An AI-Enabled Bias-Free Respiratory Disease Diagnosis Model Using Cough Audio
Abstract
1. Introduction
- In contrast to the majority of previous studies that rely on crowd-sourced cough audio databases for training AI models, this study curated a cough data set containing COVID-19 infection status. For each participant, the curated data set includes cough recordings tagged with reliable RT-PCR information and collected in a clinical setting. Hence, the data set used has extremely reliable ground truth labels, resulting in the accurate training of RBF-Net.
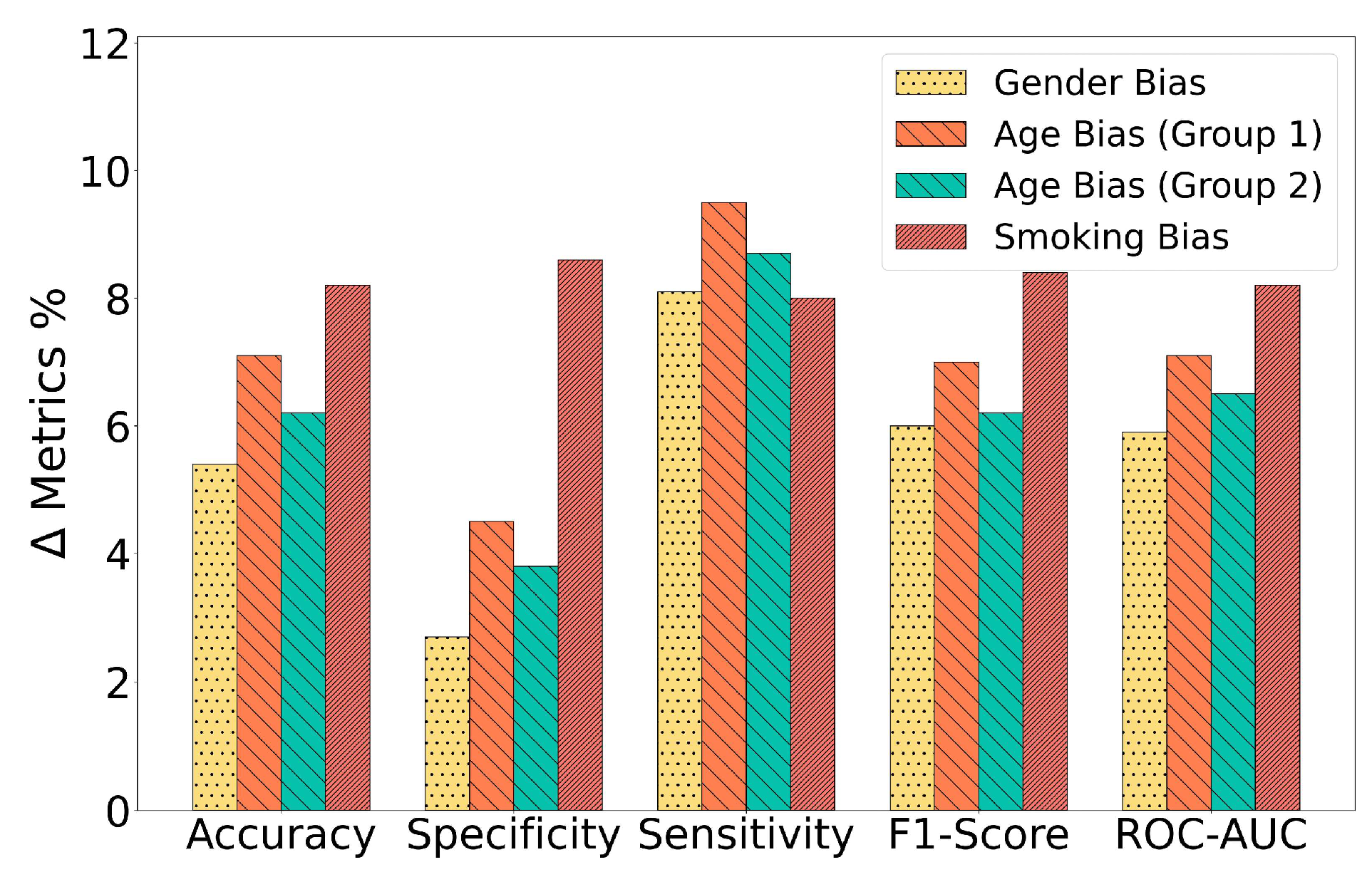
- To demonstrate the impact of confounding variables, we train a SoTA DL model on different splits of biased training scenarios from the cough data set based on gender, age, and smoking status. Moreover, we present an insightful analysis on how model performances are often overestimated due to the underlying biased distribution of the training data and the use of cross-validation technique.
