Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions
Abstract
:1. Introduction
- The main contributions of this paper are:
- To understand the activity of certain brain regions based on human emotions and to enable the interaction among brain regions and the combination of information, we propose a hierarchical neural network model structure with self-attention. The hierarchical deep neural network model with self-attention comprises a regional brain-level encoding module and a global brain-level encoding module;
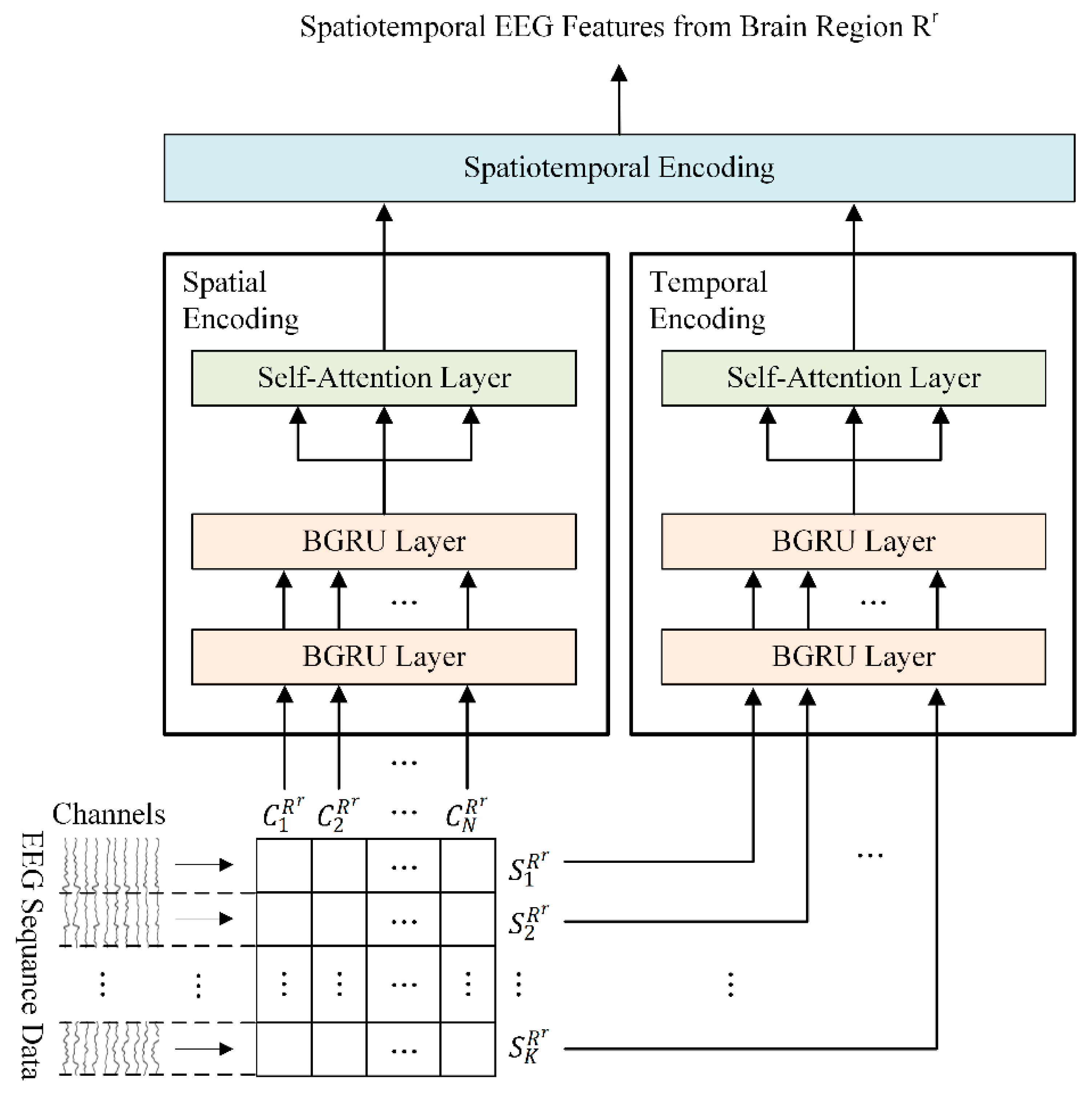
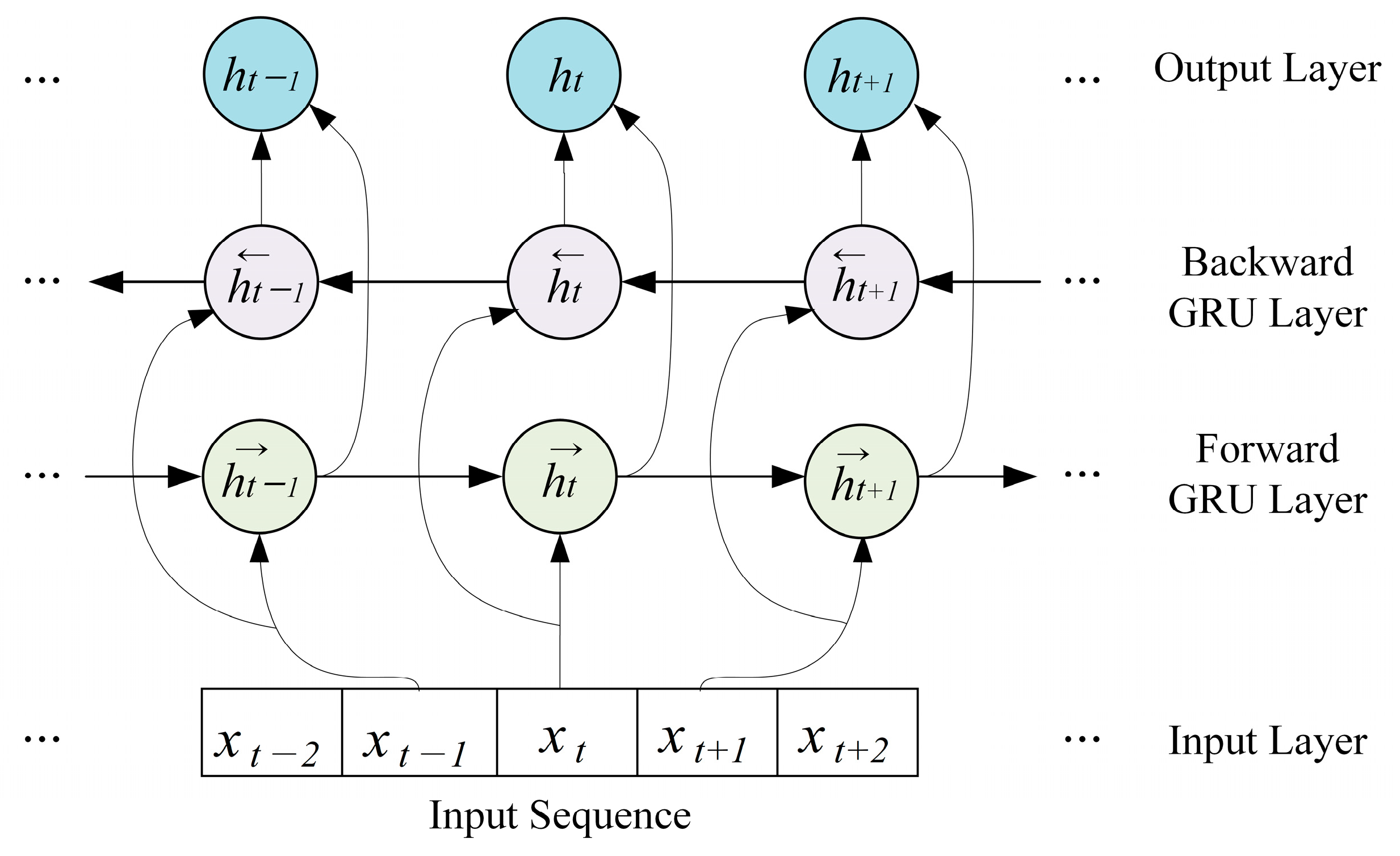
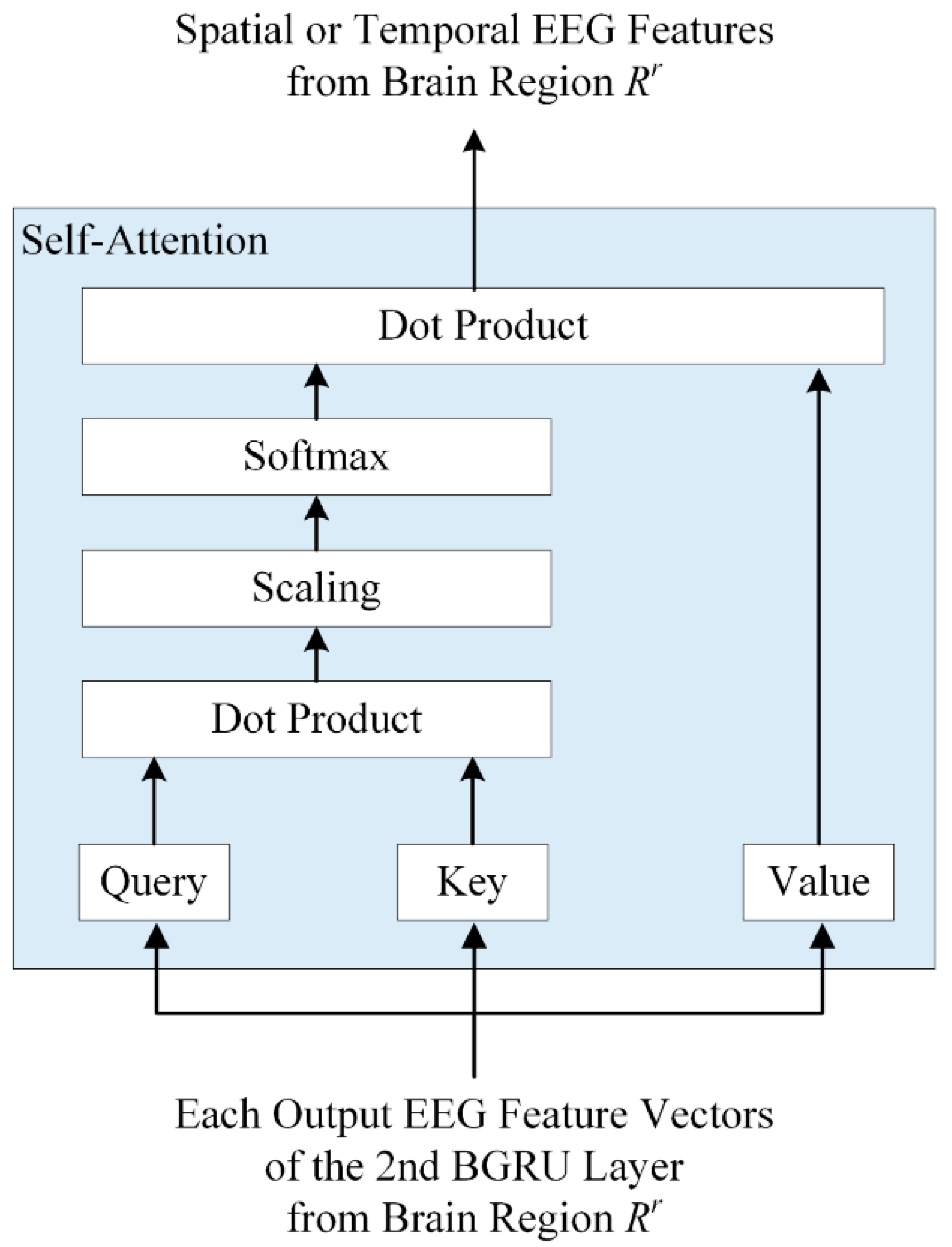
- To obtain activity weights for each brain region according to emotional state, a regional brain-level encoding module was designed based on a dual-stream parallel double BGRU encoder with self-attention (2BGRUA) for extracting spatiotemporal EEG features. In particular, the spatial encoder learns the interchannel correlations and the temporal encoder captures the temporal dependencies of the time sequence of the EEG channels. Subsequently, the output of each encoder can be integrated to obtain the local spatiotemporal EEG features;
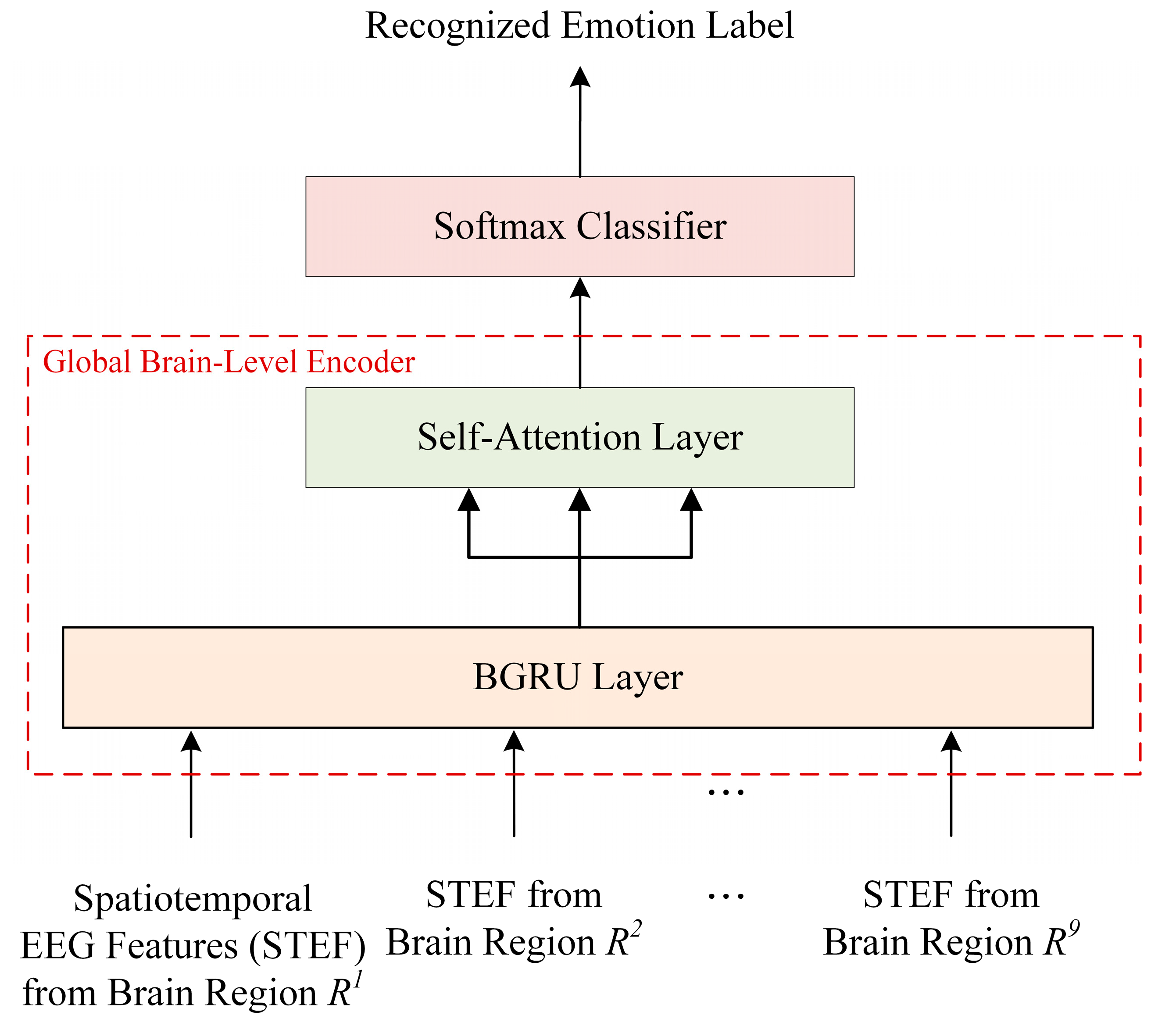
- Next, the global spatiotemporal encoding module uses a single BGRU-based self-attention (BGRUA) to integrate important information within various brain regions to improve the emotion classification performance by learning discriminative spatiotemporal EEG features from local brain regions to the entire brain region. Thus, the influence of brain regions with a high contribution is strengthened by the learned weights, and the influence of the less dependent regions is reduced.
