Optimizing EEG Signal Integrity: A Comprehensive Guide to Ocular Artifact Correction
Abstract
1. Introduction
- The power spectrum of ocular movements overwhelms informative EEG-related features, as their bandwidth (3–15 Hz) overlaps with the frequency range of important neurophysiological contents like the EEG theta and alpha bands. Ocular artifacts can also interfere with time-domain analyses, such as the extraction of Evoked Potentials [3].
- The frequency of ocular artifacts is too high to simply remove affected EEG epochs, with occurrences ranging from 12 to 18 blinks per minute.
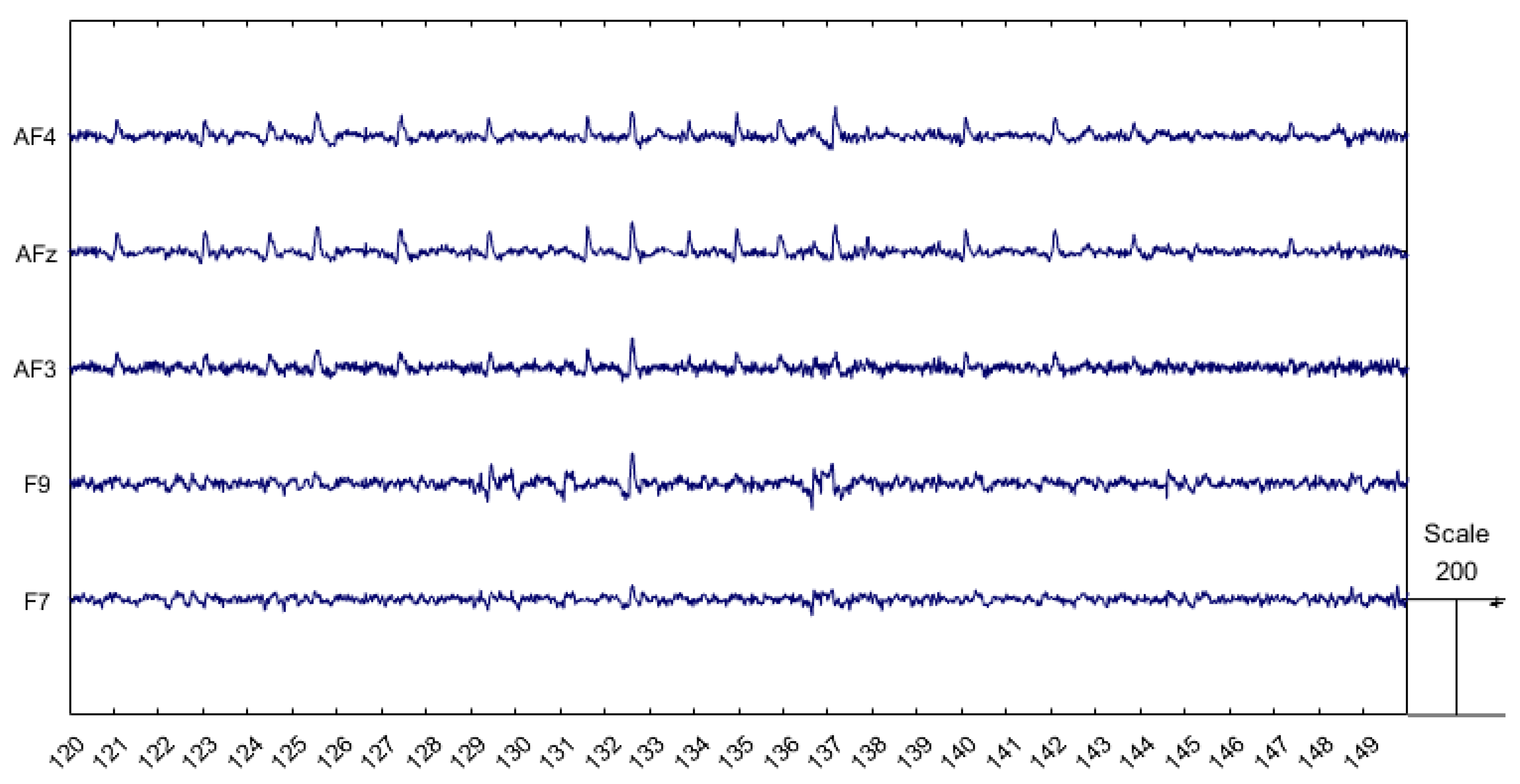
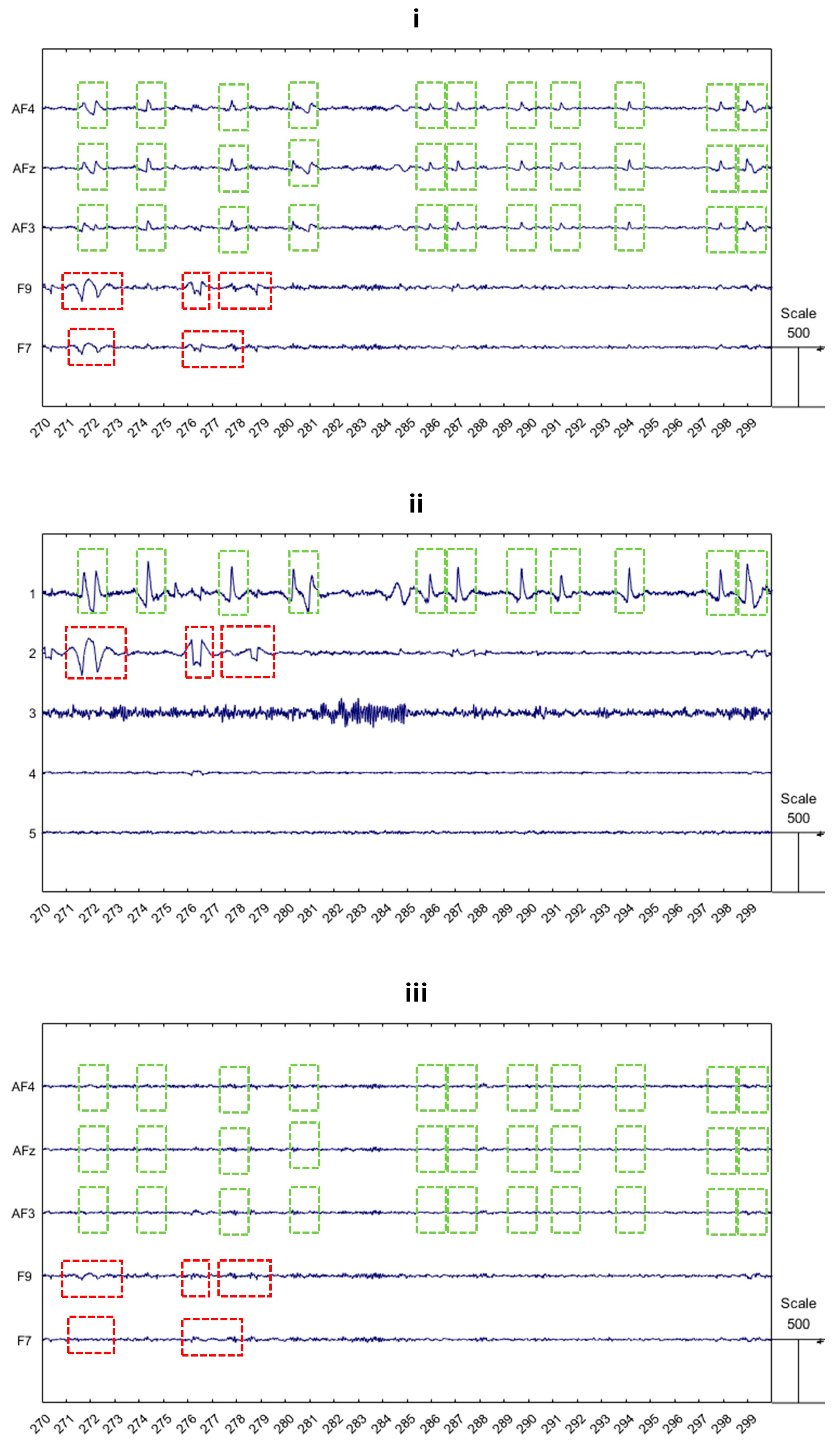
- Ocular artifacts are also characterized by much larger amplitudes compared to normal EEG signals, as shown in Figure 1. This makes them more easily identifiable, allowing expert operators to detect them visually and enabling algorithms to recognize them automatically. However, if not properly addressed, these artifacts can substantially distort the EEG signal, potentially leading to misinterpretations.