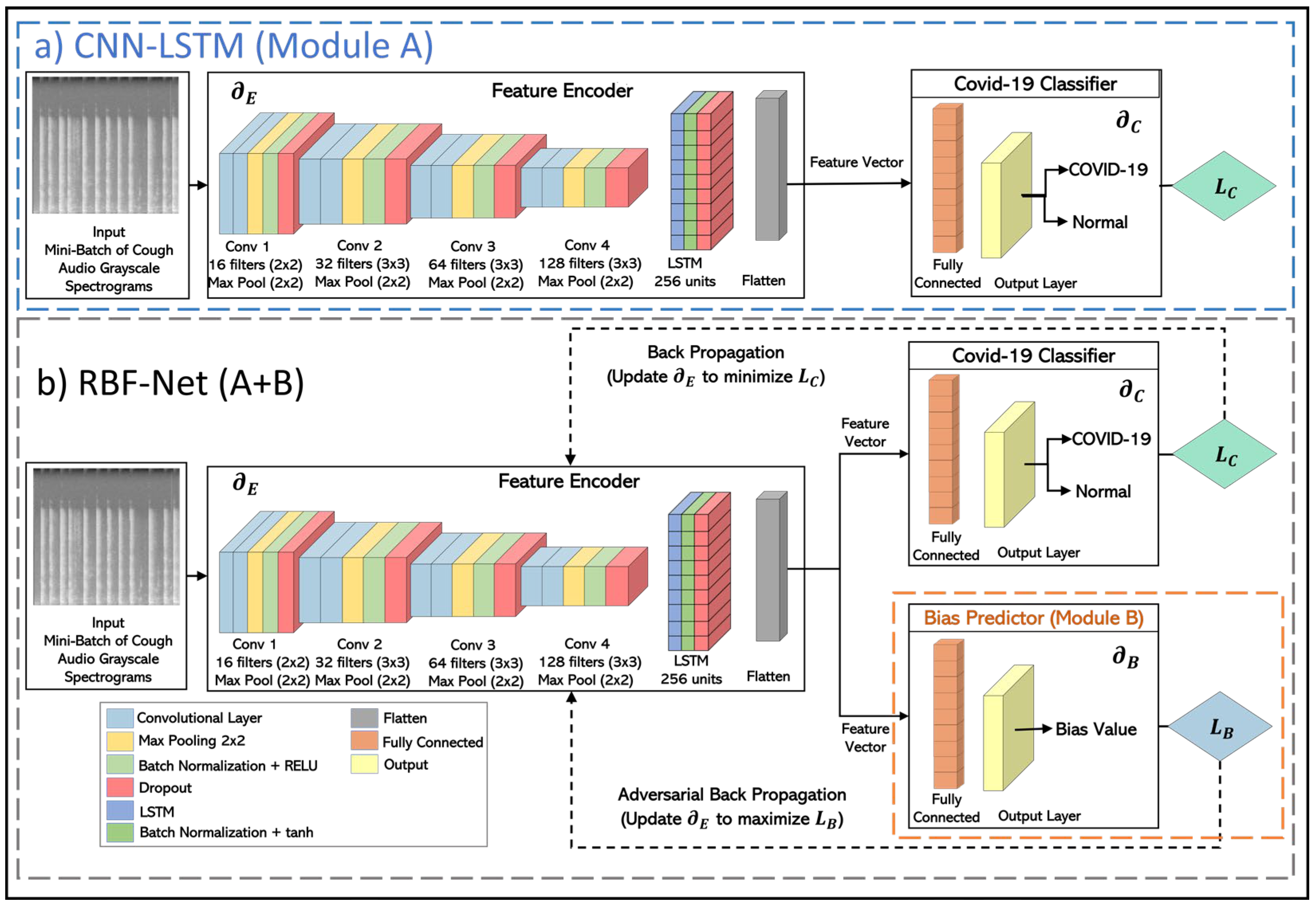
- To overcome the impact of biases, we present an RBF-Net that learns features from cough recordings that are impacted by COVID-19. We perform a comparative analysis of the existing SoTA CNN-LSTM model with RBF-Net and demonstrate the improvement achieved by the proposed RBF-Net in terms of different performance metrics.