2. Proposed Hierarchical Spatiotemporal Context Feature Learning Model for EEG-Based Emotion Recognition
2.1. Electrode Division
2.2. Preprocessing and Spectral Analysis
2.3. Regional Brain-Level Encoding Module
2.4. Global Brain-Level Encoding and Classification Module
3. Experiment and Results
3.1. Evaluation Datasets
- DEAP [37]: The DEAP dataset contains EEG and peripheral signals from 32 participants (16 males and 16 females between the ages of 19 and 37 years). EEG signals were recorded while each participant watched 40 music video clips. Each participant watched each video and rated the levels of arousal, valence, dominance, and liking on a continuous scale of 1 to 9 using a self-assessment manikin (SAM). Each trial contained a 63 s EEG signal. The first 3 s of the signal is the baseline signal. EEG signals were recorded at a sampling rate of 512 Hz using 32 electrodes. In this paper, EEG data from 24 participants (12 males and 12 females) were selected for the experiment;
- MAHNOB-HCI [38]: The MAHNOB-HCI dataset includes the EEG and peripheral signals. EEG signals were collected at a sampling rate of 256 Hz from 32 electrodes while each of the 27 participants (11 males and 16 females) watched 20 selected videos. The video clip used as the stimulus had a length of approximately 34–117 s. Each participant watched each video and self-reported the levels of arousal, valence, dominance, and predictability through the SAM on a nine-point discrete scale. For the various experiments performed in this study, 22 of the 27 participants (11 male and 11 female) were selected;
- SEED [39]: The SEED dataset provides EEG and eye-movement signals from 15 participants (7 males and 8 females). The EEG signals of each participant were collected while watching 15 Chinese movie clips, each approximately four minutes long, designed to elicit positive, neutral, and negative emotions. The sampling rate of the signal collected using the 62 electrodes was 1 kHz, which was later downsampled to 200 Hz. After watching each movie clip, each participant recorded the emotional label for each video as negative (−1), neutral (0), or positive (1). The experiment was performed using EEG data from 12 of the 15 participants (6 males and 6 females) were used.
3.2. Experimental Methods
- CNN: This method constituted two convolution layers with four 3 × 3 convolutional filters, two max-pooling layers with a 2 × 2 filter size, a dropout layer, two fully connected layers, and a softmax layer;
- LSTM: LSTM was applied rather than a CNN. It comprised two LSTM layers: a dropout, fully connected and a softmax layers. The number of hidden units of the lower- and upper-layer LSTMs were 128 and 64, respectively. The fully connected layer contained 128 hidden units;
- BGRU: BGRU was used rather than LSTM. The number of hidden units in the forward and backward GRU layers was 64;
- Convolutional recurrent neural network (CRNN) [24]: A hybrid neural network consisting of a CNN and an RNN for extracting spatiotemporal features was applied to multichannel EEG sequences. This network consisted of two convolution layers, a subsampling layer, two fully connected recurrent layers, and an output layer;
- Hierarchical-attention-based BGRU (HA-BGRU) [25]: The HA-BGRU consisted of two layers. The first layer encoded the local correlation between samples in each epoch of the EEG signal, and the second layer encoded the temporal correlation between epochs in an EEG sequence. The BGRU network and attention mechanism were applied at both sample and epoch levels;
- Region-to-global HA-BGRU (R2G HA-BGRU): the HA-BGRU network was applied to extract regional features within each brain region and global features between regions;
- R2G transformer encoder (TF-Encoder): in the R2G HA-BGRU method, transformer encoders were applied instead of BGRU with attention mechanism;
- R2G hierarchical spatial attention-based BGRU (HSA-BGRU): only temporal encoding in a regional brain-level encoding module was used, and the same method as the HSCFLM was applied for the rest;
- HSCFLM: This method mainly consisted of a regional brain-level encoding module, a global brain-level encoding, and classification module. This is the proposed method.