2. General Framework
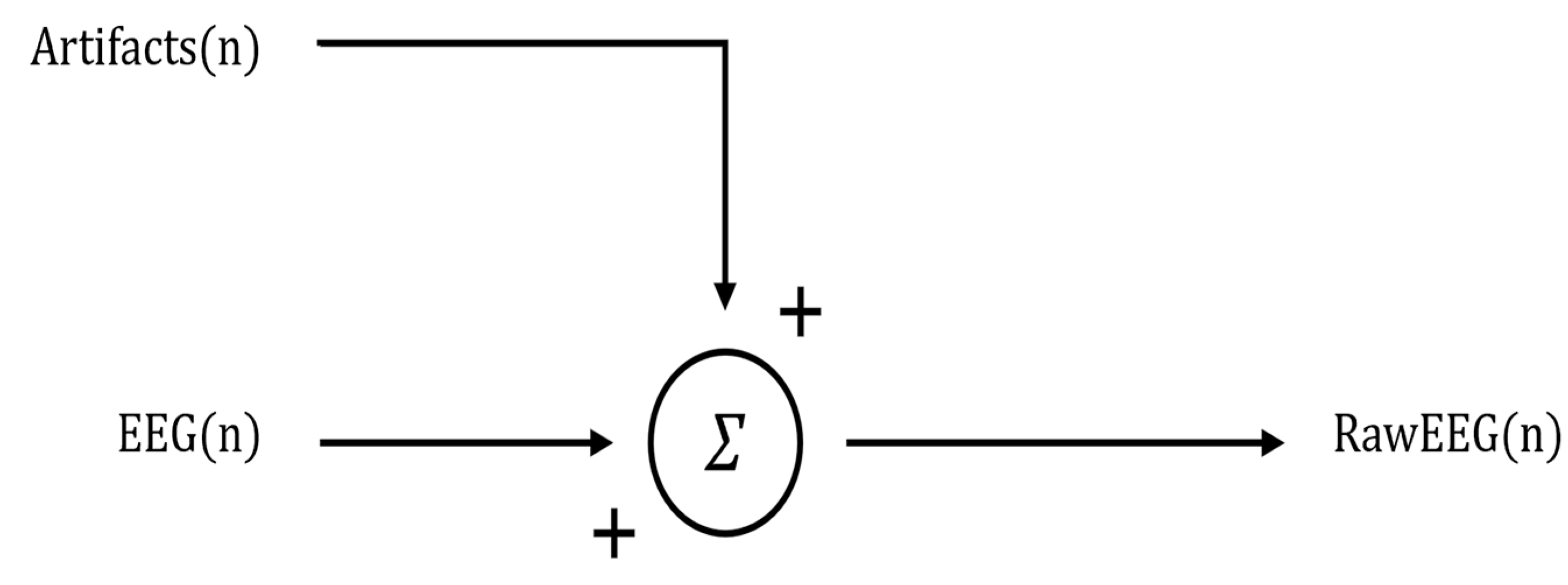
- Regression-based methods: These correspond to an approach requiring an ocular blink template, which is specific to each subject. The several regression-based methods proposed in the scientific literature rely on electrooculography (EOG) or, in case of unavailability of the EOG channel, on frontal, prefrontal, and anterofrontal EEG channels as ocular blink templates. Such methods generally require a calibration run specifically corresponding to the ocular blink template collection. Within the state of the art, these methods can be divided into linear regression-based methods, i.e., methods which foresee the modeling and subtraction of the ocular blinks’ contribution using the ocular blink template as a regressor, and adaptive filtering-based methods, i.e., methods which foresee the adjustment of dynamic model parameters for a better fit with the EEG signal to be corrected.
