Deep-Learning-Based High-Intensity Focused Ultrasound Lesion Segmentation in Multi-Wavelength Photoacoustic Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Image Preprocessing
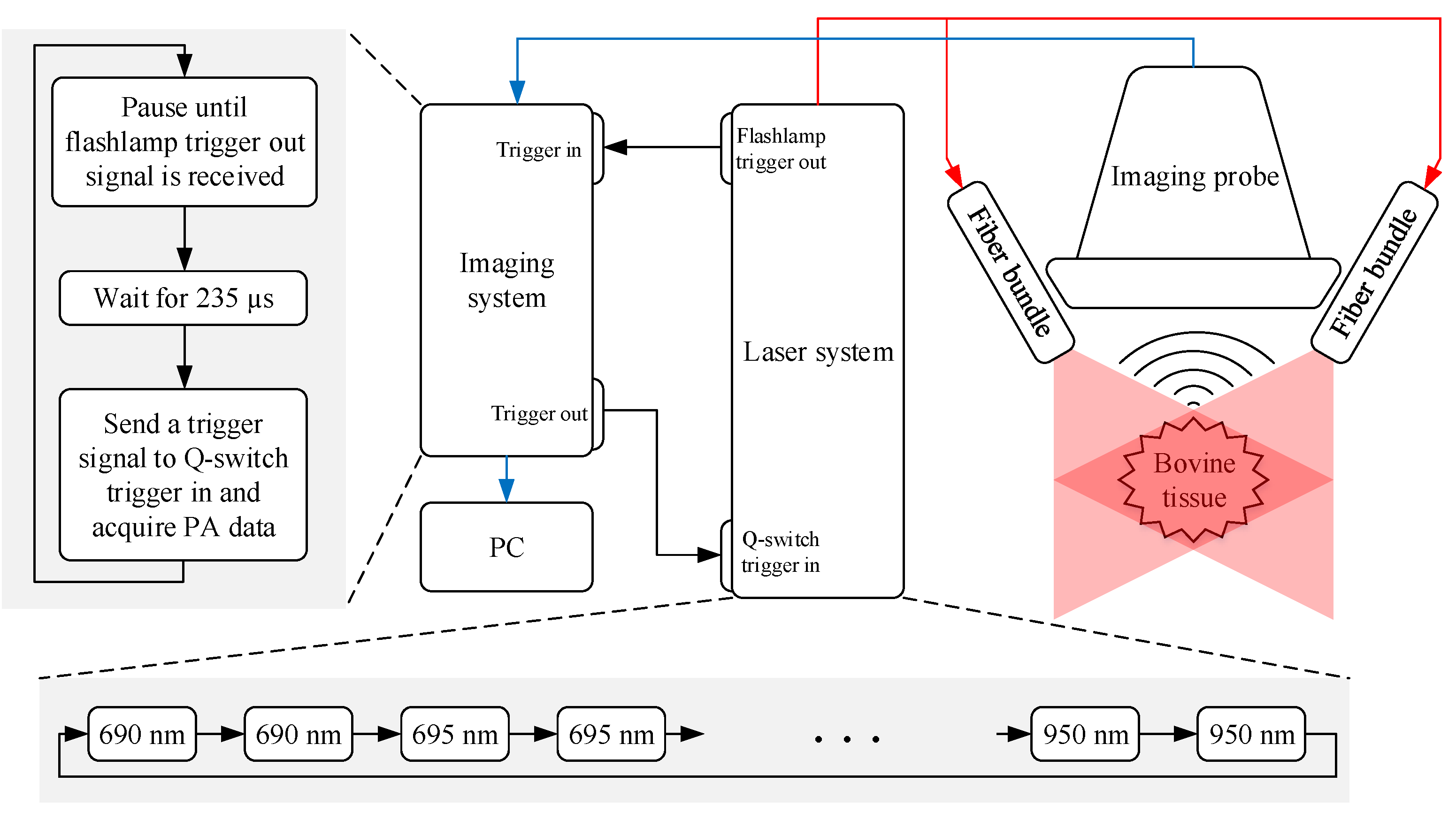
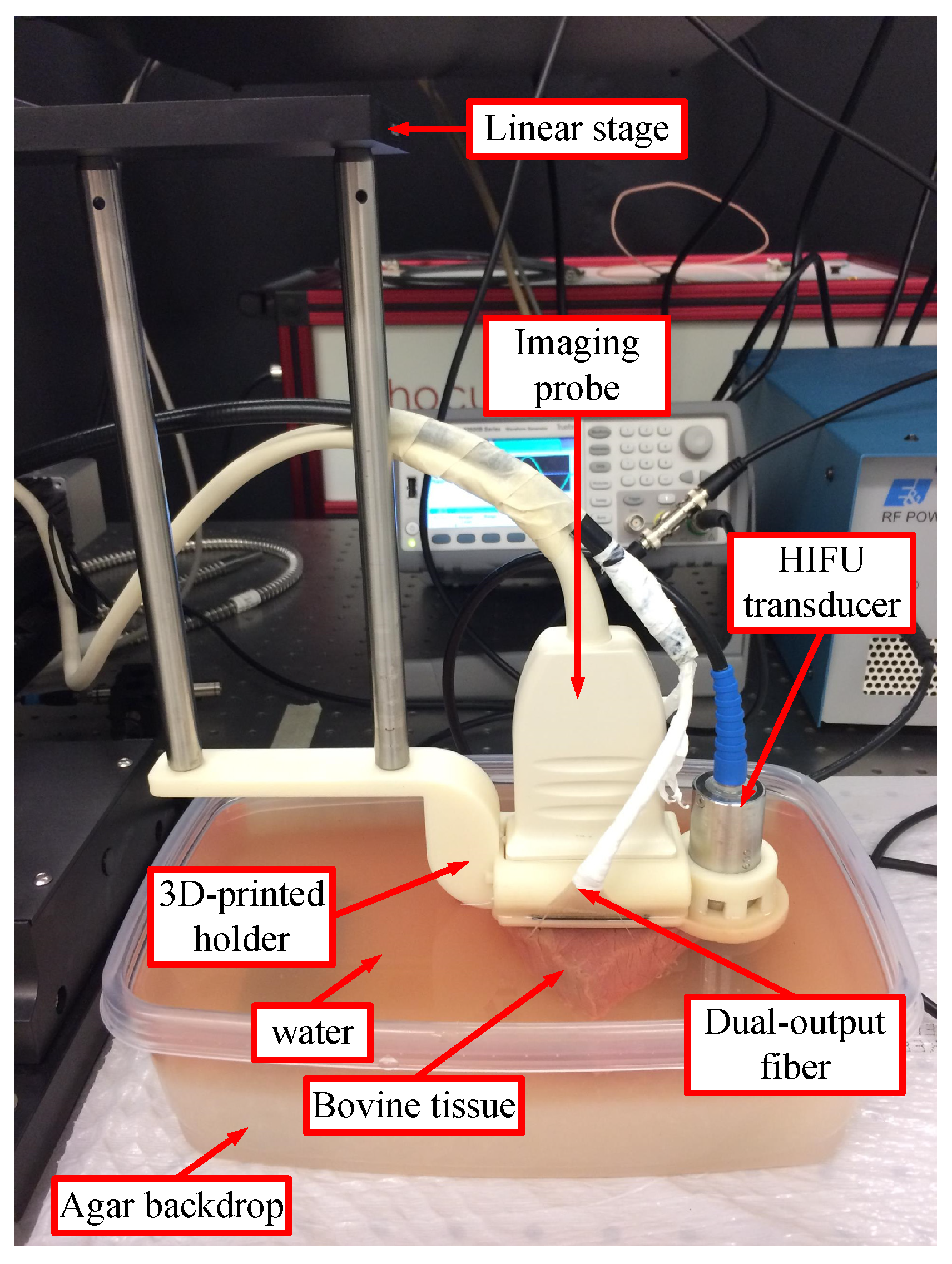
2.1.1. Experimental Setup
2.1.2. Experimental Procedure
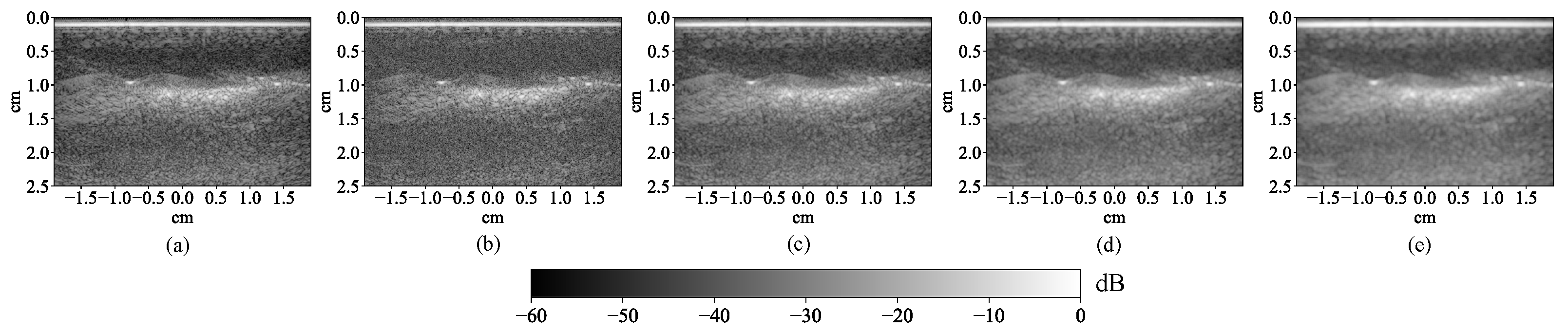
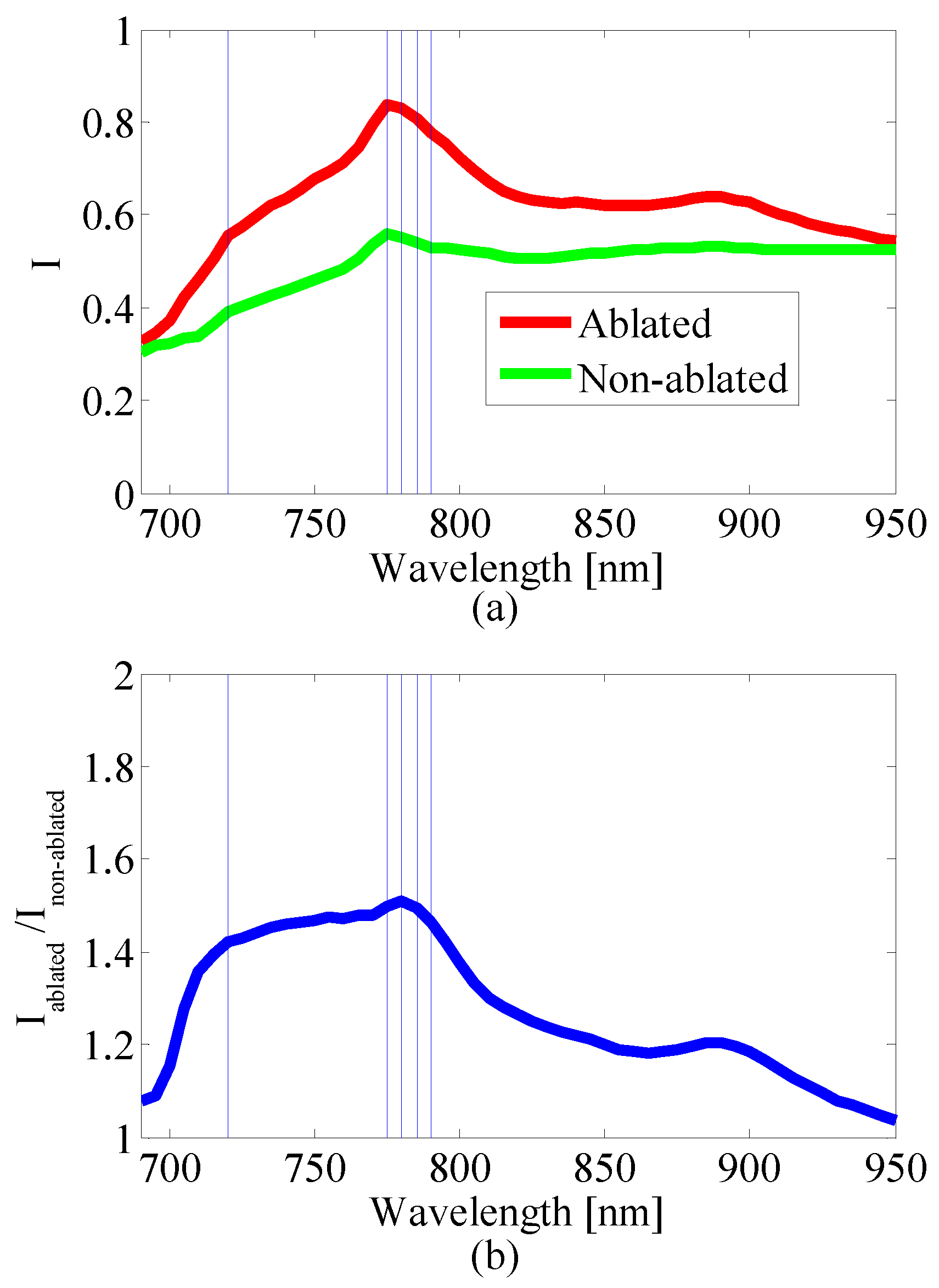
2.1.3. Multi-Wavelength Photoacoustic Image Preprocessing
2.2. Lesion Segmentation
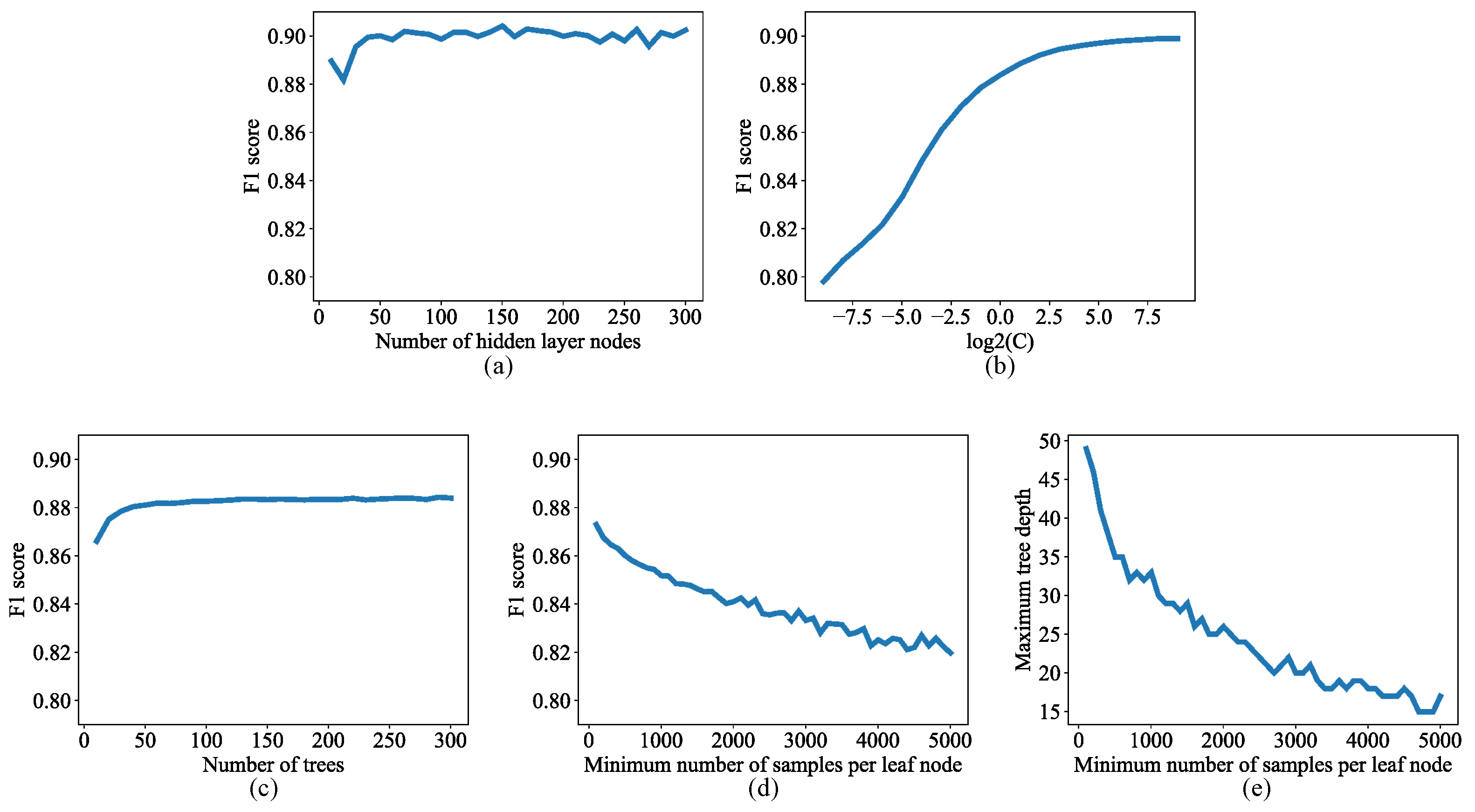
2.2.1. Lesion Segmentation with Traditional Machine Learning Algorithms
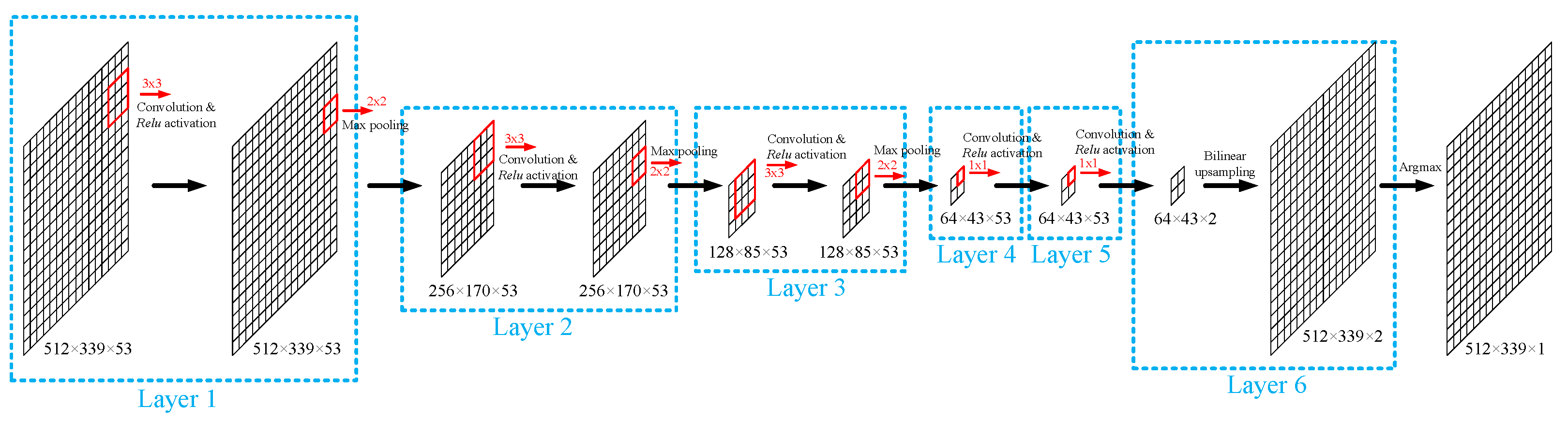
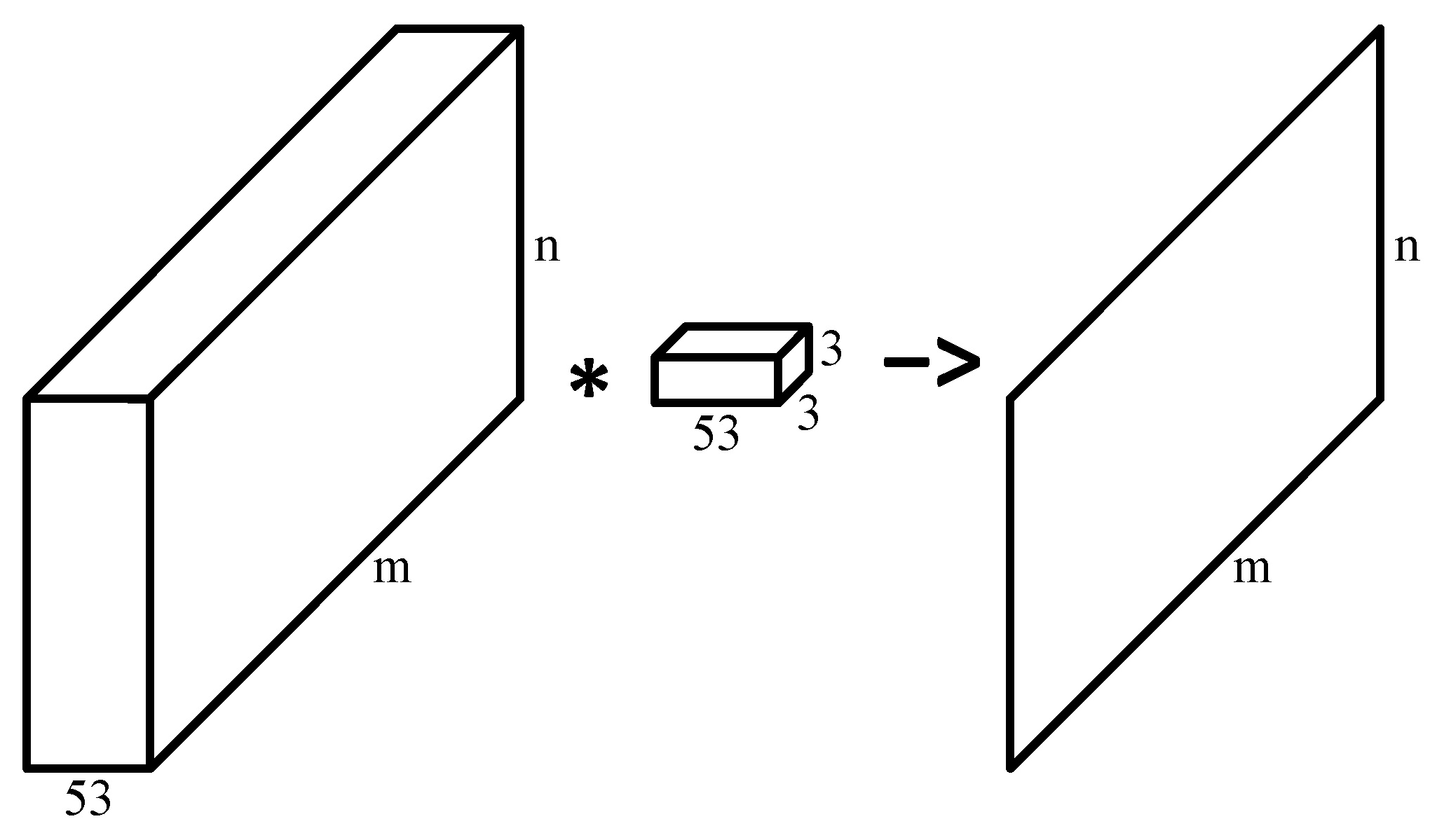
2.2.2. Lesion Segmentation with Convolutional Neural Network
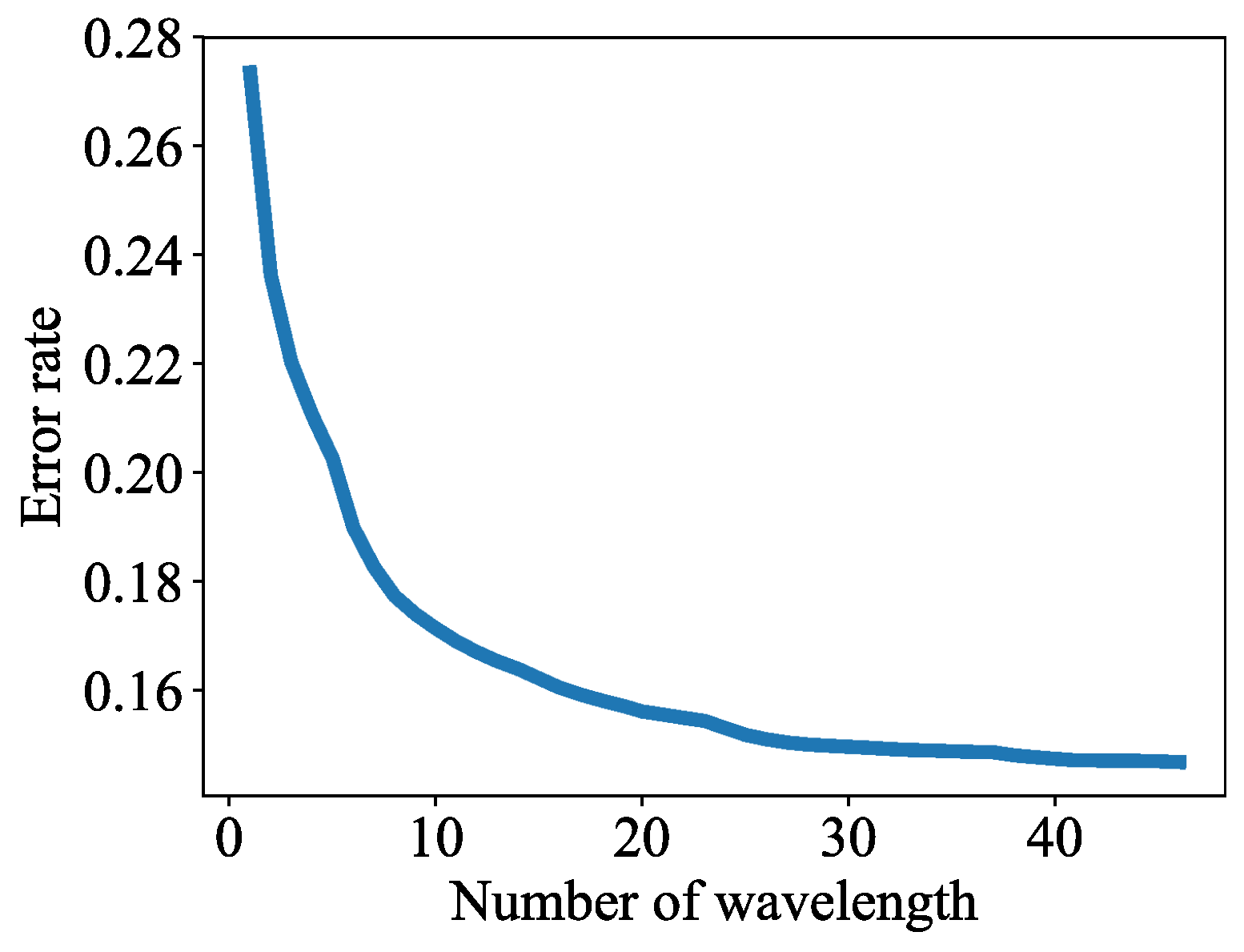
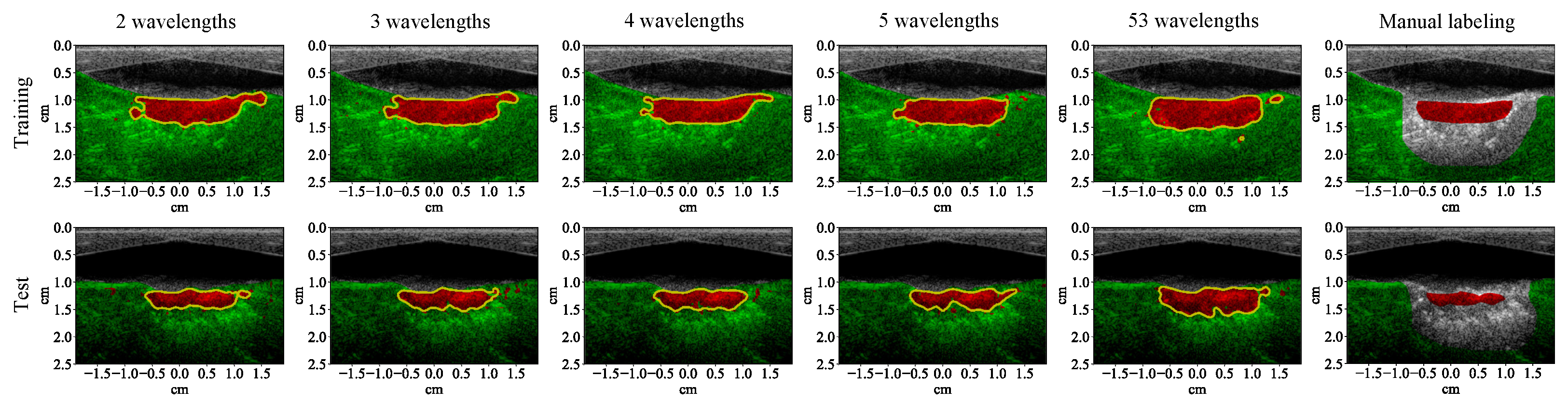
2.2.3. Wavelength Selection
3. Results
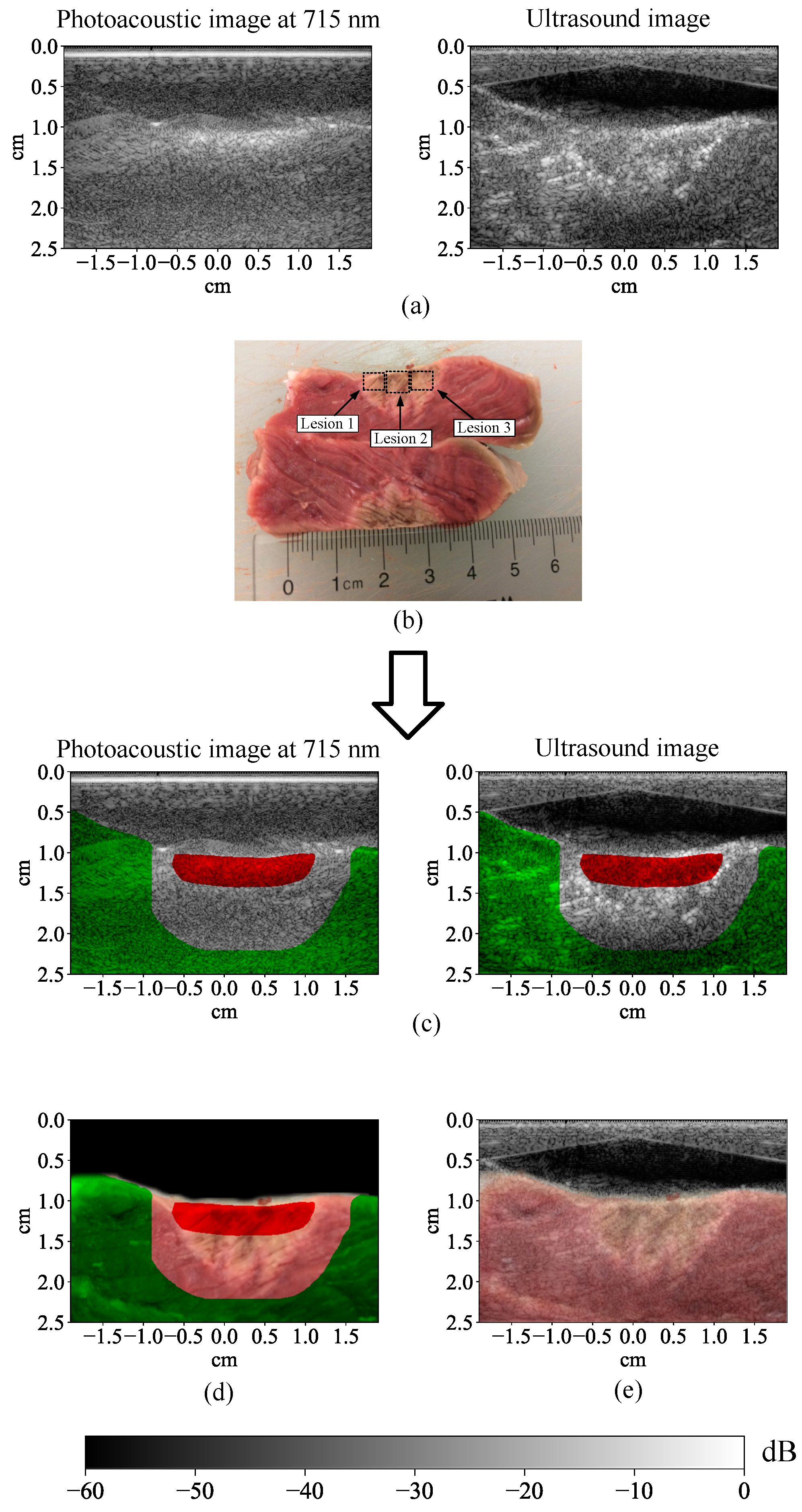
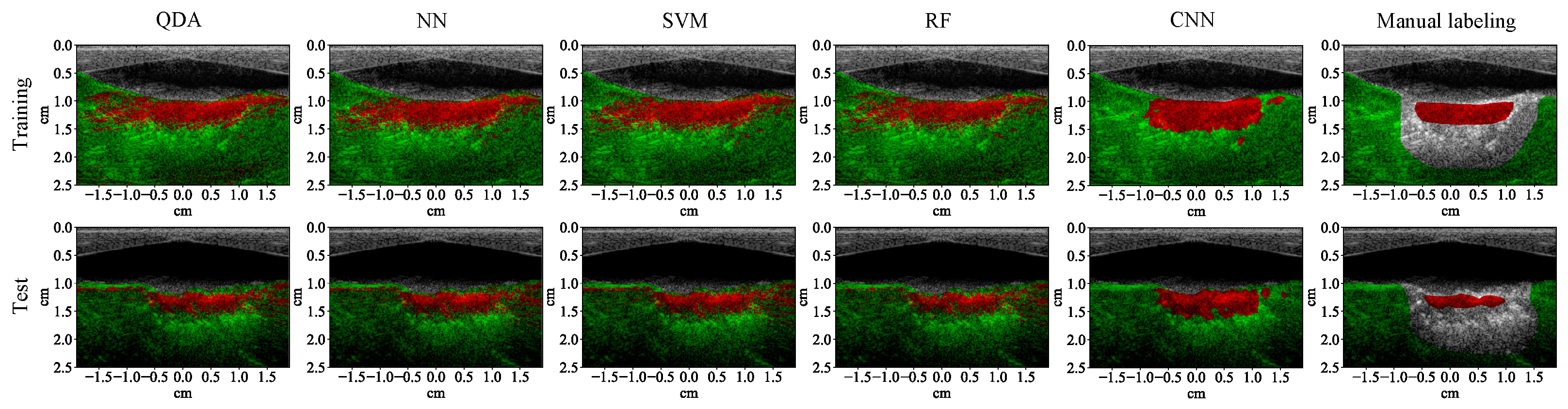
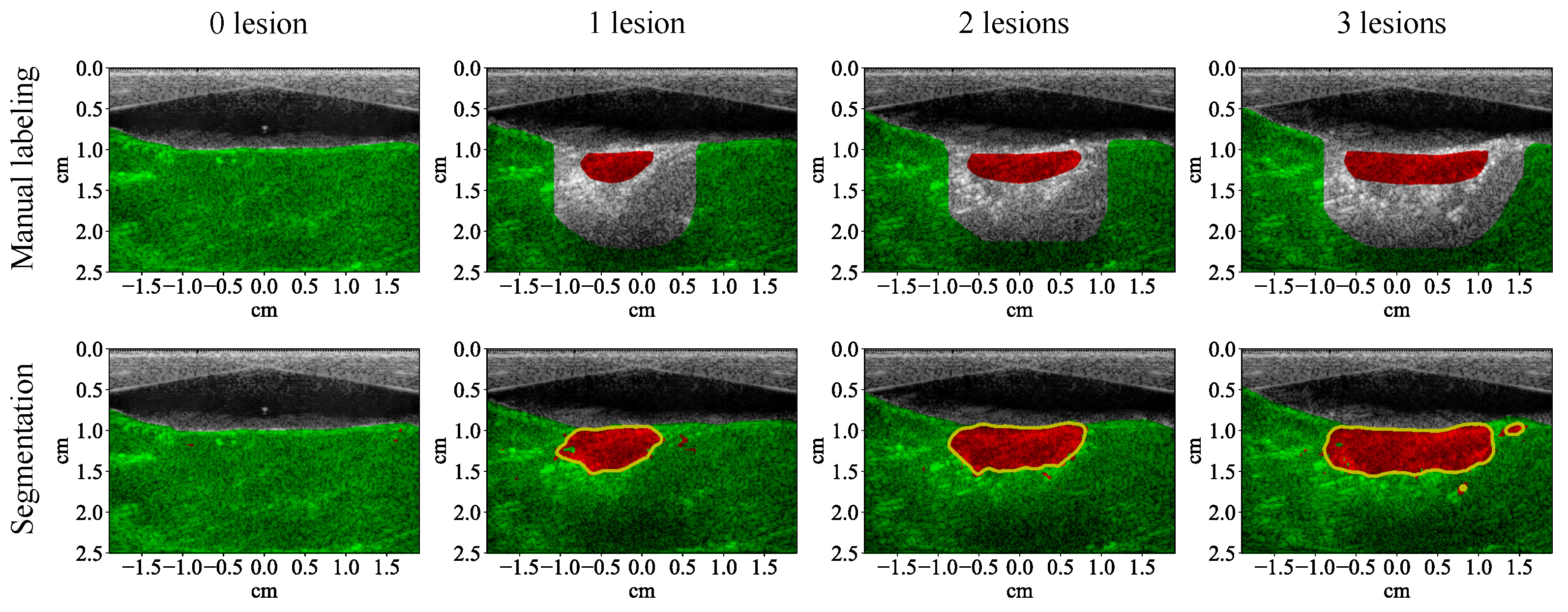
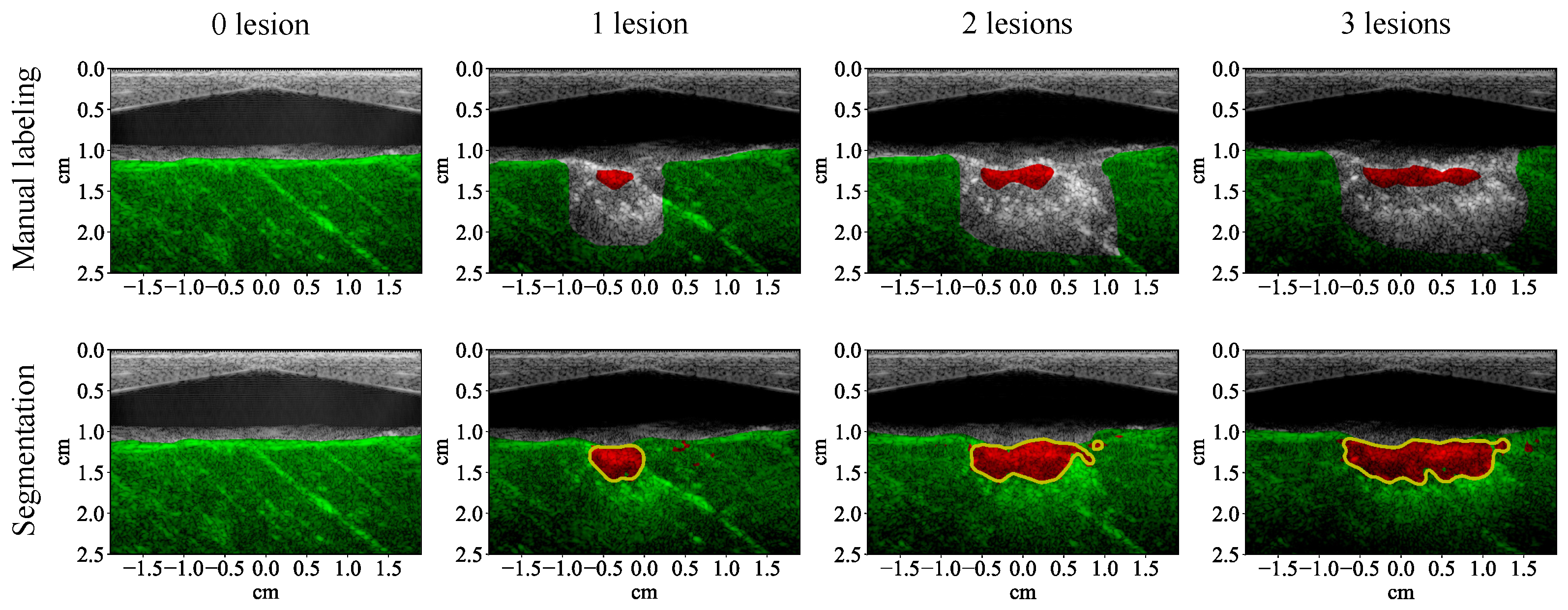
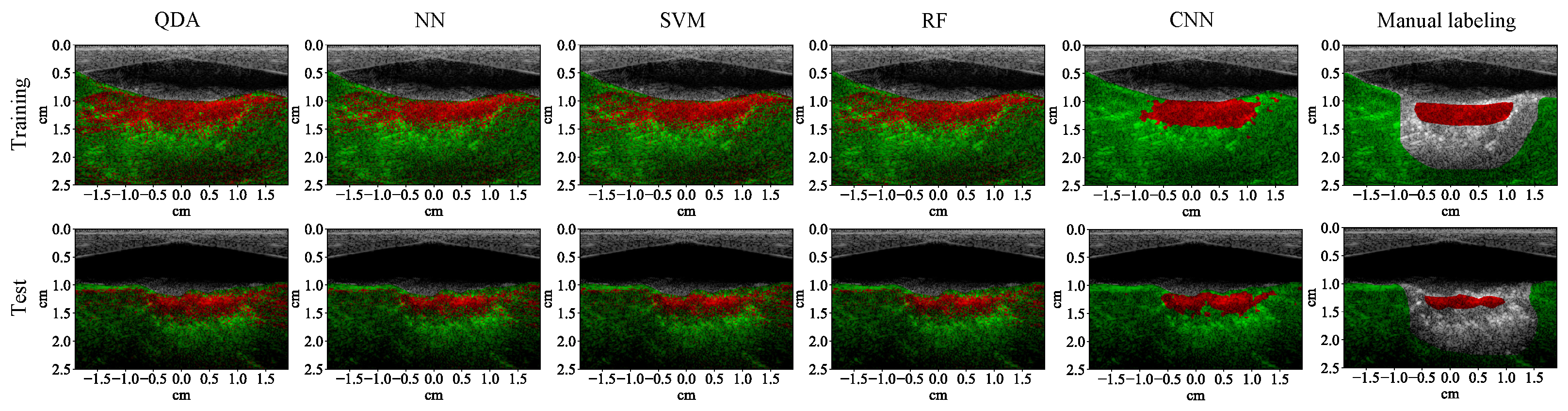
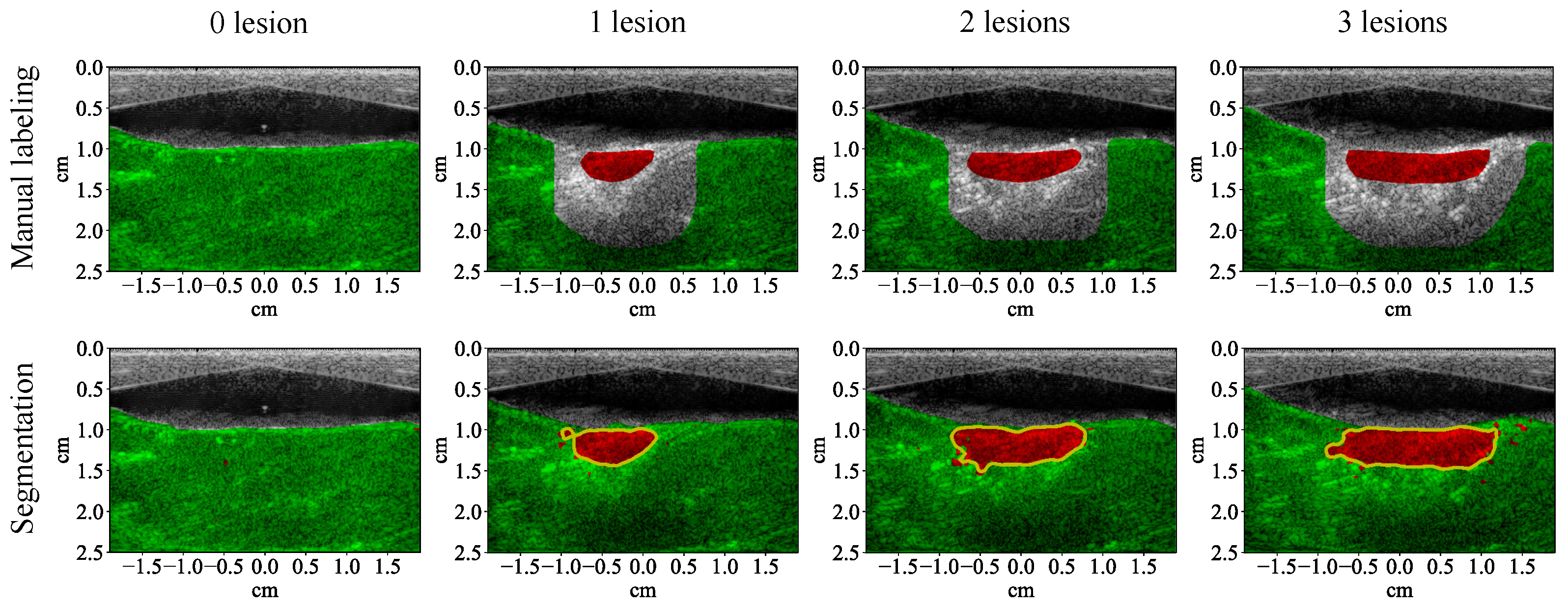
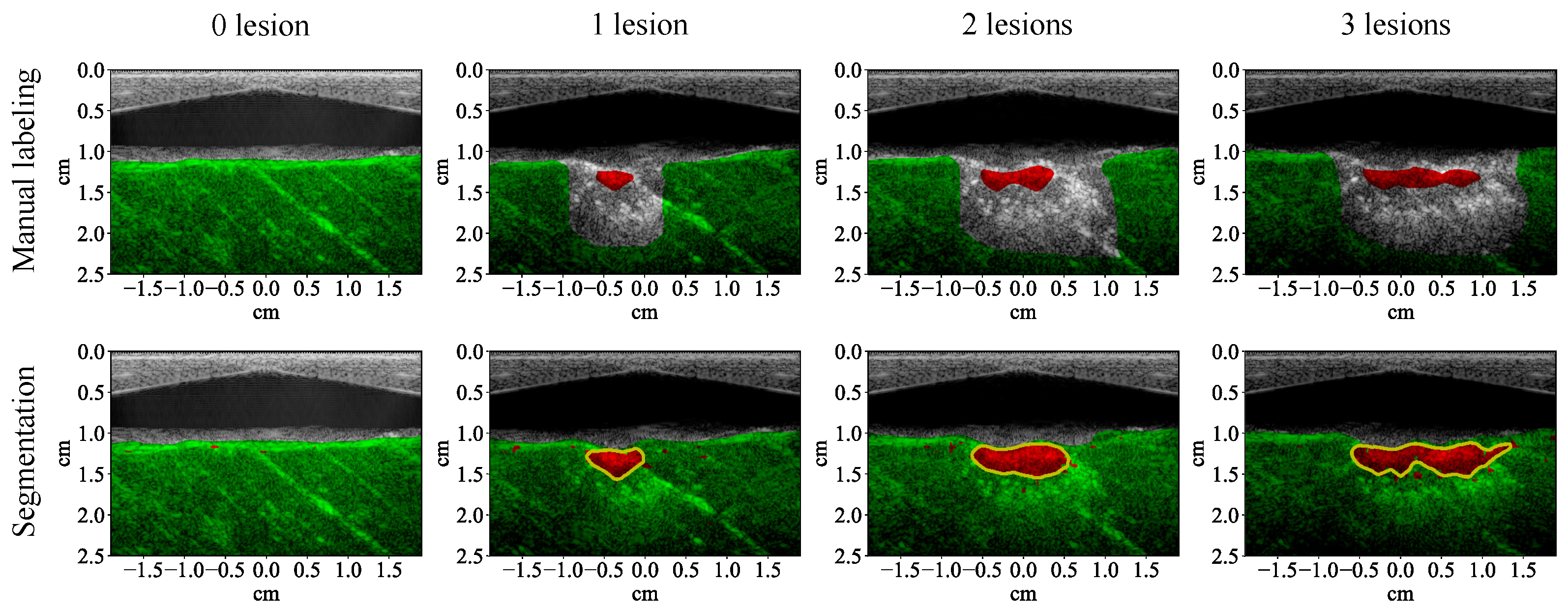
3.1. Lesion Segmentation with All Wavelengths
3.2. Lesion Segmentation with Reduced Wavelengths
4. Discussion
4.1. High-Performance Lesion Segmentation
4.2. Convenient Implementation
4.3. Advantages over Temperature-Measurement-Based Methods
4.4. Potential for Real-Time Implementation
4.5. Potential Improvement with Gold-Standard Training Examples
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kennedy, J.E. High-intensity focused ultrasound in the treatment of solid tumours. Nat. Rev. Cancer 2005, 5, 321–327. [Google Scholar] [CrossRef] [PubMed]
