MRI-Based Deep Learning Method for Classification of IDH Mutation Status
Abstract
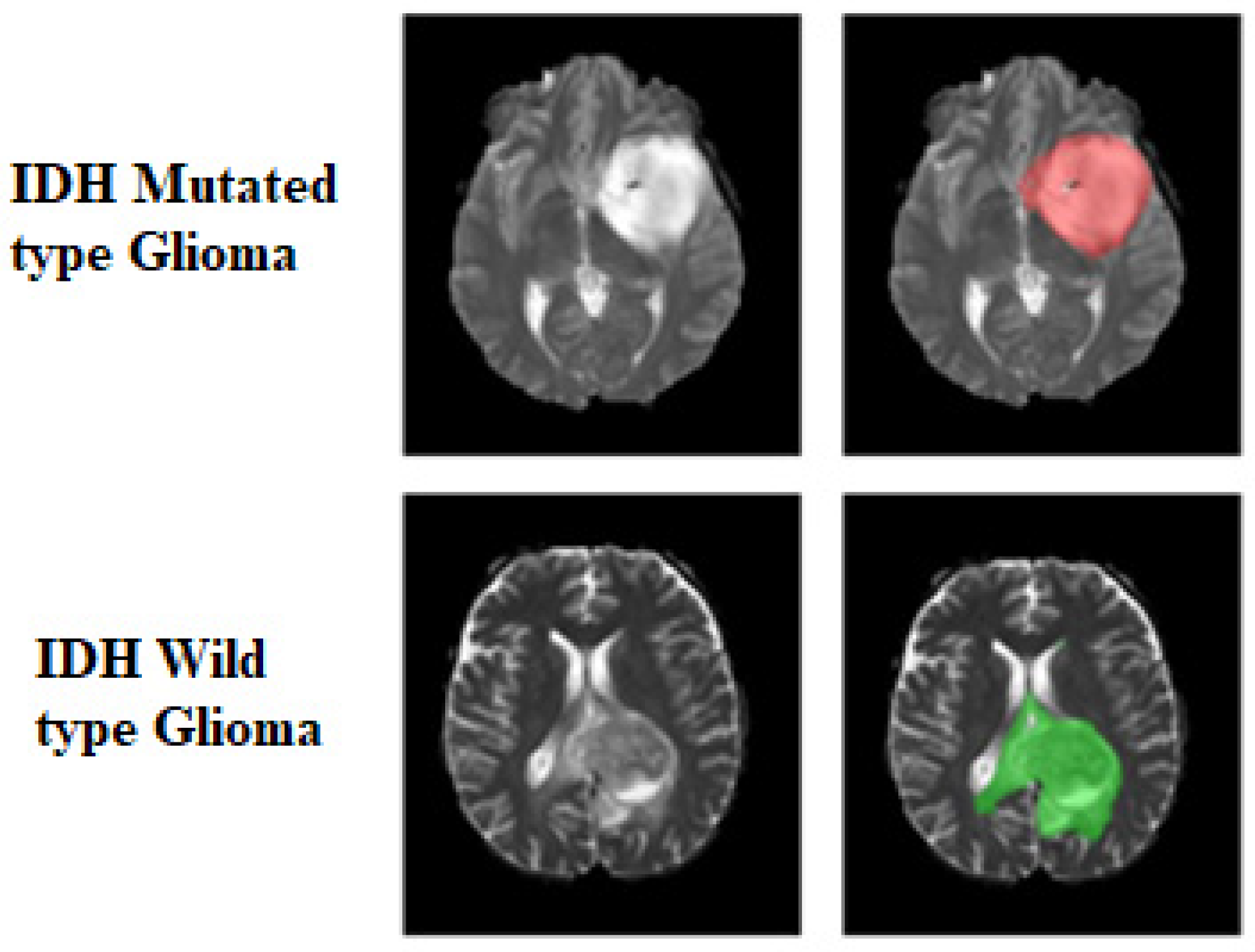
1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. Training Data
2.1.2. Testing Data
2.2. Pre-Processing
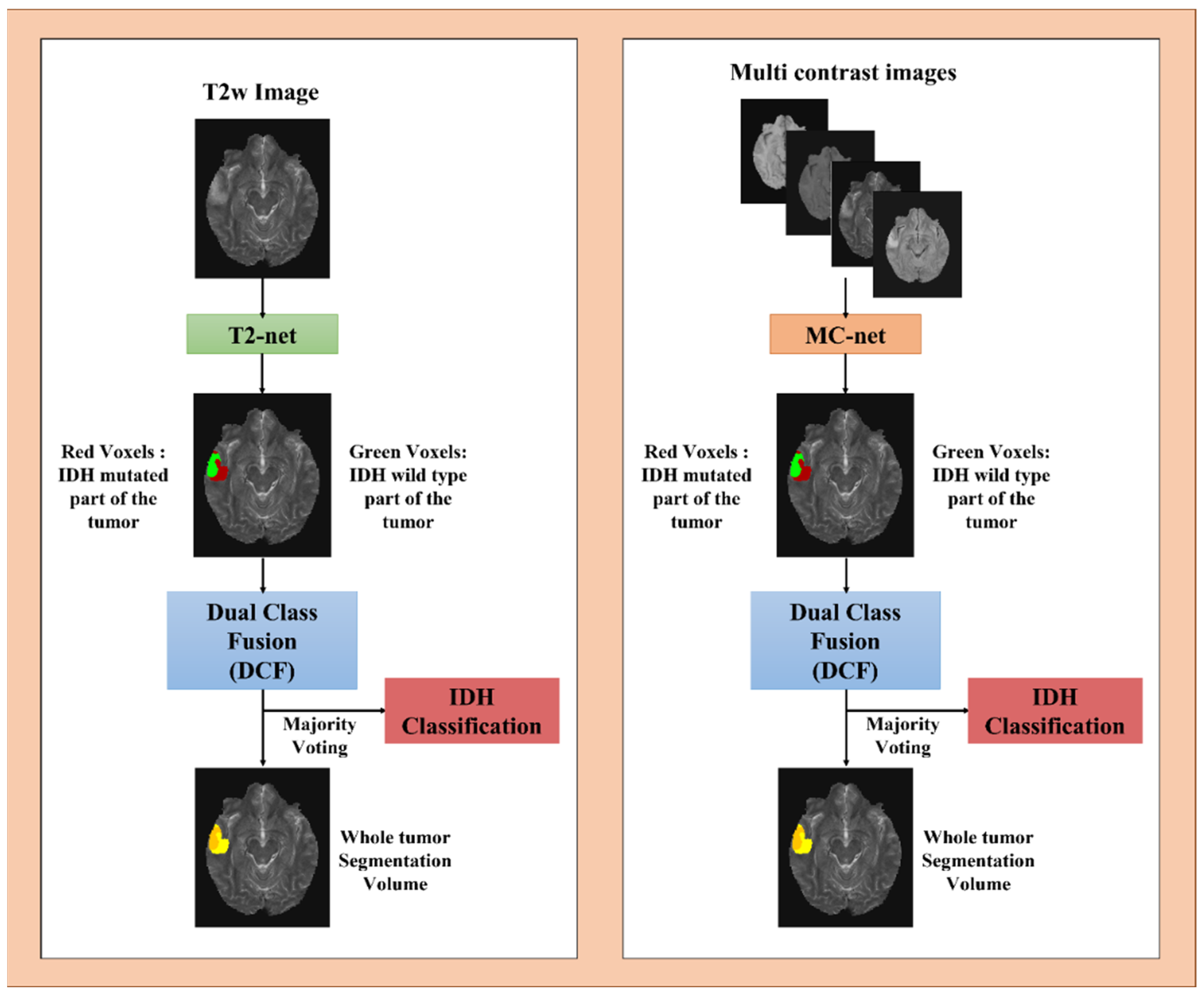
2.3. Network Details
2.4. Network Implementation and Cross-Validation
2.5. Testing Procedure
2.6. Statistical Analysis
3. Results
3.1. T2-Net
3.2. MC-Net
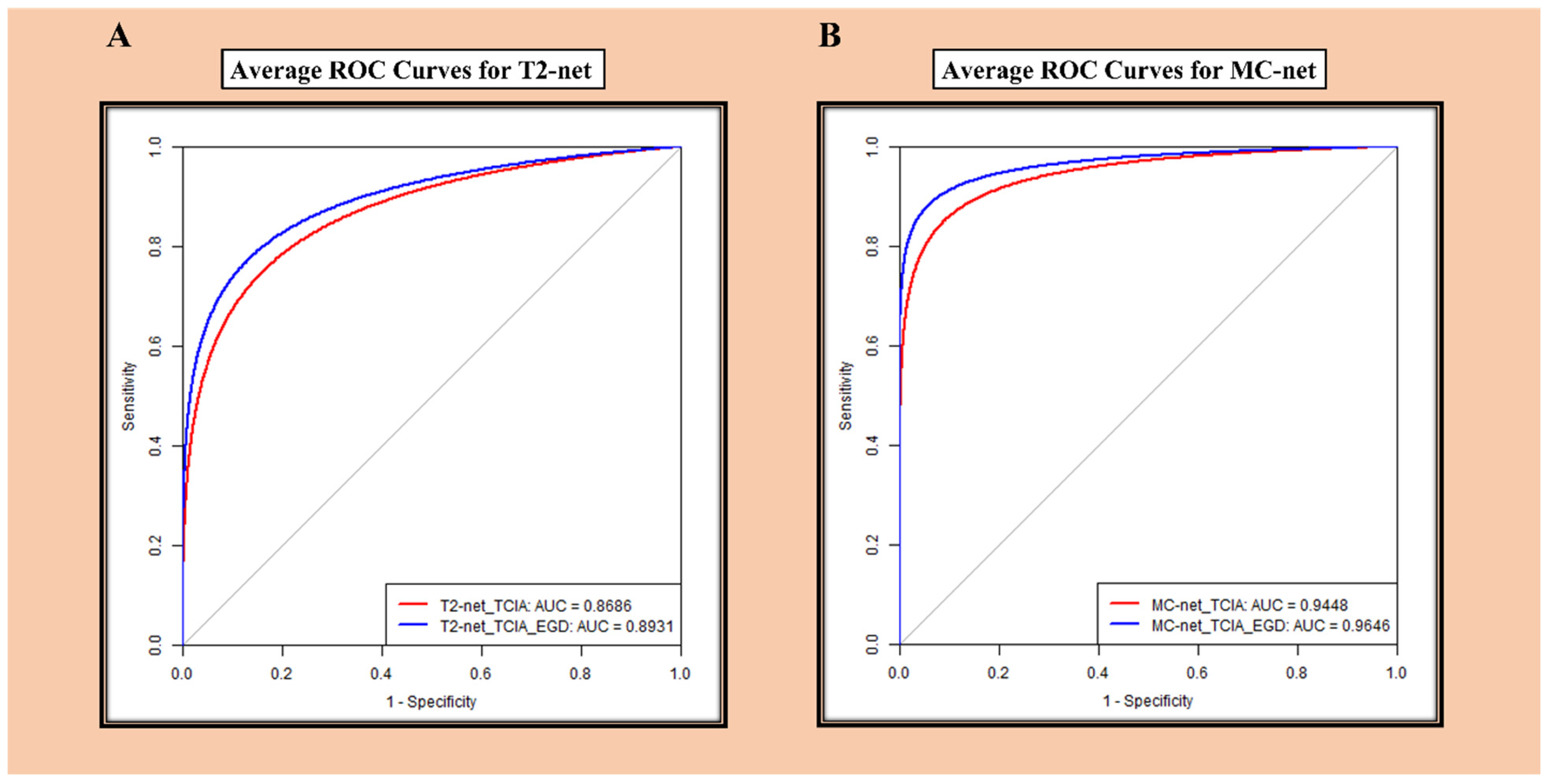
3.3. ROC Analysis
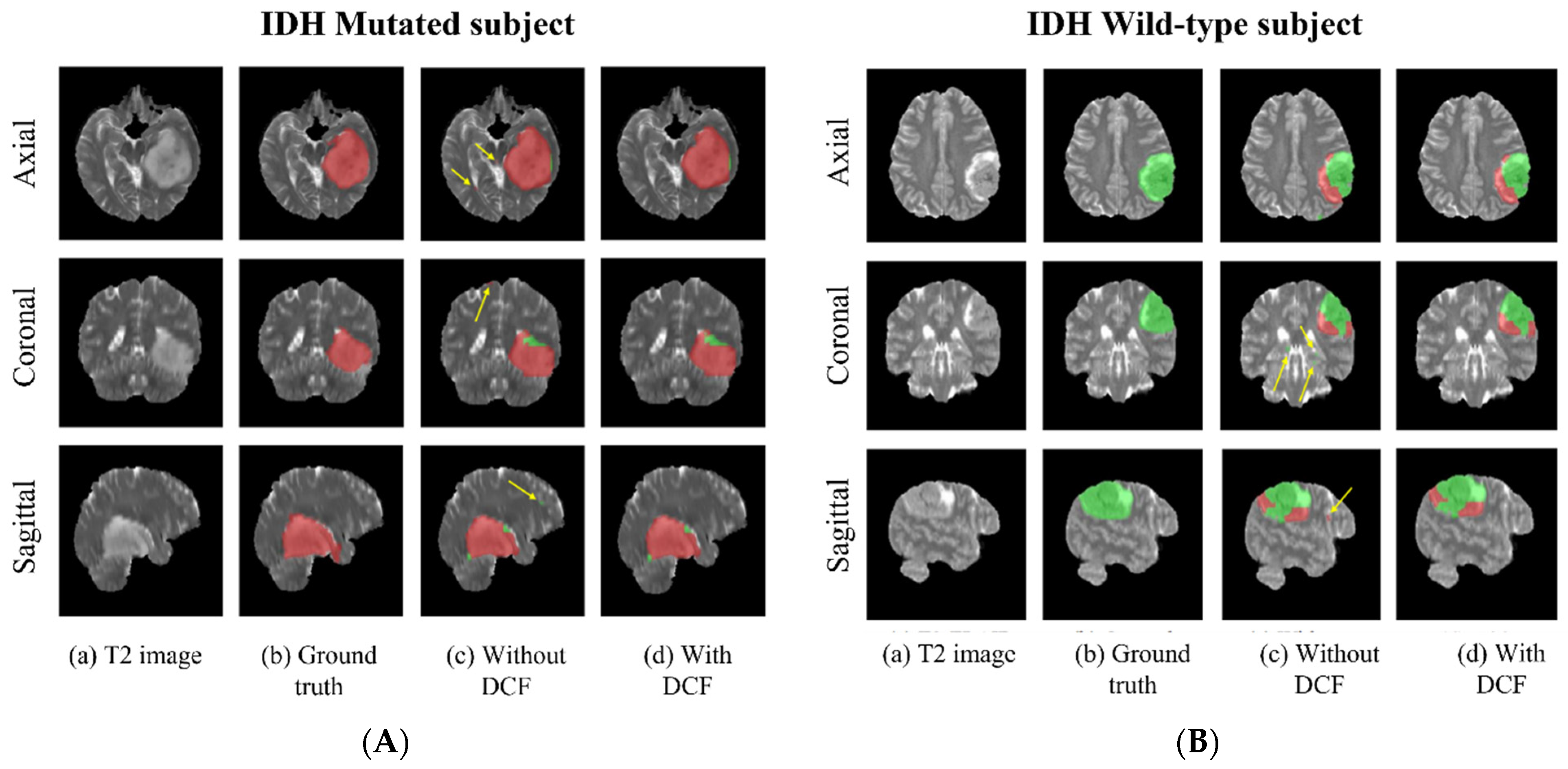
3.4. Voxel-Wise Classification
3.5. Training and Segmentation Times
4. Discussion
5. Future Work
6. Conclusions
7. Importance of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B.; Prins, R.M.; Thomas, M.A.; Nagarajan, R.; Yen, K.E.; Bittinger, M.A.; Salamon, N.; Chou, A.P.; Yong, W.H.; Soto, H. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J. Neuro-Oncol. 2012, 107, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.K.; DeBerardinis, R.J.; Hatanpaa, K.J.; Rakheja, D.; Kovacs, Z.; Yang, X.-L.; Mashimo, T.; Raisanen, J.M.; Marin-Valencia, I. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 2012, 18, 624–629. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, M.I.; Young, R.J.; Rubel, J.; Rosenblum, M.; Tisnado, J.; Briggs, S.; Arevalo-Perez, J.; Cross, J.R.; Campos, C.; Straley, K. Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro-Oncol. 2015, 18, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Tietze, A.; Choi, C.; Mickey, B.; Maher, E.A.; Parm Ulhøi, B.; Sangill, R.; Lassen-Ramshad, Y.; Lukacova, S.; Østergaard, L.; von Oettingen, G. Noninvasive assessment of isocitrate dehydrogenase mutation status in cerebral gliomas by magnetic resonance spectroscopy in a clinical setting. J. Neurosurg. 2017, 128, 391–398. [Google Scholar] [CrossRef]