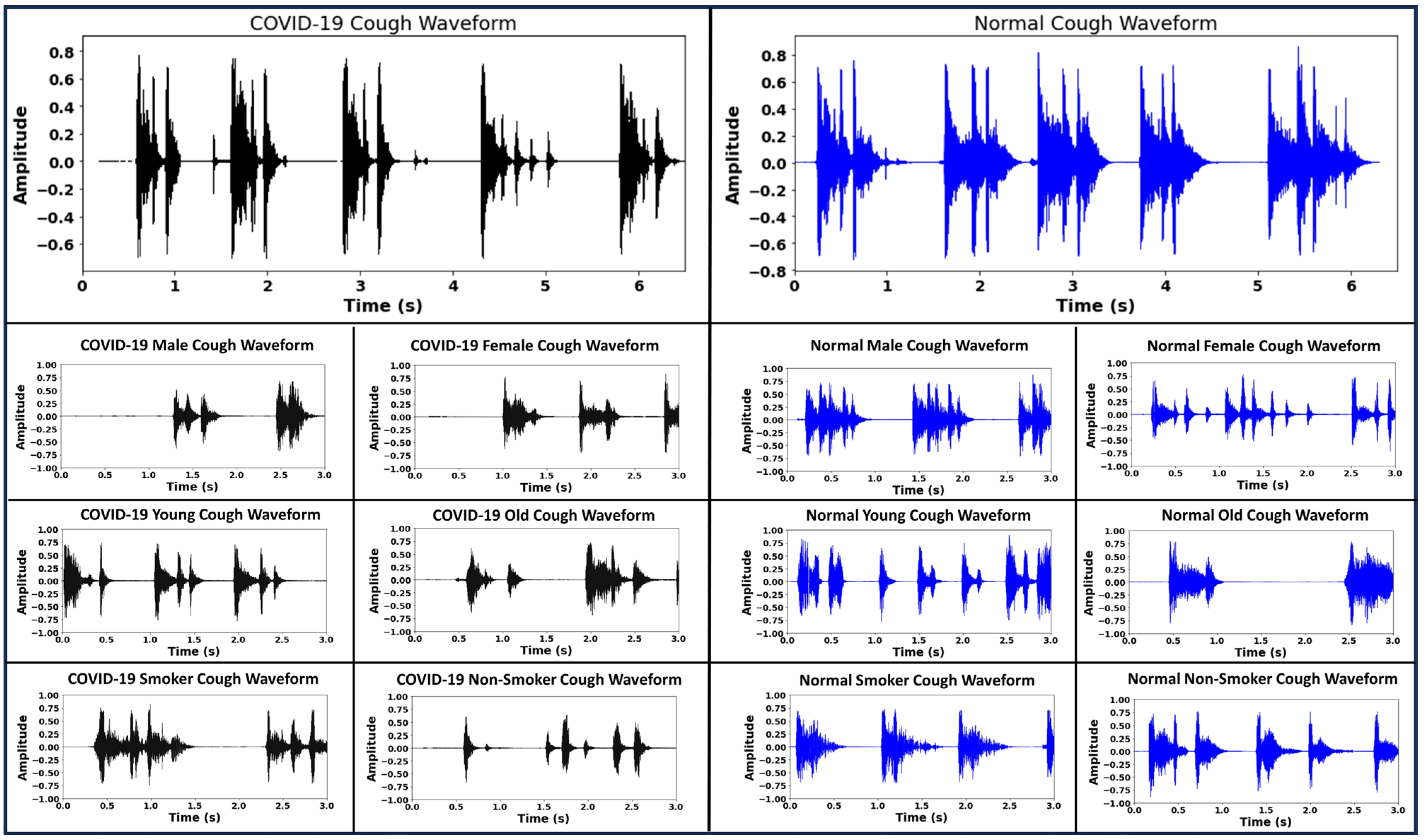
2. Cough Data Acquisition and Pre-Processing
3. RBF-Net Architecture
4. Methodology
4.1. Creation of Cough Spectrograms
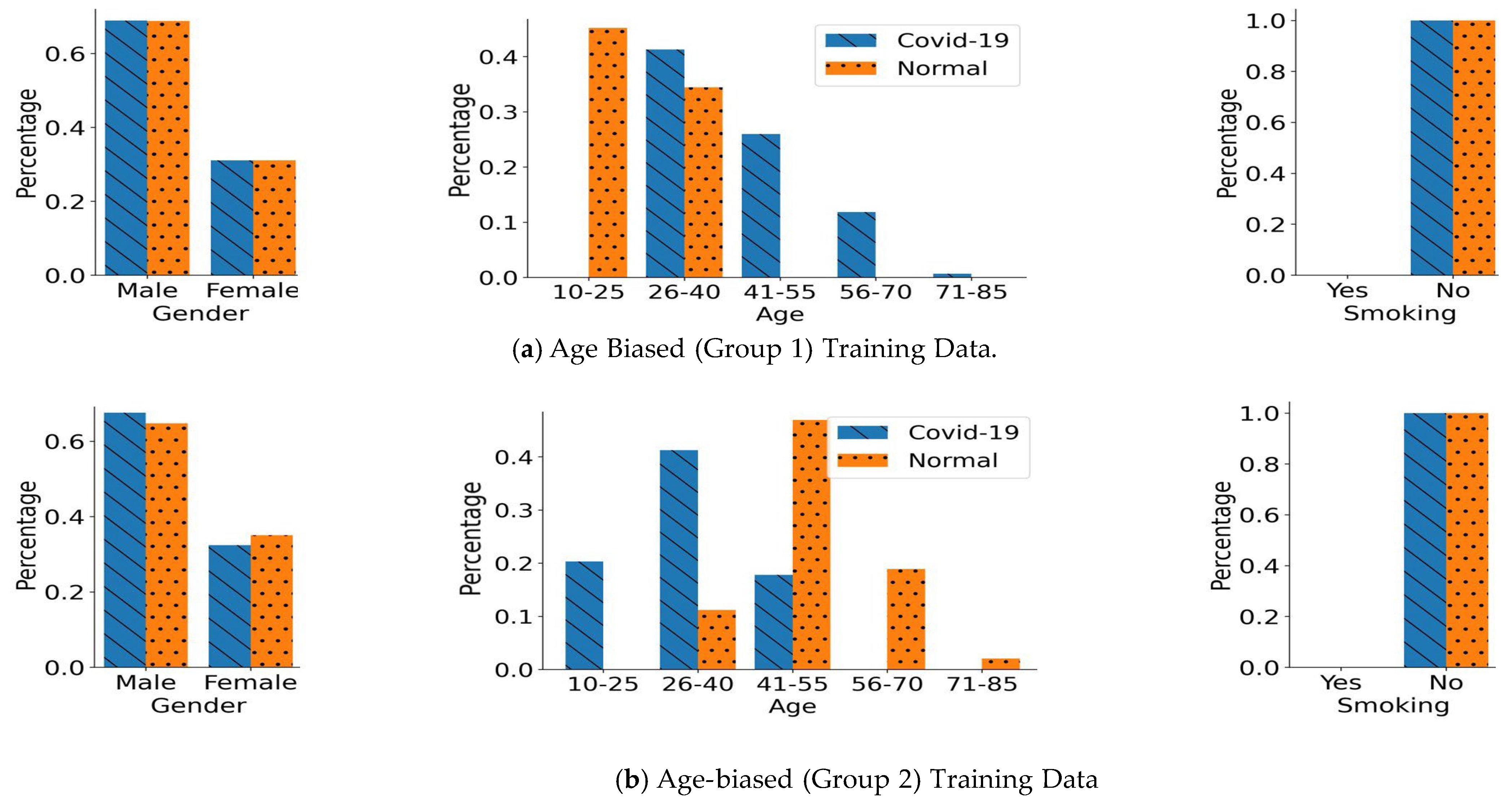
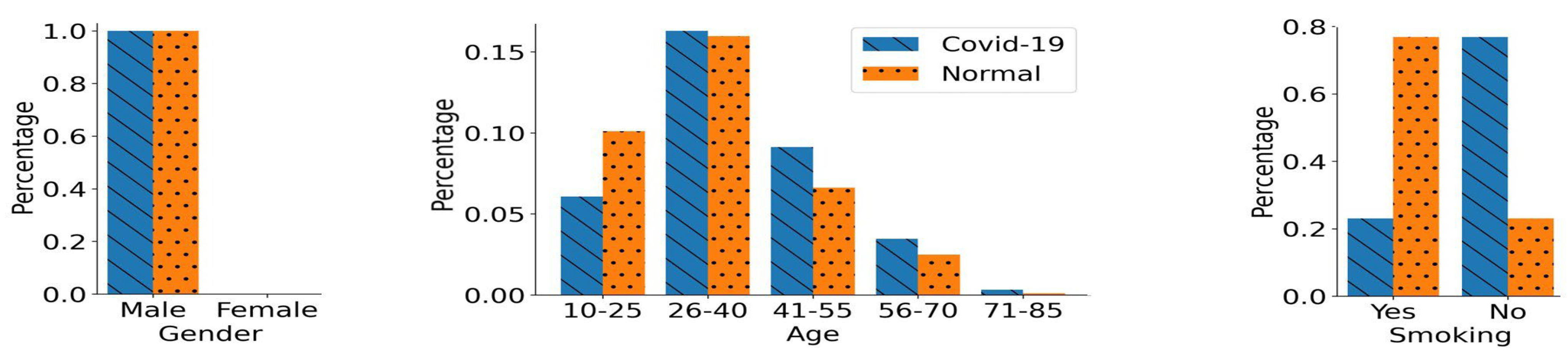
4.2. Biased Training Data Generation
- (1)
- Gender Bias:
- (2)
- Age Bias:
- (3)
- Smoking Status Bias:
4.3. Model Training
5. Results
5.1. Performance in Gender-Biased Training Scenario
5.2. Performance in the Age-Biased Training Scenarios
5.3. Performance in the Smoking Status-Biased Training Scenario
5.4. Ablation Study
6. Discussion and Limitations
7. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Coalition of Respiratory Health Organisations. Issues Recommendations to Improve Lung Health. Available online: https://www.ersnet.org/news-and-features/news/global-coalition-of-respiratory-health-organisations-issues-recommendations-to-improve-lung-health/ (accessed on 10 June 2023).