- van den Bijgaart, R.J.; Eikelenboom, D.C.; Hoogenboom, M.; Fütterer, J.J.; den Brok, M.H.; Adema, G.J. Thermal and mechanical high-intensity focused ultrasound: Perspectives on tumor ablation, immune effects and combination strategies. Cancer Immunol. Immunother. 2017, 66, 247–258. [Google Scholar] [CrossRef]
- Özsoy, Ç.; Lafci, B.; Reiss, M.; Deán-Ben, X.L.; Razansky, D. Real-time assessment of high-intensity focused ultrasound heating and cavitation with hybrid optoacoustic ultrasound imaging. Photoacoustics 2023, 31, 100508. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, W.; Fan, W.; Huang, J.; Zhang, F.; Wu, P. Noninvasive treatment of malignant bone tumors using high-intensity focused ultrasound. Cancer 2010, 116, 3934–3942. [Google Scholar] [CrossRef] [PubMed]
- Merckel, L.G.; Bartels, L.W.; Köhler, M.O.; van den Bongard, H.D.; Deckers, R.; Willem, P.T.M.; Binkert, C.A.; Moonen, C.T.; Gilhuijs, K.G.; van den Bosch, M.A. MR-guided high-intensity focused ultrasound ablation of breast cancer with a dedicated breast platform. Cardiovasc. Interv. Radiol. 2013, 36, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Klingler, H.C.; Susani, M.; Seip, R.; Mauermann, J.; Sanghvi, N.; Marberger, M.J. A novel approach to energy ablative therapy of small renal tumours: Laparoscopic high-intensity focused ultrasound. Eur. Urol. 2008, 53, 810–818. [Google Scholar] [CrossRef]
- Illing, R.; Kennedy, J.; Wu, F.; Ter Haar, G.; Protheroe, A.; Friend, P.; Gleeson, F.; Cranston, D.; Phillips, R.; Middleton, M. The safety and feasibility of extracorporeal high-intensity focused ultrasound (HIFU) for the treatment of liver and kidney tumours in a Western population. Br. J. Cancer 2005, 93, 890–895. [Google Scholar] [CrossRef]
- Wu, F. High intensity focused ultrasound: A noninvasive therapy for locally advanced pancreatic cancer. World J. Gastroenterol. 2014, 20, 16480–16488. [Google Scholar] [CrossRef]