- Suh, C.H.; Kim, H.S.; Paik, W.; Choi, C.; Ryu, K.H.; Kim, D.; Woo, D.C.; Park, J.E.; Jung, S.C.; Choi, C.G.; et al. False-Positive Measurement at 2-Hydroxyglutarate MR Spectroscopy in Isocitrate Dehydrogenase Wild-Type Glioblastoma: A Multifactorial Analysis. Radiology 2019, 291, 752–762. [Google Scholar] [CrossRef]
- SongTao, Q.; Lei, Y.; Si, G.; YanQing, D.; HuiXia, H.; XueLin, Z.; LanXiao, W.; Fei, Y. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012, 103, 269–273. [Google Scholar] [CrossRef]
- Okita, Y.; Narita, Y.; Miyakita, Y.; Ohno, M.; Matsushita, Y.; Fukushima, S.; Sumi, M.; Ichimura, K.; Kayama, T.; Shibui, S. IDH1/2 mutation is a prognostic marker for survival and predicts response to chemotherapy for grade II gliomas concomitantly treated with radiation therapy. Int. J. Oncol. 2012, 41, 1325–1336. [Google Scholar] [CrossRef][Green Version]
- Mohrenz, I.V.; Antonietti, P.; Pusch, S.; Capper, D.; Balss, J.; Voigt, S.; Weissert, S.; Mukrowsky, A.; Frank, J.; Senft, C.; et al. Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis 2013, 18, 1416–1425. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Botman, D.; Smits, M.A.; Hira, V.V.; van Lith, S.A.; Stap, J.; Henneman, P.; Khurshed, M.; Lenting, K.; Mul, A.N.; et al. Radioprotection of IDH1-Mutated Cancer Cells by the IDH1-Mutant Inhibitor AGI-5198. Cancer Res. 2015, 75, 4790–4802. [Google Scholar] [CrossRef]
- Sulkowski, P.L.; Corso, C.D.; Robinson, N.D.; Scanlon, S.E.; Purshouse, K.R.; Bai, H.; Liu, Y.; Sundaram, R.K.; Hegan, D.C.; Fons, N.R.; et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 2017, 9, eaal2463. [Google Scholar] [CrossRef] [PubMed]