- Chronic Obstructive Pulmonary Disease (COPD) Includes: Chronic Bronchitis and Emphysema. FastStats. Respiratory Disease. Available online: https://www.cdc.gov/nchs/fastats/copd.htm (accessed on 10 June 2023).
- Respiratory Diseases in the World. Available online: https://www.thoracic.org/about/global-public-health/firs/resources/firs-report-for-web.pdf (accessed on 10 June 2023).
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, A.; Nabeel, M.; Masood, U.; Mahmood, T.; Hashmi, M.S.; Posokhova, I.; Rizwan, A.; Imran, A. Towards using cough for respiratory disease diagnosis by leveraging artificial intelligence: A survey. Inform. Med. Unlocked 2022, 29, 100832. [Google Scholar] [CrossRef]
- Hemdan, E.E.-D.; El-Shafai, W.; Sayed, A. CR19: A framework for preliminary detection of COVID-19 in cough audio signals using machine learning algorithms for automated medical diagnosis applications. J. Ambient Intell. Humaniz. Comput. 2022, 14, 11715–11727. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.I.; Lozano, M.; Estrada-Petrocelli, L.; Birring, S.; Turner, R. The present and future of cough counting tools. J. Thorac. Dis. 2020, 12, 5207–5223. [Google Scholar] [CrossRef] [PubMed]
- Laguarta, J.; Hueto, F.; Subirana, B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020, 1, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.E.; Ibtehaz, N.; Rahman, T.; Mekki, Y.M.S.; Qibalwey, Y.; Mahmud, S.; Ezeddin, M.; Zughaier, S.; Al-Maadeed, S.A.S. QUCoughScope: An artificially intelligent mobile application to detect asymptomatic COVID-19 patients using cough and breathing sounds. arXiv 2021, arXiv:2103.12063. [Google Scholar]
- Imran, A.; Posokhova, I.; Qureshi, H.N.; Masood, U.; Riaz, M.S.; Ali, K.; John, C.N.; Hussain, M.I.; Nabeel, M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked 2020, 20, 100378. [Google Scholar] [CrossRef]
- Bales, C.; Nabeel, M.; John, C.N.; Masood, U.; Qureshi, H.N.; Farooq, H.; Posokhova, I.; Imran, A. Can machine learning be used to recognize and diagnose coughs? In Proceedings of the 2020 International Conference on e-Health and Bioengineering (EHB), Iasi, Romania, 29–30 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Swarnkar, V.; Abeyratne, U.; Tan, J.; Ng, T.W.; Brisbane, J.M.; Choveaux, J.; Porter, P. Stratifying asthma severity in children using cough sound analytic technology. J. Asthma 2021, 58, 160–169. [Google Scholar] [CrossRef]
- Pahar, M.; Klopper, M.; Warren, R.; Niesler, T. COVID-19 cough classification using machine learning and global smartphone recordings. Comput. Biol. Med. 2021, 135, 104572. [Google Scholar] [CrossRef]
- Brown, C.; Chauhan, J.; Grammenos, A.; Han, J.; Hasthanasombat, A.; Spathis, D.; Xia, T.; Cicuta, P.; Mascolo, C. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Online, 23–27 August 2020; pp. 3474–3484. [Google Scholar]
- Fakhry, A.; Jiang, X.; Xiao, J.; Chaudhari, G.; Han, A.; Khanzada, A. Virufy: A multi-branch deep learning network for automated detection of COVID-19. arXiv 2021, arXiv:2103.01806. [Google Scholar]
- Kumar, L.K.; Alphonse, P. Automatic Diagnosis of COVID-19 Disease using Deep Convolutional Neural Network with Multi-Feature Channel from Respiratory Sound Data: Cough, Voice, and Breath. Alex. Eng. J. 2021, 61, 1319–1334. [Google Scholar]