- Blana, A.; Walter, B.; Rogenhofer, S.; Wieland, W.F. High-intensity focused ultrasound for the treatment of localized prostate cancer: 5-year experience. Urology 2004, 63, 297–300. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.Z.; Liu, Y.J.; Hu, X.; Zhou, K.; Chen, L.; Peng, S.; Zhu, H.; Zou, H.L.; Bai, J.; et al. Feasibility of magnetic resonance imaging-guided high intensity focused ultrasound therapy for ablating uterine fibroids in patients with bowel lies anterior to uterus. Eur. J. Radiol. 2010, 73, 396–403. [Google Scholar] [CrossRef]
- Ninet, J.; Roques, X.; Seitelberger, R.; Deville, C.; Pomar, J.L.; Robin, J.; Jegaden, O.; Wellens, F.; Wolner, E.; Vedrinne, C.; et al. Surgical ablation of atrial fibrillation with off-pump, epicardial, high-intensity focused ultrasound: Results of a multicenter trial. J. Thorac. Cardiovasc. Surg. 2005, 130, 803.e1–803.e8. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Antz, M.; Wong, T.; Schmidt, B.; Ernst, S.; Ouyang, F.; Vogtmann, T.; Wu, R.; Yokoyama, K.; Lockwood, D.; et al. Initial experience using a forward directed, high-intensity focused ultrasound balloon catheter for pulmonary vein antrum isolation in patients with atrial fibrillation. J. Cardiovasc. Electrophysiol. 2007, 18, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Mitnovetski, S.; Almeida, A.A.; Goldstein, J.; Pick, A.W.; Smith, J.A. Epicardial high-intensity focused ultrasound cardiac ablation for surgical treatment of atrial fibrillation. Hear. Lung Circ. 2009, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Copelan, A.; Hartman, J.; Chehab, M.; Venkatesan, A.M. High-intensity focused ultrasound: Current status for image-guided therapy. Semin. Interv. Radiol. 2015, 32, 398–415. [Google Scholar] [CrossRef]
- Elhelf, I.S.; Albahar, H.; Shah, U.; Oto, A.; Cressman, E.; Almekkawy, M. High intensity focused ultrasound: The fundamentals, clinical applications and research trends. Diagn. Interv. Imaging 2018, 99, 349–359. [Google Scholar] [CrossRef]