- Beiko, J.; Suki, D.; Hess, K.R.; Fox, B.D.; Cheung, V.; Cabral, M.; Shonka, N.; Gilbert, M.R.; Sawaya, R.; Prabhu, S.S.; et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro-Oncol. 2014, 16, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, Q.; Wang, L.; Liu, Y.; Li, B.; Liang, Z.; Gao, P.; Zheng, K.; Zhao, B.; Lu, H. Radiomics strategy for molecular subtype stratification of lower-grade glioma: Detecting IDH and TP53 mutations based on multimodal MRI. J. Magn. Reson. Imaging 2018, 48, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chang, K.; Ramkissoon, S.; Tanguturi, S.; Bi, W.L.; Reardon, D.A.; Ligon, K.L.; Alexander, B.M.; Wen, P.Y.; Huang, R.Y. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-Oncol. 2017, 19, 109–117. [Google Scholar] [CrossRef]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef]
- Wegmayr, V.; Aitharaju, S.; Buhmann, J. Classification of brain MRI with big data and deep 3D convolutional neural networks. In Medical Imaging 2018: Computer-Aided Diagnosis; Petrick, N., Mori, K., Eds.; SPIE: Bellingham, WA, USA, 2018; Volume 1057501. [Google Scholar] [CrossRef]
- Feng, X.; Yang, J.; Lipton, Z.C.; Small, S.A.; Provenzano, F.A.; Alzheimer’s Disease Neuroimaging Initiative. Deep Learning on MRI Affirms the Prominence of the Hippocampal Formation in Alzheimer’s Disease Classification. bioRxiv 2018, 2018, 456277. [Google Scholar] [CrossRef]
- Nalawade, S.; Murugesan, G.K.; Vejdani-Jahromi, M.; Fisicaro, R.A.; Bangalore Yogananda, C.G.; Wagner, B.; Mickey, B.; Maher, E.; Pinho, M.C.; Fei, B.; et al. Classification of brain tumor isocitrate dehydrogenase status using MRI and deep learning. J. Med. Imaging 2019, 6, 046003. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Dubbink, H.J.; Vincent, A.J.P.E.; van den Bent, M.J.; French, P.J.; et al. The Erasmus Glioma Database (EGD): Structural MRI Scans, WHO 2016 Subtypes, and Segmentations of 774 Patients with Glioma. Data Brief 2021, 37, 107191. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Dubbink, H.J.; Atmodimedjo, P.N.; Kros, J.M.; French, P.J.; Sanson, M.; Idbaih, A.; Wesseling, P.; Enting, R.; Spliet, W.; Tijssen, C.; et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: A report of the prospective randomized EORTC Brain Tumor Group 26951 phase III trial. Neuro-Oncol. 2016, 18, 388–400. [Google Scholar] [CrossRef]