- Islam, M.Z.; Islam, M.M.; Asraf, A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked 2020, 20, 100412. [Google Scholar] [CrossRef] [PubMed]
- Sharan, R.V.; Abeyratne, U.R.; Swarnkar, V.R.; Porter, P. Automatic croup diagnosis using cough sound recognition. IEEE Trans. Biomed. Eng. 2018, 66, 485–495. [Google Scholar] [CrossRef]
- Danda, S.R.; Chen, B. Toward Mitigating Spreading of Coronavirus via Mobile Devices. IEEE Internet Things Mag. 2020, 3, 12–16. [Google Scholar] [CrossRef]
- Manshouri, N.M. Identifying COVID-19 by using spectral analysis of cough recordings: A distinctive classification study. Cogn. Neurodyn. 2022, 16, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Mouawad, P.; Dubnov, T.; Dubnov, S. Robust Detection of COVID-19 in Cough Sounds. SN Comput. Sci. 2021, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Bansal, V.; Pahwa, G.; Kannan, N. Cough Classification for COVID-19 based on audio mfcc features using Convolutional Neural Networks. In Proceedings of the 2020 IEEE International Conference on Computing, Power and Communication Technologies (GUCON), Greater Noida, India, 2–4 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 604–608. [Google Scholar]
- Mohammed, E.A.; Keyhani, M.; Sanati-Nezhad, A.; Hejazi, S.H.; Far, B.H. An ensemble learning approach to digital coronavirus preliminary screening from cough sounds. Sci. Rep. 2021, 11, 15404. [Google Scholar] [CrossRef] [PubMed]
- Naeem, H.; Bin-Salem, A.A. A CNN-LSTM network with multi-level feature extraction-based approach for automated detection of coronavirus from CT scan and X-ray images. Appl. Soft Comput. 2021, 113, 107918. [Google Scholar] [CrossRef]
- Dastider, A.G.; Sadik, F.; Fattah, S.A. An integrated autoencoder-based hybrid CNN-LSTM model for COVID-19 severity prediction from lung ultrasound. Comput. Biol. Med. 2021, 132, 104296. [Google Scholar] [CrossRef]
- Dutta, S.; Bandyopadhyay, S.K.; Kim, T.H. CNN-LSTM model for verifying predictions of COVID-19 cases. Asian J. Res. Comput. Sci. 2020, 5, 25–32. [Google Scholar] [CrossRef]
- Hamdi, S.; Oussalah, M.; Moussaoui, A.; Saidi, M. Attention-based hybrid CNN-LSTM and spectral data augmentation for COVID-19 diagnosis from cough sound. J. Intell. Inf. Syst. 2022, 59, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Kara, M.; Öztürk, Z.; Akpek, S.; Turupcu, A. COVID-19 Diagnosis from chest CT scans: A weakly supervised CNN-LSTM approach. AI 2021, 2, 330–341. [Google Scholar] [CrossRef]
- Rayan, A.; Alruwaili, S.H.; Alaerjan, A.S.; Alanazi, S.; Taloba, A.I.; Shahin, O.R.; Salem, M. Utilizing CNN-LSTM techniques for the enhancement of medical systems. Alex. Eng. J. 2023, 72, 323–338. [Google Scholar] [CrossRef]
- Kollias, D.; Arsenos, A.; Kollias, S. AI-MIA: COVID-19 detection and severity analysis through medical imaging. In Computer Vision—ECCV 2022 Workshops: Tel Aviv, Israel, 23–27 October 2022, Proceedings, Part VII; Springer: Berlin/Heidelberg, Germany, 2023; pp. 677–690. [Google Scholar]
- Sunitha, G.; Arunachalam, R.; Abd-Elnaby, M.; Eid, M.M.; Rashed, A.N.Z. A comparative analysis of deep neural network architectures for the dynamic diagnosis of COVID-19 based on acoustic cough features. Int. J. Imaging Syst. Technol. 2022, 32, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Han, J.; Xia, T.; Spathis, D.; Bondareva, E.; Siegele-Brown, C.; Chauhan, J.; Grammenos, A.; Hasthanasombat, A.; Floto, R.A.; et al. Exploring longitudinal cough, breath, and voice data for COVID-19 progression prediction via sequential deep learning: Model development and validation. J. Med. Internet Res. 2022, 24, e37004. [Google Scholar] [CrossRef] [PubMed]