- Independent Component Analysis (ICA): This includes a wide range of methodologies based on the decomposition of the EEG signal to its independent components. Therefore, this approach foresees independent component analysis by identifying the ones associated with the ocular blinks and removing them.
- Artifact Subspace Reconstruction (ASR): This is an advanced technique that operates by detecting and reconstructing the subspace of the EEG data contaminated by artifacts. This method leverages the statistical properties of the EEG signal to differentiate between neural activity and artifacts. By identifying the subspace where artifacts dominate, ASR can reconstruct the clean EEG signal by removing the contributions from this subspace.
3. Methods and Principles
3.1. Regression-Based Methods
- The rawEEG signal is typically filtered to pass only interesting frequencies (for example, between 1 and 50 Hz) to eliminate slow fluctuations and high-frequency disturbances in order to remove artifacts whose frequencies do not overlap with the EEG spectrum.
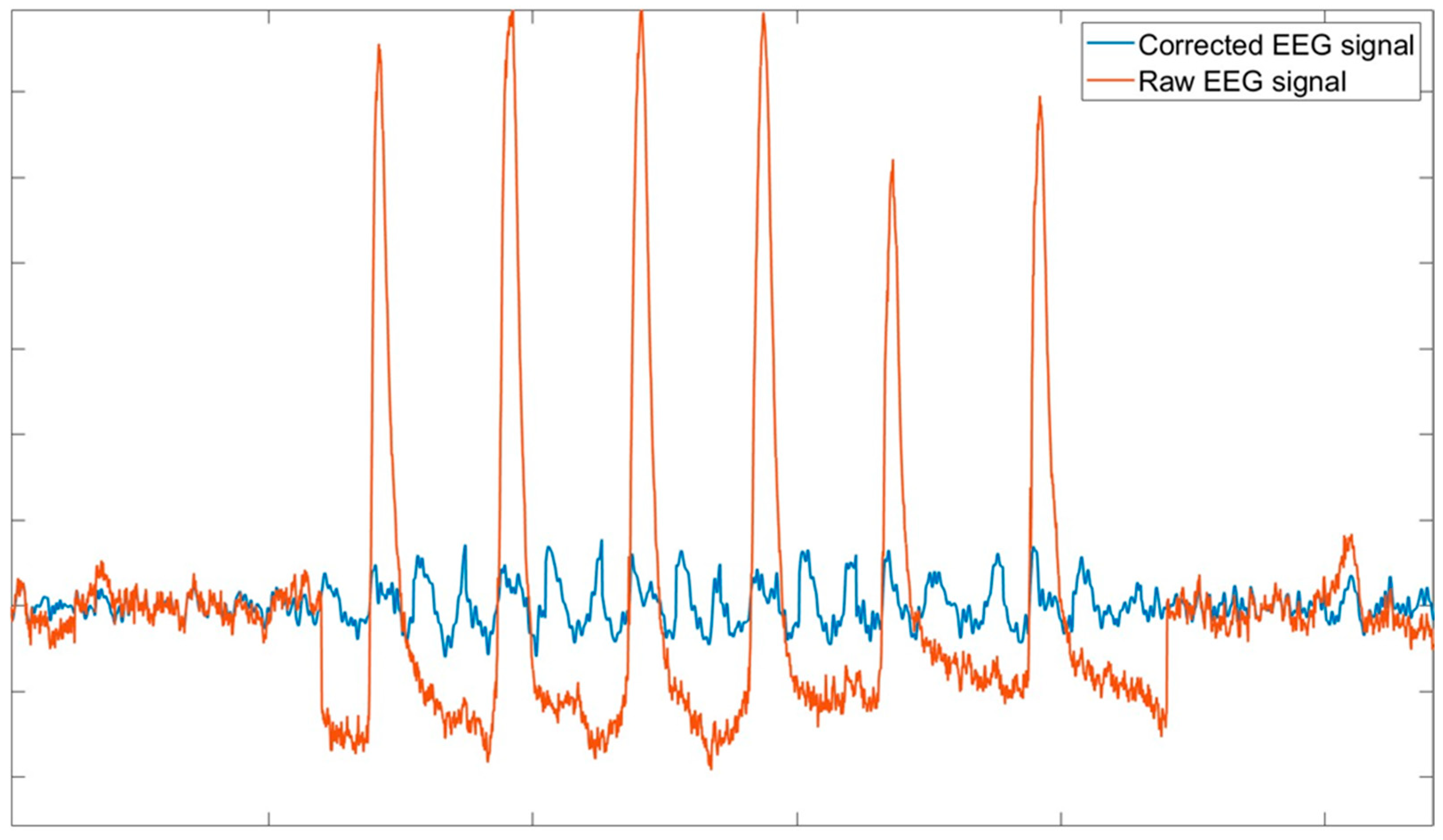
- The EOG signal is low-pass filtered (cut-off frequency 15 Hz) to eliminate high-frequency ailments and to increase method accuracy, since it has been demonstrated that the main spectral content of ocular blinks is up to 15 Hz (Figure 3).