- Cryan, J.B.; Haidar, S.; Ramkissoon, L.A.; Bi, W.L.; Knoff, D.S.; Schultz, N.; Abedalthagafi, M.; Brown, L.; Wen, P.Y.; Reardon, D.A.; et al. Clinical multiplexed exome sequencing distinguishes adult oligodendroglial neoplasms from astrocytic and mixed lineage gliomas. Oncotarget 2014, 5, 8083–8092. [Google Scholar] [CrossRef]
- Gutman, D.A.; Dunn, W.D., Jr.; Grossmann, P.; Cooper, L.A.; Holder, C.A.; Ligon, K.L.; Alexander, B.M.; Aerts, H.J. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology 2015, 57, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.; Villanueva-Meyer, J.E.; Rudie, J.D.; Rauschecker, A.M.; Baid, U.; Bakas, S.; Cha, S.; Mongan, J.T.; Hess, C.P. The University of California San Francisco Preoperative Diffuse Glioma MRI Dataset. Radiol. Artif. Intell. 2022, 4, e220058. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.V.; Pfefferbaum, A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 2010, 31, 798–819. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Tustison, N.J.; Cook, P.A.; Klein, A.; Song, G.; Das, S.R.; Duda, J.T.; Kandel, B.M.; van Strien, N.; Stone, J.R.; Gee, J.C.; et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage 2014, 99, 166–179. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Zhou, H.; Chang, K.; Bai, H.X.; Xiao, B.; Su, C.; Bi, W.L.; Zhang, P.J.; Senders, J.T.; Vallieres, M.; Kavouridis, V.K.; et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neurooncol. 2019, 142, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Yu, J.; Guo, Y.; Cao, W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci. Rep. 2017, 7, 5467. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-Oncol. 2021, 23, 304–313. [Google Scholar] [CrossRef]