- Coppock, H.; Gaskell, A.; Tzirakis, P.; Baird, A.; Jones, L.; Schuller, B. End-to-end convolutional neural network enables COVID-19 detection from breath and cough audio: A pilot study. BMJ Innov. 2021, 7, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Andreu-Perez, J.; Perez-Espinosa, H.; Timonet, E.; Kiani, M.; Giron-Perez, M.I.; Benitez-Trinidad, A.B.; Jarchi, D.; Rosales-Perez, A.; Gkatzoulis, N.; Reyes-Galaviz, O.F.; et al. A generic deep learning based cough analysis system from clinically validated samples for point-of-need COVID-19 test and severity levels. IEEE Trans. Serv. Comput. 2021, 15, 1220–1232. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Xia, T.; Spathis, D.; Bondareva, E.; Brown, C.; Chauhan, J.; Dang, T.; Grammenos, A.; Hasthanasombat, A.; Floto, A.; et al. Sounds of COVID-19: Exploring realistic performance of audio-based digital testing. NPJ Digit. Med. 2022, 5, 16. [Google Scholar] [CrossRef]
- Dentamaro, V.; Giglio, P.; Impedovo, D.; Moretti, L.; Pirlo, G. AUCO ResNet: An end-to-end network for COVID-19 pre-screening from cough and breath. Pattern Recognit. 2022, 127, 108656. [Google Scholar] [CrossRef]
- Pourhoseingholi, M.A.; Baghestani, A.R.; Vahedi, M. How to control confounding effects by statistical analysis. Gastroenterol. Hepatol. Bed Bench 2012, 5, 79. [Google Scholar]
- Jager, K.; Zoccali, C.; Macleod, A.; Dekker, F. Confounding: What it is and how to deal with it. Kidney Int. 2008, 73, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Adeli, E.; Pohl, K.M. Training confounder-free deep learning models for medical applications. Nat. Commun. 2020, 11, 6010. [Google Scholar] [CrossRef] [PubMed]
- AI4COVID-19: An Artificial Intelligence Powered App for Detecting COVID-19 from Cough Sound. 2021. Available online: https://ai4networks.com/covid19.php (accessed on 16 February 2022).
- Audacity Free Open Source. Cross-Platform Audio Software, Version 3.4.2. Available online: https://www.audacityteam.org/ (accessed on 10 July 2022).
- Mushtaq, Z.; Su, S.-F. Environmental sound classification using a regularized deep convolutional neural network with data augmentation. Appl. Acoust. 2020, 167, 107389. [Google Scholar] [CrossRef]
- Song, I. Diagnosis of pneumonia from sounds collected using low cost cell phones. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–8. [Google Scholar]
- Jayalakshmy, S.; Sudha, G.F. Conditional GAN based augmentation for predictive modeling of respiratory signals. Comput. Biol. Med. 2021, 138, 104930. [Google Scholar] [CrossRef] [PubMed]
- Pramono, R.X.A.; Imtiaz, S.A.; Rodriguez-Villegas, E. Evaluation of features for classification of wheezes and normal respiratory sounds. PLoS ONE 2019, 14, e0213659. [Google Scholar] [CrossRef] [PubMed]
- Sejdic, E.; Djurovic, I.; Jiang, J. Time–frequency feature representation using energy concentration: An overview of recent advances. Digit. Signal Process. 2009, 19, 153–183. [Google Scholar] [CrossRef]
- AutoML.org. Available online: https://www.automl.org/automl/ (accessed on 10 March 2023).