- Hynynen, K.; Pomeroy, O.; Smith, D.N.; Huber, P.E.; McDannold, N.J.; Kettenbach, J.; Baum, J.; Singer, S.; Jolesz, F.A. MR imaging-guided focused ultrasound surgery of fibroadenomas in the breast: A feasibility study. Radiology 2001, 219, 176–185. [Google Scholar] [CrossRef]
- Tempany, C.M.; Stewart, E.A.; McDannold, N.; Quade, B.J.; Jolesz, F.A.; Hynynen, K. MR imaging–guided focused ultrasound surgery of uterine leiomyomas: A feasibility study. Radiology 2003, 226, 897–905. [Google Scholar] [CrossRef]
- Rieke, V.; Butts Pauly, K. MR thermometry. J. Magn. Reson. Imaging 2008, 27, 376–390. [Google Scholar] [CrossRef]
- Li, S.; Wu, P.H. Magnetic resonance image-guided versus ultrasound-guided high-intensity focused ultrasound in the treatment of breast cancer. Chin. J. Cancer 2013, 32, 441. [Google Scholar] [CrossRef]
- Steiner, P.; Botnar, R.; Dubno, B.; Zimmermann, G.G.; Gazelle, G.S.; Debatin, J.F. Radio-frequency-induced thermoablation: Monitoring with T1-weighted and proton-frequency-shift MR imaging in an interventional 0.5-T environment. Radiology 1998, 206, 803–810. [Google Scholar] [CrossRef]
- Wu, X.; Sanders, J.L.; Stephens, D.N.; Oralkan, Ö. Photoacoustic-imaging-based temperature monitoring for high-intensity focused ultrasound therapy. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3235–3238. [Google Scholar]
- Vaezy, S.; Shi, X.; Martin, R.W.; Chi, E.; Nelson, P.I.; Bailey, M.R.; Crum, L.A. Real-time visualization of high-intensity focused ultrasound treatment using ultrasound imaging. Ultrasound Med. Biol. 2001, 27, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Zhou, F.; Wang, S.; Wan, M. Evaluate thermal lesion using Nakagami imaging for monitoring of high-intensity focused ultrasound. AIP Conf. Proc. 2017, 1816, 050002. [Google Scholar]