- Temporal alignment and segmentation of EEG and EOG signals is conducted, which constitute a preliminary step necessary in order to accurately correct ocular artifact contributions in EEG signals.
- coefficient estimation is the most important step of the regression algorithm. In fact, once these coefficients are estimated, the algorithm can be considered calibrated and the EEG signal can also be correct in real time. This step is composed of three sub-steps:
- The raw EEG and raw EOG signals are firstly averaged across epochs; these averages represent the “signal baseline” [17].
- The averages evaluated in the previous step are then subtracted from each epoch of the EEG and EOG signals, respectively. After this subtraction, the resulting signals represent the “deviation from the baseline”.
- Then, the “deviation from the baseline” signals serve as variables for the correlation analysis. The correlation is computed considering the EOG as the independent variable and the EEG as the dependent variable. Finally, the correlation coefficient is the estimation of (Figure 4).
3.2. Independent Component Analysis (ICA)
- The raw EEG signal is often band-pass filtered (e.g., 1–50 Hz) to remove slow drifts and high-frequency noise. This step ensures that the signal primarily contains frequencies of interest, removing artifacts that do not overlap with the EEG spectrum.
- The EEG signal can be optionally segmented in epochs of a specific time duration. This step is especially indicated in the case of time-locked EEG feature computation (e.g., event-related potentials).
- The step corresponding to the independent components analysis is the most relevant within the ICA application. In fact, the independent components of the EEG signal must be analyzed for identifying those that correspond to the ocular blink contribution. In this regard, two approaches have been extensively observed and validated by previous scientific works:
- The components are visually inspected for the purpose of identifying which of them are related to the ocular blink artifacts. For this approach, an expert operator is required to perform the signal processing, who must be able to correctly recognize visual ocular blink patterns.
- Automated methods can also be used, such as correlation with electrooculogram (EOG) signals, kurtosis, or power spectral density analysis [24].
- Once the ocular blink components are identified, such components must be removed.
3.3. Artifact Subspace Reconstruction (ASR)
- The raw EEG signal is preliminary band-pass filtered to remove both high-frequency and low-frequency noise (1–50 Hz). This step ensures that the signal mainly includes only the artifacts that overlap the EEG spectrum in order to improve ASR accuracy.
- The ASR algorithm is calibrated using the reference data, which should be free from artifacts. As presented before, this step may differ depending on which version of the algorithm is chosen.
- For online applications, it is essential to acquire reference data before the experiment during a calibration run, which typically consists of 1–2 min of EEG recording with eyes closed. Although 1–2 min is the recommended duration for optimal algorithm calibration, even 30 s can be sufficient. It is crucial that the algorithm is calibrated for each subject using their respective calibration run to ensure accuracy and effectiveness.
- For offline applications, ASR can automatically extract artifact-free portions from the EEG signal (i.e., not from the calibration run) and concatenate them to create 1–2 min of “calibration” data.
- In order to determine the threshold, ASR firstly computes the mixing matrix from the calibration data. Next, the matrix is obtained using Singular Value Decomposition (SVD) from the mixing matrix. Then, the reference EEG data are projected into the principal component space using Formula 4. The principal components (Y) are segmented into 0.5 s windows, and the mean (μ) and standard deviation (σ) are evaluated across the windows for each component. Finally, the threshold is defined for each i-th component. As discussed above, the usual value of parameter k is between 10 and 30.
3.4. Deep Learning-Based Algorithms
- Offline step. The offline step consists of two sub-steps, focusing on extracting the training dataset from the EEG signal. Once the training dataset is created, the model is trained accordingly.
- Normalizing data and building the training dataset. In this step, the EEG signal is normalized, and the training dataset is built from the raw EEG data. Artifactual samples are removed using statistical thresholds, resulting in a clean EEG dataset without artifacts.
- Training the chosen deep learning model on the clean data. After this step, the model will need to tune its own parameters and will be capable of recognizing features attributed to uncontaminated EEG.
- Online step. The online step is the real cleaning process of the algorithm. In this step, the EEG signal is normalized, and the training dataset is built from the raw EEG data. Artifactual samples are removed using statistical thresholds, resulting in a clean EEG dataset without artifacts.
4. Discussion
4.1. Regression-Based Methods
- Pros: Regression-based methods are simple and effective when an external template, such as an EOG channel, is available. They can be implemented easily with a small number of EEG channels and are particularly useful for real-time applications.
- Cons: The main limitation is that they rely heavily on the quality of the template. If the template is noisy or not perfectly aligned with the artifact in the EEG, the correction may be inaccurate. Additionally, these methods might not fully eliminate artifacts, especially when the EOG signal is strongly correlated with the EEG.