- Matsui, Y.; Maruyama, T.; Nitta, M.; Saito, T.; Tsuzuki, S.; Tamura, M.; Kusuda, K.; Fukuya, Y.; Asano, H.; Kawamata, T. Prediction of lower-grade glioma molecular subtypes using deep learning. J. Neuro-Oncol. 2020, 146, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Karami, G.; Pascuzzo, R.; Figini, M.; Del Gratta, C.; Zhang, H.; Bizzi, A. Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers 2023, 15, 482. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Tagliente, E.; Dellepiane, F.; Lucignani, M.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Moltoni, G.; Nicolai, M.; et al. Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. J. Pers. Med. 2021, 11, 290. [Google Scholar] [CrossRef]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. arXiv 2017, arXiv:1709.00382. [Google Scholar]
- Delfanti, R.L.; Piccioni, D.E.; Handwerker, J.; Bahrami, N.; Krishnan, A.; Karunamuni, R.; Hattangadi-Gluth, J.A.; Seibert, T.M.; Srikant, A.; Jones, K.A.; et al. Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: Implications for IDH, 1p/19q and ATRX status. J. Neurooncol. 2017, 135, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.; Sowmya, A.; Oliver, R. Automated Brain Tumor Segmentation Using Multimodal Brain Scans: A Survey Based on Models Submitted to the BraTS 2012–2018 Challenges. IEEE Rev. Biomed. Eng. 2020, 13, 156–168. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S. The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Pusch, S.; Sahm, F.; Meyer, J.; Mittelbronn, M.; Hartmann, C.; von Deimling, A. Glioma IDH1 mutation patterns off the beaten track. Neuropathol. Appl. Neurobiol. 2011, 37, 428–430. [Google Scholar] [CrossRef]