- Gelet, A.; Chapelon, J.; Bouvier, R.; Rouviere, O.; Lasne, Y.; Lyonnet, D.; Dubernard, J. Transrectal high-intensity focused ultrasound: Minimally invasive therapy of localized prostate cancer. J. Endourol. 2000, 14, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva, R.; Guglielmina, J.N.; Abehsera, M.; Boulanger, L.; Laurent, N.; Poncelet, E. Ultrasound-guided high-intensity focused ultrasound treatment of breast fibroadenoma-a multicenter experience. J. Ther. Ultrasound 2015, 3, 1–8. [Google Scholar] [CrossRef]
- Lee, J.S.; Hong, G.Y.; Park, B.J.; Kim, T.E. Ultrasound-guided high-intensity focused ultrasound treatment for uterine fibroid & adenomyosis: A single center experience from the Republic of Korea. Ultrason. Sonochem. 2015, 27, 682–687. [Google Scholar]
- Zhang, C.; Jacobson, H.; Ngobese, Z.; Setzen, R. Efficacy and safety of ultrasound-guided high intensity focused ultrasound ablation of symptomatic uterine fibroids in black women: A preliminary study. BJOG Int. J. Obstet. Gynaecol. 2017, 124, 12–17. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, F.; Wan, M.; Wei, M.; Fu, Q.; Wang, X.; Wang, S. Feasibility of using Nakagami distribution in evaluating the formation of ultrasound-induced thermal lesions. J. Acoust. Soc. Am. 2012, 131, 4836–4844. [Google Scholar] [CrossRef]
- Cui, H.; Staley, J.; Yang, X. Integration of photoacoustic imaging and high-intensity focused ultrasound. J. Biomed. Opt. 2010, 15, 021312. [Google Scholar] [CrossRef]
- Xu, M.; Wang, L.V. Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 2006, 77, 041101. [Google Scholar] [CrossRef]
- Wang, L.V.; Wu, H.i. Biomedical Optics: Principles and Imaging; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Li, C.; Wang, L.V. Photoacoustic tomography and sensing in biomedicine. Phys. Med. Biol. 2009, 54, R59–R97. [Google Scholar] [CrossRef]