- Aytekin, I.; Dalmaz, O.; Gonc, K.; Ankishan, H.; Saritas, E.U.; Bagci, U.; Celik, H.; Cukur, T. COVID-19 Detection from Respiratory Sounds with Hierarchical Spectrogram Transformers. arXiv 2022, arXiv:2207.09529. [Google Scholar] [CrossRef]
- Orlandic, L.; Teijeiro, T.; Atienza, D. The coughvid crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Sci. Data 2021, 8, 156. [Google Scholar] [CrossRef]


| Hyperparameters | Best Value |
|---|---|
| Number of Convolutional Blocks | 4 |
| LSTM Units | 256 |
| LSTM Activation | tanh |
| CNN Activation | ReLU |
| Epochs | 1000 |
| Optimizer | ADAM |
| Batch Size | 256 |
| Learning Rate | 0.0001 |
| Model | Accuracy | Specificity | Sensitivity | F-1 Score | ROC-AUC | ||
|---|---|---|---|---|---|---|---|
| Cross-Validation | CNN-LSTM | 0.890 | 0.913 | 0.866 | 0.892 | 0.893 | |
| Gender Bias | Unseen Testing Data | CNN-LSTM | 0.787 | 0.860 | 0.715 | 0.785 | 0.787 |
| RBF-Net | 0.841 | 0.887 | 0.796 | 0.845 | 0.846 | ||
| Model | Accuracy | Specificity | Sensitivity | F-1 Score | ROC-AUC | ||
|---|---|---|---|---|---|---|---|
| Cross-Validation | CNN-LSTM | 0.887 | 0.908 | 0.867 | 0.894 | 0.892 | |
| Age-biased Group 1 | Unseen Testing Data | CNN-LSTM | 0.774 | 0.843 | 0.706 | 0.775 | 0.776 |
| RBF-Net | 0.845 | 0.888 | 0.801 | 0.845 | 0.846 | ||
| Cross-Validation | CNN-LSTM | 0.884 | 0.901 | 0.867 | 0.884 | 0.881 | |
| Age-biased Group 2 | Unseen Testing Data | CNN-LSTM | 0.756 | 0.825 | 0.687 | 0.757 | 0.756 |
| RBF-Net | 0.818 | 0.863 | 0.774 | 0.819 | 0.821 | ||
| Model | Accuracy | Specificity | Sensitivity | F-1 Score | ROC-AUC | ||
|---|---|---|---|---|---|---|---|
| Cross-Validation | CNN-LSTM | 0.862 | 0.881 | 0.844 | 0.867 | 0.862 | |
| Smoking Bias | Unseen Testing Data | CNN-LSTM | 0.723 | 0.750 | 0.694 | 0.727 | 0.723 |
| RBF-Net | 0.805 | 0.836 | 0.774 | 0.811 | 0.805 |
| Model | Unbiased | Gender-Biased | Age-Biased | Smoking Status-Biased | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | F1-Score | ROC-AUC | Acc | F1-Score | ROC-AUC | Acc | F1-Score | ROC-AUC | Acc | F1-Score | F1-Score | |
| CNN (baseline) | 0.821 | 0.822 | 0.822 | 0.751 | 0.751 | 0.751 | 0.746 | 0.741 | 0.746 | 0.683 | 0.679 | 0.684 |
| CNN-LSTM (A) | 0.874 | 0.877 | 0.874 | 0.787 | 0.785 | 0.787 | 0.774 | 0.775 | 0.776 | 0.723 | 0.727 | 0.723 |
| RBF-Net (A + B) | 0.879 | 0.883 | 0.878 | 0.841 | 0.845 | 0.846 | 0.845 | 0.845 | 0.846 | 0.805 | 0.811 | 0.805 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeed, T.; Ijaz, A.; Sadiq, I.; Qureshi, H.N.; Rizwan, A.; Imran, A. An AI-Enabled Bias-Free Respiratory Disease Diagnosis Model Using Cough Audio. Bioengineering 2024, 11, 55. https://doi.org/10.3390/bioengineering11010055
Saeed T, Ijaz A, Sadiq I, Qureshi HN, Rizwan A, Imran A. An AI-Enabled Bias-Free Respiratory Disease Diagnosis Model Using Cough Audio. Bioengineering. 2024; 11(1):55. https://doi.org/10.3390/bioengineering11010055
Chicago/Turabian StyleSaeed, Tabish, Aneeqa Ijaz, Ismail Sadiq, Haneya Naeem Qureshi, Ali Rizwan, and Ali Imran. 2024. "An AI-Enabled Bias-Free Respiratory Disease Diagnosis Model Using Cough Audio" Bioengineering 11, no. 1: 55. https://doi.org/10.3390/bioengineering11010055
APA StyleSaeed, T., Ijaz, A., Sadiq, I., Qureshi, H. N., Rizwan, A., & Imran, A. (2024). An AI-Enabled Bias-Free Respiratory Disease Diagnosis Model Using Cough Audio. Bioengineering, 11(1), 55. https://doi.org/10.3390/bioengineering11010055