- Best use cases: These methods are most effective in controlled laboratory environments where EOG recordings are available, and the primary concern is the removal of blink artifacts with minimal computational complexity. Additionally, the evolutions of such methods, such as the one provided by Reblinca [16], could be indicated for out-of-the-lab application [35,36,37,38,39], where EEG data collection from the frontal or anterofrontal channels is possible.
4.2. Independent Component Analysis (ICA)
- Pros: ICA is a powerful technique for decomposing EEG signals into independent sources, allowing for precise identification and removal of ocular artifacts without needing additional channels. It is particularly effective in separating overlapping artifacts and neural activity.
- Cons: ICA requires a relatively large number of EEG channels to be effective, and its success depends on the quality of the decomposition. It also assumes that the sources are statistically independent, which may not always hold true. Additionally, it is computationally intensive and not ideal for real-time processing.
- Best use cases: ICA is best suited for offline analysis in studies with high-density EEG setups, where the goal is to achieve a clean separation of neural and artifact signals for in-depth analysis. In terms of experimental settings, it appears to be clear that the ICA-based techniques are the most indicated when the EEG data collection is performed in laboratory settings [40,41,42,43,44].
4.3. Artifact Subspace Reconstruction (ASR)
- Pros: ASR is highly effective at removing a wide range of artifacts by reconstructing the EEG signal from a subspace that excludes the contaminated components. It is adaptive and can be used both online and offline, making it versatile.
- Cons: ASR’s effectiveness depends on the quality of the initial calibration data. Poor calibration can lead to overcorrection, where some neural signals might be mistakenly removed. It also requires more computational resources compared to simpler methods, like regression.
4.4. Deep Learning-Based Algorithms
- Pros: Deep learning models, particularly Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs), can outperform traditional methods like ICA and regression-based techniques in terms of accuracy. These models can automatically learn intricate, non-linear patterns in EEG signals, which allows them to separate ocular artifacts from neural activity with higher precision. Such methods do not need any manual and/or visual intervention, like ICA, and they are robust in terms of handling large and complex datasets, making them ideal for high-density EEG data. Furthermore, such models can generalize well to new and unseen datasets. This makes these methods highly adaptable across different individuals, experimental conditions, and EEG system configurations.
- Cons: One of the possible limitations of deep learning-based methods consists of the need for large, annotated training datasets to achieve high performance. Moreover, such models are computationally expensive, especially during the training phase. A further limitation to consider corresponds to the overfitting risk, which could occur if the training dataset is not large or diverse enough. Finally, one of the major concerns with deep learning models is the “black box” nature of neural networks. Unlike traditional methods like ICA, which offer interpretable components corresponding to underlying brain processes, deep learning models do not easily offer insights into how the correction is being performed.
- Best use cases: As suggested by the positive aspects associated with these methods, deep learning approaches are ideal for scenarios where large-scale datasets with high-density EEG are available [49,50,51,52]. Therefore, they could be implemented for real-time artifact correction in BCIs [53,54,55,56]. In these cases, the need for immediate feedback requires robust artifact detection and removal, which deep learning methods can provide, especially when artifact patterns are non-linear or difficult to model with traditional methods. In parallel, these approaches are particularly indicated when dealing with EEG data collected through wearable and mobile systems. As this kind of equipment becomes more prevalent for in-field research, deep learning methods offer the potential for on-device, real-time processing.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aricò, P.; Reynal, M.; Di Flumeri, G.; Borghini, G.; Sciaraffa, N.; Imbert, J.-P.; Hurter, C.; Terenzi, M.; Ferreira, A.; Pozzi, S.; et al. How Neurophysiological Measures Can be Used to Enhance the Evaluation of Remote Tower Solutions. Front. Hum. Neurosci. 2019, 13, 303. [Google Scholar] [CrossRef] [PubMed]
- Capotorto, R.; Ronca, V.; Sciaraffa, N.; Borghini, G.; Di Flumeri, G.; Mezzadri, L.; Vozzi, A.; Giorgi, A.; Germano, D.; Babiloni, F.; et al. Cooperation objective evaluation in aviation: Validation and comparison of two novel approaches in simulated environment. Front. Neurosci. 2024, 18, 1409322. [Google Scholar] [CrossRef] [PubMed]
- Aloise, F.; Aricò, P.; Schettini, F.; Salinari, S.; Mattia, D.; Cincotti, F. Asynchronous gaze-independent event-related potential-based brain-computer interface. Artif. Intell. Med. 2013, 59, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, M.A.; Kizilisik, S.; Guren, O. Removal of Ocular Artifacts in EEG Using Deep Learning. In Proceedings of the 2022 Medical Technologies Congress (TIPTEKNO), Antalya, Turkey, 31 October–2 November 2022. [Google Scholar] [CrossRef]