- Wang, L.V.; Hu, S. Photoacoustic tomography: In vivo imaging from organelles to organs. Science 2012, 335, 1458–1462. [Google Scholar] [CrossRef]
- Alhamami, M.; Kolios, M.C.; Tavakkoli, J. Photoacoustic detection and optical spectroscopy of high-intensity focused ultrasound-induced thermal lesions in biologic tissue. Med. Phys. 2014, 41, 053502. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Sanders, J.; Dundar, M.; Oralkan, Ö. Multi-wavelength photoacoustic imaging for monitoring lesion formation during high-intensity focused ultrasound therapy. Proc. SPIE 2017, 10064, 100644A. [Google Scholar]
- Sun, Y.; O’Neill, B. Imaging high-intensity focused ultrasound-induced tissue denaturation by multispectral photoacoustic method: An ex vivo study. Appl. Opt. 2013, 52, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Iskander-Rizk, S.; Kruizinga, P.; van der Steen, A.F.; van Soest, G. Spectroscopic photoacoustic imaging of radiofrequency ablation in the left atrium. Biomed. Opt. Express 2018, 9, 1309–1322. [Google Scholar] [CrossRef]
- Gray, J.; Dana, N.; Dextraze, K.; Maier, F.; Emelianov, S.; Bouchard, R. Multi-wavelength photoacoustic visualization of high intensity focused ultrasound lesions. Ultrason. Imaging 2016, 38, 96–112. [Google Scholar] [CrossRef]
- Vezhnevets, A.; Buhmann, J.M. Towards weakly supervised semantic segmentation by means of multiple instance and multitask learning. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; pp. 3249–3256. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Lin, G.; Milan, A.; Shen, C.; Reid, I. Refinenet: Multi-path refinement networks for high-resolution semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Kim, K.S.; Choi, H.H.; Moon, C.S.; Mun, C.W. Comparison of k-nearest neighbor, quadratic discriminant and linear discriminant analysis in classification of electromyogram signals based on the wrist-motion directions. Curr. Appl. Phys. 2011, 11, 740–745. [Google Scholar] [CrossRef]
