LTBP4 | LTBP4_HUMAN | Latent-transforming growth factor beta-binding protein 4 | Binds to and regulates the activation of transforming growth factor beta (TGF-β), influencing cell growth, differentiation, and extracellular matrix production. |
| TGFBI | BGH3_HUMAN | Transforming growth factor-beta-induced protein ig-h3 | Induced by TGF-β and involved in cell adhesion, migration, and extracellular matrix organization. It plays a role in tissue repair and fibrosis. |
| THBS1 | TSP1_HUMAN | Thrombospondin-1 | A glycoprotein involved in cell–cell and cell–matrix interactions, thrombospondin-1 regulates processes such as angiogenesis, wound healing, and tissue remodeling. It can influence cell adhesion and migration by interacting with various cell surface receptors. |
| TNXB | TENX_HUMAN | Tenascin-X | A large extracellular matrix glycoprotein that affects collagen fibril organization and tissue elasticity. It is involved in connective tissue structure and has roles in tissue repair and development. |
| FBN1 | FBN1_HUMAN | Fibrillin-1 | A key component of microfibrils in the extracellular matrix, fibrillin-1 provides structural support and elasticity to connective tissues. It is crucial for the integrity of tissues such as skin, lungs, and blood vessels. |
| CRISPLD2 | CRLD2_HUMAN | Cysteine-rich secretory protein LCCL domain-containing 2 | This protein is involved in cellular processes such as adhesion and migration. |
| NID1 | CO3A1_HUMAN | Nidogen-1 | Nidogen-1 is a glycoprotein that links laminin and collagen IV in the basement membrane. It plays a critical role in basement membrane stability and cell–matrix interactions. |
| ABI3BP | LTBP4_HUMAN | Target of Nesh-SH3 | This protein interacts with various cytoskeletal and signaling proteins, influencing cell adhesion, migration, and the cytoskeleton’s organization. It is involved in processes such as cell motility and signal transduction. |
Table 5.
Category—collagens (total identified = 27).
Table 5.
Category—collagens (total identified = 27).
| Annotated Gene | Protein Name | Protein Description | Protein Function |
|---|
| COL1A2 | CO1A2_HUMAN | Collagen alpha 2(I) chain | Part of type I collagen, this chain, along with the alpha 1(I) chain, forms type I collagen fibrils. Type I collagen provides tensile strength and structural support to connective tissues such as skin, tendons, and bones. |
| COL1A1 | CO1A1_HUMAN | Collagen alpha 1(I) chain | This is another component of type I collagen, working with the alpha 2(I) chain to form type I collagen fibrils that provide structural integrity and strength to various connective tissues. |
| COL3A1 | CO3A1_HUMAN | Collagen alpha 1(III) chain | Part of type III collagen, this chain combines with the alpha 1(III) chain to form type III collagen, which provides structural support and flexibility to tissues such as skin, blood vessels, and internal organs. |
| COL2A1 | CO2A1_HUMAN | Collagen alpha 1(II) chain | This chain is a component of type II collagen, which is primarily found in cartilage. Type II collagen provides tensile strength and elasticity to cartilage, essential for joint function and support. |
| COL6A3 | CO6A3_HUMAN | Collagen alpha 3(VI) chain | Part of type VI collagen, this chain contributes to the formation of collagen VI fibrils, which are important for anchoring and organizing other matrix proteins and maintaining the structural integrity of tissues such as muscle and skin. |
| COL5A2 | CO5A2_HUMAN | Collagen alpha 2(V) chain | This chain is part of type V collagen, which works with type I and type III collagens to regulate fibril diameter and organization, influencing tissue strength and flexibility. |
| COL17A1 | COHA1_HUMAN | Collagen alpha 1(XVII) chain | A component of type XVII collagen, also known as BP180, which is a key component of hemidesmosomes, important for the adhesion of the epidermis to the underlying dermis, providing stability to the skin. |
| COL7A1 | CO7A1_HUMAN | Collagen alpha 1(VII) chain | This chain is part of type VII collagen, which forms anchoring fibrils that connect the epidermis to the dermis, contributing to skin stability. |
| COL6A1 | CO6A1_HUMAN | Collagen alpha 1(VI) chain | This chain is a component of type VI collagen, which forms microfibrils that support and stabilize the extracellular matrix and connect to other collagen types, influencing tissue integrity and elasticity. |
| COL5A1 | CO5A1_HUMAN | Collagen alpha 1(V) chain | A part of type V collagen, which works with other collagen types to regulate collagen fibril formation and organization, affecting tissue structure and function. |
Table 6.
Category—proteoglycans (total identified = 18).
Table 6.