- Yang, B.; Duan, K.; Fan, C.; Hu, C.; Wang, J. Automatic ocular artifacts removal in EEG using deep learning. Biomed. Signal Process. Control 2018, 43, 148–158. [Google Scholar] [CrossRef]
- Mashhadi, N.; Khuzani, A.Z.; Heidari, M.; Khaledyan, D. Deep learning denoising for EOG artifacts removal from EEG signals. In Proceedings of the 2020 IEEE Global Humanitarian Technology Conference, GHTC 2020, Seattle, WA, USA, 29 October–1 November 2020. [Google Scholar] [CrossRef]
- Jiang, X.; Bian, G.B.; Tian, Z. Removal of Artifacts from EEG Signals: A Review. Sensors 2019, 19, 987. [Google Scholar] [CrossRef]
- Hillyard, S.A.; Galambos, R. Eye movement artifact in the CNV. Electroencephalogr. Clin. Neurophysiol. 1970, 28, 173–182. [Google Scholar] [CrossRef]
- Romero, S.; Mañanas, M.A.; Barbanoj, M.J. A comparative study of automatic techniques for ocular artifact reduction in spontaneous EEG signals based on clinical target variables: A simulation case. Comput. Biol. Med. 2008, 38, 348–360. [Google Scholar] [CrossRef]
- Sweeney, K.T.; Ward, T.E.; McLoone, S.F. Artifact removal in physiological signals--practices and possibilities. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 488–500. [Google Scholar] [CrossRef]
- Corby, J.C.; Kopell, B.S. Differential Contributions of Blinks and Vertical Eye Movements as Artifacts in EEG Recording. Psychophysiology 1972, 9, 640–644. [Google Scholar] [CrossRef]
- Gratton, G.; Coles, M.G.H.; Donchin, E. A new method for off-line removal of ocular artifact. Electroencephalogr. Clin. Neurophysiol. 1983, 55, 468–484. [Google Scholar] [CrossRef]
- Joyce, C.A.; Gorodnitsky, I.F.; Kutas, M. Automatic removal of eye movement and blink artifacts from EEG data using blind component separation. Psychophysiology 2004, 41, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Kenemans, J.L.; Molenaar, P.C.M.; Verbaten, M.N.; Slangen, J.L. Removal of the Ocular Artifact from the EEG: A Comparison of Time and Frequency Domain Methods with Simulated and Real Data. Psychophysiology 1991, 28, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Woestenburg, J.C.; Verbaten, M.N.; Slangen, J.L. The removal of the eye-movement artifact from the EEG by regression analysis in the frequency domain. Biol. Psychol. 1983, 16, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Di Flumeri, G.; Arico, P.; Borghini, G.; Colosimo, A.; Babiloni, F. A new regression-based method for the eye blinks artifacts correction in the EEG signal, without using any EOG channel. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Orlando, FL, USA, 16–20 August 2016; pp. 3187–3190. [Google Scholar] [CrossRef]
- Kumaravel, V.P.; Kartsch, V.; Benatti, S.; Vallortigara, G.; Farella, E.; Buiatti, M. Efficient Artifact Removal from Low-Density Wearable EEG using Artifacts Subspace Reconstruction. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico City, Mexico, 1–5 November 2021; Volume 2021, p. 333336. [Google Scholar] [CrossRef]
- mne.preprocessing.EOGRegression—MNE 1.9.0.dev32+g670330a1e Documentation. Available online: https://mne.tools/dev/generated/mne.preprocessing.EOGRegression.html#mne.preprocessing.EOGRegression (accessed on 16 September 2024).
- Ronca, V.; Di Flumeri, G.; Giorgi, A.; Vozzi, A.; Capotorto, R.; Germano, D.; Sciaraffa, N.; Borghini, G.; Babiloni, F.; Aricò, P.; et al. o-CLEAN: A novel multi-stage algorithm for the ocular artifacts’ correction from EEG data in out-of-the-lab applications. J. Neural Eng. 2024, 21, 056023. [Google Scholar] [CrossRef]
- Somers, B.; Francart, T.; Bertrand, A. A generic EEG artifact removal algorithm based on the multi-channel Wiener filter. J. Neural Eng. 2018, 15, 036007. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.A.; Kreutz-Delgado, K.; Makeig, S. Super-Gaussian Mixture Source Model for ICA. In Lecture Notes in Computer Science, Proceedings of the 6th International Conference, ICA 2006, Charleston, SC, USA, 5–8 March 2006; Springer: Berlin/Heidelberg, Germany, 2006; Volume 3889, pp. 854–861. [Google Scholar] [CrossRef]
- Artoni, F.; Delorme, A.; Makeig, S. Applying dimension reduction to EEG data by Principal Component Analysis reduces the quality of its subsequent Independent Component decomposition. NeuroImage 2018, 175, 176–187. [Google Scholar] [CrossRef]
- Delorme, A.; Makeig, S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Oja, E. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Onton, J.; Westerfield, M.; Townsend, J.; Makeig, S. Imaging human EEG dynamics using independent component analysis. Neurosci. Biobehav. Rev. 2006, 30, 808–822. [Google Scholar] [CrossRef]
- Miyakoshi, M. Artifact subspace reconstruction: A candidate for a dream solution for EEG studies, sleep or awake. Sleep 2023, 46, zsad241. [Google Scholar] [CrossRef]
- Chang, C.Y.; Hsu, S.H.; Pion-Tonachini, L.; Jung, T.P. Evaluation of Artifact Subspace Reconstruction for Automatic Artifact Components Removal in Multi-Channel EEG Recordings. IEEE Trans. Biomed. Eng. 2020, 67, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Anders, P.; Müller, H.; Skjæret-Maroni, N.; Vereijken, B.; Baumeister, J. The influence of motor tasks and cut-off parameter selection on artifact subspace reconstruction in EEG recordings. Med. Biol. Eng. Comput. 2020, 58, 2673–2683. [Google Scholar] [CrossRef]
- Kothe, C.A.; Makeig, S. BCILAB: A platform for brain-computer interface development. J. Neural Eng. 2013, 10, 056014. [Google Scholar] [CrossRef]
- DiGyt/asrpy: Artifact Subspace Reconstruction for Python. Available online: https://github.com/DiGyt/asrpy (accessed on 16 September 2024).