- Wilamowski, B.M. Neural network architectures and learning algorithms. IEEE Ind. Electron. Mag. 2009, 3, 56–63. [Google Scholar] [CrossRef]
- Suykens, J.A.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Yoon, H.; Chang, C.; Jang, J.H.; Bhuyan, A.; Choe, J.W.; Nikoozadeh, A.; Watkins, R.D.; Stephens, D.N.; Butts Pauly, K.; Khuri-Yakub, B.T. Ex Vivo HIFU Experiments Using a 32×32-Element CMUT Array. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2016, 63, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Wittkampf, F.H.; Hauer, R.N.; Robles de Medina, E. Control of radiofrequency lesion size by power regulation. Circulation 1989, 80, 962–968. [Google Scholar] [CrossRef]
- Thomenius, K.E. Evolution of ultrasound beamformers. In Proceedings of the 1996 IEEE Ultrasonics Symposium, San Antonio, TX, USA, 3–6 November 1996; Volume 2, pp. 1615–1622. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Powers, D.M. Evaluation: From precision, recall and f-measure to roc., informedness, markedness & correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Cherkassky, V.; Mulier, F.M. Learning from Data: Concepts, Theory, and Methods; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for Activation Functions. In Proceedings of the ICLR 2018 Conference, Vancouver, BC, USA, 30 April–3 May 2018. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the Symposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; Volume 16, pp. 265–283. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Sequential Feature Selection—MATLAB Sequentialfs. 2018. Available online: https://www.mathworks.com/help/stats/sequentialfs.html (accessed on 1 January 2008).