- Lee, D.; Suh, Y.L.; Kang, S.Y.; Park, T.I.; Jeong, J.Y.; Kim, S.H. IDH1 mutations in oligodendroglial tumors: Comparative analysis of direct sequencing, pyrosequencing, immunohistochemistry, nested PCR and PNA-mediated clamping PCR. Brain Pathol. 2013, 23, 285–293. [Google Scholar] [CrossRef]
- Agarwal, S.; Sharma, M.C.; Jha, P.; Pathak, P.; Suri, V.; Sarkar, C.; Chosdol, K.; Suri, A.; Kale, S.S.; Mahapatra, A.K.; et al. Comparative study of IDH1 mutations in gliomas by immunohistochemistry and DNA sequencing. Neuro-Oncol. 2013, 15, 718–726. [Google Scholar] [CrossRef]
- Preusser, M.; Wohrer, A.; Stary, S.; Hoftberger, R.; Streubel, B.; Hainfellner, J.A. Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J. Neuropathol. Exp. Neurol. 2011, 70, 715–723. [Google Scholar] [CrossRef]
- Tanboon, J.; Williams, E.A.; Louis, D.N. The Diagnostic Use of Immunohistochemical Surrogates for Signature Molecular Genetic Alterations in Gliomas. J. Neuropathol. Exp. Neurol. 2015, 75, 4–18. [Google Scholar] [CrossRef]
- Horbinski, C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013, 125, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.D.; Tang, L.F.; Zerbe, B.; Kvale, M.N.; Kwok, P.Y.; Schaefer, C.; Risch, N. Estimating genotype error rates from high-coverage next-generation sequence data. Genome Res. 2014, 24, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, E.; Sandhu, G.S.; Jesberger, J.; Blackham, K.A.; Hsu, D.P.; Griswold, M.A.; Sunshine, J.L. Comparison of brain MR images at 1.5T using BLADE and rectilinear techniques for patients who move during data acquisition. AJNR Am. J. Neuroradiol. 2012, 33, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Korfiatis, P.; Kline, T.L.; Coufalova, L.; Lachance, D.H.; Parney, I.F.; Carter, R.E.; Buckner, J.C.; Erickson, B.J. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 2016, 43, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
| UTSW | NYU | UWM | EGD | UCSF | Total | |