Category—proteoglycans (total identified = 18).
| Annotated Gene | Protein Name | Protein Description | Protein Function |
|---|
| DCN | PGS2_HUMAN | Decorin | A small leucine-rich proteoglycan involved in collagen fibril formation and matrix organization. It binds to collagen, modulating its assembly and stability, and plays a role in tissue repair and fibrosis. |
| LUM | LUM_HUMAN | Lumican | A member of the small leucine-rich proteoglycan family, lumican is involved in collagen fibril organization and contributes to corneal transparency and tissue repair. It affects the alignment of collagen fibers in connective tissues. |
| OGN | MIME_HUMAN | Mimecan | Also known as osteoglycin, mimecan is a small leucine-rich proteoglycan that regulates collagen fibrillogenesis and contributes to bone and cartilage matrix organization. It plays a role in tissue repair and the structural integrity of connective tissues. |
| PRELP | PRELP_HUMAN | Prolargin | A small leucine-rich proteoglycan that influences collagen fibril formation and tissue repair. It is involved in maintaining the structural integrity of connective tissues by interacting with other matrix proteins. |
| BGN | PGS1_HUMAN | Biglycan | A small leucine-rich proteoglycan involved in regulating collagen fibril formation and matrix organization. It binds to various matrix proteins and growth factors, influencing tissue repair and cellular processes. |
| VCAN | CSPG2_HUMAN | Versican core protein | A large chondroitin sulfate proteoglycan that is important for cell adhesion, migration, and tissue hydration. Versican plays a role in extracellular matrix organization and can influence tissue development and repair. |
| OMD | OMD_HUMAN | Osteomodulin | A small leucine-rich proteoglycan involved in bone matrix organization. It regulates collagen fibril assembly and mineralization, contributing to bone strength and structure. |
| HSPG2 | PGBM_HUMAN | Basement membrane-specific heparan sulfate proteoglycan core protein | A key component of basement membranes, this proteoglycan binds to various matrix proteins and growth factors, influencing cell adhesion, proliferation, and matrix organization. |
| PRG2 | PRG2_HUMAN | Bone marrow proteoglycan | Also known as osteoglycan, it is involved in bone matrix organization and mineralization, playing a role in bone development and repair. |
| ASPN | ASPN_HUMAN | Asporin | A small leucine-rich proteoglycan that modulates collagen fibril formation and influences tissue repair. It is involved in the regulation of matrix organization and cellular interactions within connective tissues. |
Table 7.
Category—ECM regulators (total identified = 135).
Table 7.
Category—ECM regulators (total identified = 135).
| Annotated Gene | Protein Name | Protein Description | Protein Function |
|---|
| CTSD | CATD_HUMAN | Cathepsin D | A lysosomal aspartic protease is involved in the degradation of proteins within lysosomes. It plays a role in various cellular processes, including antigen processing, apoptosis, and tissue remodeling |
| A2ML1 | A2ML1_HUMAN | Alpha-2-macroglobulin-like protein 1 | This protein is part of the alpha-2-macroglobulin family, which functions as a broad-spectrum protease inhibitor. It inhibits a wide range of proteases, playing a role in regulating proteinase activity and modulating inflammatory responses |
| PLG | PLMN_HUMAN | Plasminogen | The precursor of plasmin, a protease involved in fibrinolysis. Plasminogen is converted to plasmin, which breaks down fibrin in blood clots, playing a critical role in the regulation of blood clotting and tissue remodeling. |
| CTSB | CATB_HUMAN | Cathepsin B | A lysosomal cysteine protease is involved in the degradation of proteins and peptides. It participates in various processes including protein turnover, antigen processing, and tissue remodeling. Cathepsin B is also implicated in certain pathological conditions such as cancer. |
| PLOD3 | PLOD3_HUMAN | Multifunctional procollagen lysine hydroxylase and glycosyltransferase LH3 | An enzyme involved in collagen biosynthesis. It performs lysyl hydroxylation and glycosylation of collagen, which are critical for the stability and function of collagen fibers. |
| SERPINH1 | SERPH_HUMAN | Serpin H1 | This protein is a molecular chaperone specific for collagen. It is involved in the proper folding and assembly of collagen molecules in the endoplasmic reticulum. |
| P4HA1 | P4HA1_HUMAN | Prolyl 4-hydroxylase subunit alpha-1 | A component of the prolyl 4-hydroxylase enzyme complex, which is essential for collagen synthesis. It hydroxylates proline residues in collagen, a modification crucial for collagen stability and function. |
| SERPINB2 | PAI2_HUMAN | Plasminogen activator inhibitor 2 | A protein that inhibits tissue plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA), thereby regulating fibrinolysis and modulating tissue remodeling and repair processes. |
| F13A1 | F13A_HUMAN | Coagulation factor XIII A chain | A subunit of factor XIII, which is involved in blood clot stabilization. It crosslinks fibrin polymers, strengthening and stabilizing blood clots to prevent excessive bleeding. |
| PLOD1 | PLOD1_HUMAN | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | An enzyme involved in the hydroxylation of lysine residues in collagen precursors. This modification is important for the formation of stable collagen fibers and proper collagen matrix assembly. |