- Blum, S.; Jacobsen, N.S.J.; Bleichner, M.G.; Debener, S. A riemannian modification of artifact subspace reconstruction for EEG artifact handling. Front. Hum. Neurosci. 2019, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, M.; Wei, C.; Mantini, D.; Li, Z.; Liu, Q. EEGdenoiseNet: A benchmark dataset for deep learning solutions of EEG denoising. J. Neural Eng. 2021, 18, 056057. [Google Scholar] [CrossRef]
- Sawangjai, P.; Trakulruangroj, M.; Boonnag, C.; Piriyajitakonkij, M.; Tripathy, R.K.; Sudhawiyangkul, T.; Wilaiprasitporn, T. EEGANet: Removal of Ocular Artifacts From the EEG Signal Using Generative Adversarial Networks. IEEE J. Biomed. Health Inform. 2022, 26, 4913–4924. [Google Scholar] [CrossRef]
- Ronca, V.; Di Flumeri, G.; Vozzi, A.; Giorgi, A.; Arico, P.; Sciaraffa, N.; Babiloni, F.; Borghini, G. Validation of an EEG-based Neurometric for online monitoring and detection of mental drowsiness while driving. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; Volume 2022, pp. 3714–3717. [Google Scholar] [CrossRef]
- Di Flumeri, G.; Ronca, V.; Giorgi, A.; Vozzi, A.; Aricò, P.; Sciaraffa, N.; Zeng, H.; Dai, G.; Kong, W.; Babiloni, F.; et al. EEG-Based Index for Timely Detecting User’s Drowsiness Occurrence in Automotive Applications. Front. Hum. Neurosci. 2022, 16, 866118. [Google Scholar] [CrossRef] [PubMed]
- Ronca, V.; Brambati, F.; Napoletano, L.; Marx, C.; Trösterer, S.; Vozzi, A.; Aricò, P.; Giorgi, A.; Capotorto, R.; Borghini, G.; et al. A Novel EEG-Based Assessment of Distraction in Simulated Driving under Different Road and Traffic Conditions. Brain Sci. 2024, 14, 193. [Google Scholar] [CrossRef]
- Ronca, V.; Uflaz, E.; Turan, O.; Bantan, H.; MacKinnon, S.N.; Lommi, A.; Pozzi, S.; Kurt, R.E.; Arslan, O.; Kurt, Y.B.; et al. Neurophysiological Assessment of An Innovative Maritime Safety System in Terms of Ship Operators’ Mental Workload, Stress, and Attention in the Full Mission Bridge Simulator. Brain Sci. 2023, 13, 1319. [Google Scholar] [CrossRef]
- Di Flumeri, G.; Giorgi, A.; Germano, D.; Ronca, V.; Vozzi, A.; Borghini, G.; Tamborra, L.; Simonetti, I.; Capotorto, R.; Ferrara, S.; et al. A Neuroergonomic Approach Fostered by Wearable EEG for the Multimodal Assessment of Drivers Trainees. Sensors 2023, 23, 8389. [Google Scholar] [CrossRef]
- Borghini, G.; Ronca, V.; Vozzi, A.; Aricò, P.; Di Flumeri, G.; Babiloni, F. Monitoring performance of professional and occupational operators. In Handbook of Clinical Neurology; Elsevier B.V.: Amsterdam, The Netherlands, 2020; Volume 168, pp. 199–205. [Google Scholar] [CrossRef]
- Inguscio, B.M.S.; Cartocci, G.; Sciaraffa, N.; Nicastri, M.; Giallini, I.; Greco, A.; Babiloni, F.; Mancini, P. Gamma-Band Modulation in Parietal Area as the Electroencephalographic Signature for Performance in Auditory-Verbal Working Memory: An Exploratory Pilot Study in Hearing and Unilateral Cochlear Implant Children. Brain Sci. 2022, 12, 1291. [Google Scholar] [CrossRef] [PubMed]
- Cartocci, G.; Inguscio, B.M.S.; Giliberto, G.; Vozzi, A.; Giorgi, A.; Greco, A.; Babiloni, F.; Attanasio, G. Listening Effort in Tinnitus: A Pilot Study Employing a Light EEG Headset and Skin Conductance Assessment during the Listening to a Continuous Speech Stimulus under Different SNR Conditions. Brain Sci. 2023, 13, 1084. [Google Scholar] [CrossRef] [PubMed]
- Giambra, L.M. Task-unrelated-thought frequency as a function of age: A laboratory study. Psychol. Aging 1989, 4, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, M.; Di Flumeri, G.; Aricò, P.; Sciaraffa, N.; Babiloni, F.; Borghini, G. Neurophysiological Vigilance Characterisation and Assessment: Laboratory and Realistic Validations Involving Professional Air Traffic Controllers. Brain Sci. 2020, 10, 48. [Google Scholar] [CrossRef]
- Di Flumeri, G.; Arico, P.; Borghini, G.; Sciaraffa, N.; Maglione, A.G.; Rossi, D.; Modica, E.; Trettel, A.; Babiloni, F.; Colosimo, A.; et al. EEG-based Approach-Withdrawal index for the pleasantness evaluation during taste experience in realistic settings. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Jeju Island, Republic of Korea, 11–15 July2017; pp. 3228–3231. [Google Scholar] [CrossRef]