|---|---|---|---|---|---|---|
| Mutated | 104 | 23 | 19 | 150 | 103 | 399 |
| Wildtype | 256 | 113 | 156 | 306 | 392 | 1223 |
| Total | 360 | 136 | 175 | 456 | 495 | 1622 |
| Network Type | Training Group | Metrics | UTSW | NYU | UWM | EGD | UCSF | Overall Accuracy | Overall AUC |
|---|---|---|---|---|---|---|---|---|---|
| T2-net | TCIA | Accuracy | 83.9 | 77.9 | 85.1 | 85.3 | 88.7 | 85.4 | 0.8686 |
| Sensitivity | 75.0 | 60.9 | 78.9 | 76.7 | 76.7 | 75.4 | |||
| Specificity | 87.5 | 81.4 | 85.9 | 89.5 | 91.8 | 88.6 | |||
| Dice Score | 0.83 ± 0.18 | 0.87 ± 0.14 | 0.84 ± 0.16 | 0.73 ± 0.19 | 0.82 ± 0.16 | 0.80 ± 0.18 | |||
| TCIA + EGD | Accuracy | 86.1 | 81.6 | 89.7 | - | 89.5 | 87.6 | 0.8931 | |
| Sensitivity | 81.7 | 60.9 | 84.2 | - | 70.9 | 75.5 | |||
| Specificity | 87.9 | 85.8 | 90.4 | - | 94.4 | 90.8 | |||
| Dice Score | 0.85 ± 0.15 | 0.88 ± 0.13 | 0.85 ± 0.15 | - | 0.83 ± 0.15 | 0.85 ± 0.15 |
| Network Type | Training Group | Metrics | UTSW | NYU | UWM | EGD | UCSF | Overall Accuracy | Overall AUC |
|---|---|---|---|---|---|---|---|---|---|
| MC-net | TCIA | Accuracy | 87.2 | 91.9 | 87.4 | 92.3 | 93.5 | 91.0 | 0.9448 |
| Sensitivity | 73.1 | 78.3 | 84.2 | 86.0 | 89.3 | 83.0 | |||
| Specificity | 93.0 | 94.7 | 87.8 | 95.4 | 94.5 | 93.6 | |||
| Dice Score | 0.90 ± 0.13 | 0.92 ± 0.10 | 0.91 ± 0.11 | 0.77 ± 0.17 | 0.87 ± 0.14 | 0.86 ± 0.15 | |||
| TCIA + EGD | Accuracy | 90.0 | 92.6 | 92.6 | - | 94.9 | 92.8 | 0.9646 | |
| Sensitivity | 80.8 | 79.3 | 68.4 | - | 88.3 | 82.3 | |||
| Specificity | 93.8 | 96.5 | 95.5 | - | 96.7 | 95.6 | |||
| Dice Score | 0.90 ± 0.13 | 0.92 ± 0.13 | 0.92 ± 0.07 | - | 0.87 ± 0.13 | 0.89 ± 0.13 |
| Network Type | Training Group | IDH Type | UTSW | NYU | UWM | EGD | UCSF | Overall |
|---|---|---|---|---|---|---|---|---|
| T2-net | TCIA | Mutant | 71.05 | 56.95 | 76.40 | 74.97 | 73.49 | 72.60 |