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Shi, S.; Wang, Q.; Xu, P.; Chu, X. Benchmarking state-of-the-art deep learning software tools. In Proceedings of the 7th International Conference on Cloud Computing and Big Data, Macau, China, 16–18 November 2016; pp. 99–104. [Google Scholar]
- Zhang, C.; Li, P.; Sun, G.; Guan, Y.; Xiao, B.; Cong, J. Optimizing FPGA-based accelerator design for deep convolutional neural networks. In Proceedings of the 2015 ACM/SIGDA International Symposium on Field-Programmable Gate Arrays, Monterey, CA, USA, 22–24 February 2015; pp. 161–170. [Google Scholar]
- Abdiansah, A.; Wardoyo, R. Time complexity analysis of support vector machines (SVM) in LibSVM. Int. J. Comput. Appl. 2015, 128, 28–34. [Google Scholar] [CrossRef]
- Kothapalli, S.; Ma, T.; Vaithilingam, S.; Oralkan, Ö.; Khuri-Yakub, B.T.; Gambhir, S.S. Deep Tissue Photoacoustic Imaging Using a Miniaturized 2-D Capacitive Micromachined Ultrasonic Transducer Array. IEEE Trans. Biomed. Eng. 2012, 59, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Chang, C.; Rasmussen, M.F.; Moini, A.; Brenner, K.; Stephens, D.N.; Oralkan, Ö.; Khuri-Yakub, B. Integration of a dual-mode catheter for ultrasound image guidance and HIFU ablation using a 2-D CMUT array. In Proceedings of the 2017 IEEE International Ultrasonics Symposium, Washington, DC, USA, 6–9 September 2017; pp. 1–4. [Google Scholar] [CrossRef]
| Linear array: | |
| Center frequency | 5.2 MHz |
| Number of elements | 128 |
| Size (azimuth) | 38 mm |
| Imaging system: | |
| Sampling rate | 62.5 MS/s |
| Number of receive channels | 64 |
| Multiplexing capability | 2:1 |
| Number of acquisitions for average | 100 |
| Laser system: | |
| Pulse width | 5 ns |
| Repetition rate | 20 Hz * |
| Imaging wavelength | 690 nm to 950 nm |
| Scanning step size | 5 nm |
| Number of wavelengths | 53 |
| Maximum pulse energy | 14.74 mJ (715 nm) ** |
| Minimum pulse energy | 7.93 mJ (950 nm) ** |
| Dual-output fiber: | |
| Aperture (per branch) | 0.25 mm × 40 mm |
| Imaging frequency | 5.2 MHz |
| Distance between pixels | 73.92 m ( wavelength) |
| Size of the image view | 3.78 (width) cm × 2.5 (height) cm |
| Number of pixels | 512 (width) × 339 (height) |
| Layer 1–3: | |
| Convolution kernel size | 3 × 3 |
| Convolution stride | 1 × 1 |
| Activation function | relu |
| Max-pooling kernel size | 2 × 2 |
| Max-pooling stride | 2 × 2 |
| Layer 4–5: | |
| Convolution kernel size | 1 × 1 |
| Convolution stride | 1 × 1 |
| Activation function | relu |
| Number of Wavelengths | QDA | NN | SVM | RF | CNN |
|---|---|---|---|---|---|
| 53 | 91.34 | 92.62 | 92.65 | 89.93 | 99.63 |
| 5 | 84.77 | 85.42 | 85.51 | 85.13 | 96.26 |
| 4 | 84.64 | 84.68 | 84.76 | 84.67 | 94.05 |
| 3 | 82.71 | 82.77 | 83.08 | 82.60 | 96.60 |
| 2 | 78.45 | 78.16 | 78.74 | 78.18 | 92.79 |
| Number of Wavelengths | QDA | NN | SVM | RF | CNN |
|---|---|---|---|---|---|
| from 53 to 5 | 6.57 | 7.20 | 7.14 | 4.80 | 3.37 |
| from 5 to 4 | 0.13 | 0.74 | 0.75 | 0.46 | 2.21 |
| from 4 to 3 | 1.93 | 1.91 | 1.68 | 2.07 | −2.55 |
| from 3 to 2 | 4.26 | 4.61 | 4.34 | 4.42 | 3.81 |
| Number of Wavelengths | QDA | NN | SVM | RF | CNN |
|---|---|---|---|---|---|
| 53 | 377.99 | 357.57 | >1000 | 322.55 | 504.20 |
| 5 | 45.96 | 127.61 | >1000 | 307.75 | 170.21 |
| 4 | 42.29 | 118.73 | >1000 | 303.97 | 163.19 |
| 3 | 34.99 | 112.87 | >1000 | 271.98 | 161.82 |
| 2 | 33.65 | 108.06 | >1000 | 248.31 | 157.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Sanders, J.L.; Dundar, M.M.; Oralkan, Ö. Deep-Learning-Based High-Intensity Focused Ultrasound Lesion Segmentation in Multi-Wavelength Photoacoustic Imaging. Bioengineering 2023, 10, 1060. https://doi.org/10.3390/bioengineering10091060
Wu X, Sanders JL, Dundar MM, Oralkan Ö. Deep-Learning-Based High-Intensity Focused Ultrasound Lesion Segmentation in Multi-Wavelength Photoacoustic Imaging. Bioengineering. 2023; 10(9):1060. https://doi.org/10.3390/bioengineering10091060
Chicago/Turabian StyleWu, Xun, Jean L. Sanders, M. Murat Dundar, and Ömer Oralkan. 2023. "Deep-Learning-Based High-Intensity Focused Ultrasound Lesion Segmentation in Multi-Wavelength Photoacoustic Imaging" Bioengineering 10, no. 9: 1060. https://doi.org/10.3390/bioengineering10091060
APA StyleWu, X., Sanders, J. L., Dundar, M. M., & Oralkan, Ö. (2023). Deep-Learning-Based High-Intensity Focused Ultrasound Lesion Segmentation in Multi-Wavelength Photoacoustic Imaging. Bioengineering, 10(9), 1060. https://doi.org/10.3390/bioengineering10091060