- Giorgi, A.; Ronca, V.; Vozzi, A.; Aricò, P.; Borghini, G.; Capotorto, R.; Tamborra, L.; Simonetti, I.; Sportiello, S.; Petrelli, M.; et al. Neurophysiological mental fatigue assessment for developing user-centered Artificial Intelligence as a solution for autonomous driving. Front. Neurorobot. 2023, 17, 1240933. [Google Scholar] [CrossRef]
- Gargiulo, G.; Bifulco, P.; Calvo, R.A.; Cesarelli, M.; Jin, C.; Van Schaik, A. A mobile EEG system with dry electrodes. In Proceedings of the 2008 IEEE-BIOCAS Biomedical Circuits and Systems Conference, BIOCAS 2008, Baltimore, MD, USA, 20–22 November 2008; pp. 273–276. [Google Scholar] [CrossRef]
- Chi, Y.M.; Wang, Y.T.; Wang, Y.; Maier, C.; Jung, T.P.; Cauwenberghs, G. Dry and Noncontact EEG Sensors for Mobile Brain-Computer Interfaces. IEEE Trans. Neural Syst. Rehabilitation Eng. 2011, 20, 228–235. [Google Scholar] [CrossRef]
- Borghini, G.; Aricò, P.; Di Flumeri, G.; Sciaraffa, N.; Colosimo, A.; Herrero, M.-T.; Bezerianos, A.; Thakor, N.V.; Babiloni, F. A new perspective for the training assessment: Machine learning-based neurometric for augmented user’s evaluation. Front. Neurosci. 2017, 11, 325. [Google Scholar] [CrossRef]
- Sciaraffa, N.; Liu, J.; Aricò, P.; Di Flumeri, G.; Inguscio, B.M.S.; Borghini, G.; Babiloni, F. Multivariate model for cooperation: Bridging social physiological compliance and hyperscanning. Soc. Cogn. Affect. Neurosci. 2021, 16, 193–209. [Google Scholar] [CrossRef]
- Toppi, J.; Borghini, G.; Petti, M.; He, E.J.; De Giusti, V.; He, B.; Astolfi, L.; Babiloni, F. Investigating Cooperative Behavior in Ecological Settings: An EEG Hyperscanning Study. PLoS ONE 2016, 11, e0154236. [Google Scholar] [CrossRef]
- Koike, T.; Tanabe, H.C.; Sadato, N. Hyperscanning neuroimaging technique to reveal the ‘two-in-one’ system in social interactions. Neurosci. Res. 2015, 90, 25–32. [Google Scholar] [CrossRef]
- Arico, P.; Borghini, G.; Di Flumeri, G.; Sciaraffa, N.; Babiloni, F. Passive BCI beyond the lab: Current trends and future directions. Physiol. Meas. 2018, 39, 08TR02. [Google Scholar] [CrossRef] [PubMed]
- Douibi, K.; Le Bars, S.; Lemontey, A.; Nag, L.; Balp, R.; Breda, G. Toward EEG-Based BCI Applications for Industry 4.0: Challenges and Possible Applications. Front. Hum. Neurosci. 2021, 15, 705064. [Google Scholar] [CrossRef] [PubMed]
- Belo, J.; Clerc, M.; Schön, D. EEG-Based Auditory Attention Detection and Its Possible Future Applications for Passive BCI. Front. Comput. Sci. 2021, 3, 661178. [Google Scholar] [CrossRef]
- Grozea, C.; Voinescu, C.D.; Fazli, S. Bristle-sensors—Low-cost flexible passive dry EEG electrodes for neurofeedback and BCI applications. J. Neural Eng. 2011, 8, 025008. [Google Scholar] [CrossRef]
- Vecchiato, G.; Borghini, G.; Aricò, P.; Graziani, I.; Maglione, A.G.; Cherubino, P.; Babiloni, F. Investigation of the effect of EEG-BCI on the simultaneous execution of flight simulation and attentional tasks. Med. Biol. Eng. Comput. 2016, 54, 1503–1513. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ronca, V.; Capotorto, R.; Di Flumeri, G.; Giorgi, A.; Vozzi, A.; Germano, D.; Virgilio, V.D.; Borghini, G.; Cartocci, G.; Rossi, D.; et al. Optimizing EEG Signal Integrity: A Comprehensive Guide to Ocular Artifact Correction. Bioengineering 2024, 11, 1018. https://doi.org/10.3390/bioengineering11101018
Ronca V, Capotorto R, Di Flumeri G, Giorgi A, Vozzi A, Germano D, Virgilio VD, Borghini G, Cartocci G, Rossi D, et al. Optimizing EEG Signal Integrity: A Comprehensive Guide to Ocular Artifact Correction. Bioengineering. 2024; 11(10):1018. https://doi.org/10.3390/bioengineering11101018
Chicago/Turabian StyleRonca, Vincenzo, Rossella Capotorto, Gianluca Di Flumeri, Andrea Giorgi, Alessia Vozzi, Daniele Germano, Valerio Di Virgilio, Gianluca Borghini, Giulia Cartocci, Dario Rossi, and et al. 2024. "Optimizing EEG Signal Integrity: A Comprehensive Guide to Ocular Artifact Correction" Bioengineering 11, no. 10: 1018. https://doi.org/10.3390/bioengineering11101018
APA StyleRonca, V., Capotorto, R., Di Flumeri, G., Giorgi, A., Vozzi, A., Germano, D., Virgilio, V. D., Borghini, G., Cartocci, G., Rossi, D., Inguscio, B. M. S., Babiloni, F., & Aricò, P. (2024). Optimizing EEG Signal Integrity: A Comprehensive Guide to Ocular Artifact Correction. Bioengineering, 11(10), 1018. https://doi.org/10.3390/bioengineering11101018