| Wildtype | 84.05 | 80.59 | 85.20 | 87.07 | 88.71 | 86.13 | ||
| TCIA + EGD | Mutant | 78.48 | 62.10 | 81.94 | - | 70.18 | 73.79 | |
| Wildtype | 84.63 | 83.03 | 87.65 | - | 91.99 | 88.09 | ||
| MC-net | TCIA | Mutant | 71.17 | 69.43 | 81.11 | 81.57 | 84.92 | 79.00 |
| Wildtype | 90.57 | 91.37 | 85.26 | 93.11 | 91.18 | 90.80 | ||
| TCIA + EGD | Mutant | 80.01 | 73.64 | 69.65 | - | 85.68 | 80.98 | |
| Wildtype | 91.71 | 94.15 | 93.33 | - | 93.50 | 93.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bangalore Yogananda, C.G.; Wagner, B.C.; Truong, N.C.D.; Holcomb, J.M.; Reddy, D.D.; Saadat, N.; Hatanpaa, K.J.; Patel, T.R.; Fei, B.; Lee, M.D.; et al. MRI-Based Deep Learning Method for Classification of IDH Mutation Status. Bioengineering 2023, 10, 1045. https://doi.org/10.3390/bioengineering10091045
Bangalore Yogananda CG, Wagner BC, Truong NCD, Holcomb JM, Reddy DD, Saadat N, Hatanpaa KJ, Patel TR, Fei B, Lee MD, et al. MRI-Based Deep Learning Method for Classification of IDH Mutation Status. Bioengineering. 2023; 10(9):1045. https://doi.org/10.3390/bioengineering10091045
Chicago/Turabian StyleBangalore Yogananda, Chandan Ganesh, Benjamin C. Wagner, Nghi C. D. Truong, James M. Holcomb, Divya D. Reddy, Niloufar Saadat, Kimmo J. Hatanpaa, Toral R. Patel, Baowei Fei, Matthew D. Lee, and et al. 2023. "MRI-Based Deep Learning Method for Classification of IDH Mutation Status" Bioengineering 10, no. 9: 1045. https://doi.org/10.3390/bioengineering10091045
APA StyleBangalore Yogananda, C. G., Wagner, B. C., Truong, N. C. D., Holcomb, J. M., Reddy, D. D., Saadat, N., Hatanpaa, K. J., Patel, T. R., Fei, B., Lee, M. D., Jain, R., Bruce, R. J., Pinho, M. C., Madhuranthakam, A. J., & Maldjian, J. A. (2023). MRI-Based Deep Learning Method for Classification of IDH Mutation Status. Bioengineering, 10(9), 1045. https://doi.org/10.3390/bioengineering10091045