3.3. Experimental Results
4. Conclusions, Limitations, and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Powers, J.P.; LaBar, K.S. Regulating emotion through distancing: A taxonomy, neurocognitive model, and supporting meta-analysis. Neurosci. Biobehav. Rev. 2019, 96, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Nagesh, B.; Routray, A.; Sarma, M. A human–computer interaction framework for emotion recognition through time-series thermal video sequences. Comput. Electr. Eng. 2021, 93, 107280. [Google Scholar] [CrossRef]
- Fei, Z.; Yang, E.; Li, D.D.U.; Butler, S.; Ijomah, W.; Li, X.; Zhou, H. Deep convolution network based emotion analysis towards mental health care. Neurocomputing 2020, 388, 212–227. [Google Scholar] [CrossRef]
- McDuff, D.; El Kaliouby, R.; Cohn, J.F.; Picard, R.W. Predicting ad liking and purchase intent: Large-scale analysis of facial responses to ads. IEEE Trans. Affect. Comput. 2014, 6, 223–235. [Google Scholar] [CrossRef]
- Picard, R.W. Affective computing: Challenges. Int. J. Hum. Comput. 2003, 59, 55–64. [Google Scholar] [CrossRef]
- Tian, W. Personalized emotion recognition and emotion prediction system based on cloud computing. Math. Probl. Eng. 2021, 2021, 9948733. [Google Scholar] [CrossRef]
- Schirmer, A.; Adolphs, R. Emotion perception from face, voice, and touch: Comparisons and convergence. Trends Cogn. Sci. 2017, 21, 216–228. [Google Scholar] [CrossRef]
- Marinoiu, E.; Zanfir, M.; Olaru, V.; Sminchisescu, C. 3D Human Sensing, Action and Emotion Recognition in Robot Assisted Therapy of Children with Autism. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 2158–2167. [Google Scholar] [CrossRef]
- Sönmez, Y.Ü.; Varol, A. A speech emotion recognition model based on multi-level local binary and local ternary patterns. IEEE Access 2020, 8, 190784–190796. [Google Scholar] [CrossRef]
- Karnati, M.; Seal, A.; Bhattacharjee, D.; Yazidi, A.; Krejcar, O. Understanding deep learning techniques for recognition of human emotions using facial expressions: A comprehensive survey. IEEE Trans. Instrum. Meas. 2023, 72, 5006631. [Google Scholar] [CrossRef]
- Li, L.; Chen, J.H. Emotion recognition using physiological signals. In Proceedings of the International Conference on Artificial Reality and Telexistence (ICAT), Hangzhou, China, 28 November–1 December 2006; pp. 437–446. [Google Scholar] [CrossRef]
- Leelaarporn, P.; Wachiraphan, P.; Kaewlee, T.; Udsa, T.; Chaisaen, R.; Choksatchawathi, T.; Laosirirat, R.; Lakhan, P.; Natnithikarat, P.; Thanontip, K.; et al. Sensor-driven achieving of smart living: A review. IEEE Sens. J. 2021, 21, 10369–10391. [Google Scholar] [CrossRef]
- Qing, C.; Qiao, R.; Xu, X.; Cheng, Y. Interpretable emotion recognition using EEG signals. IEEE Access 2019, 7, 94160–94170. [Google Scholar] [CrossRef]
- Goshvarpour, A.; Abbasi, A.; Goshvarpour, A. An accurate emotion recognition system using ECG and GSR signals and matching pursuit method. Biomed. J. 2017, 40, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhou, W.; Cai, H.; Su, Y.; Wang, J.; Zhang, Z.; Lei, T. Ubiquitous depression detection of sleep physiological data by using combination learning and functional networks. IEEE Access 2020, 8, 94220–94235. [Google Scholar] [CrossRef]
- Alarcao, S.M.; Fonseca, M.J. Emotions recognition using EEG signals: A survey. IEEE Trans. Affect. 2017, 10, 374–393. [Google Scholar] [CrossRef]
- Gómez, A.; Quintero, L.; López, N.; Castro, J. An approach to emotion recognition in single-channel EEG signals: A mother child interaction. J. Phys. Conf. Ser. 2016, 705, 012051. [Google Scholar] [CrossRef]
- Li, P.; Liu, H.; Si, Y.; Li, C.; Li, F.; Zhu, X.; Xu, P. EEG based emotion recognition by combining functional connectivity network and local activations. IEEE Trans. Biomed. Eng. 2019, 66, 2869–2881. [Google Scholar] [CrossRef]
- Wang, X.W.; Nie, D.; Lu, B.L. Emotional state classification from EEG data using machine learning approach. Neurocomputing 2014, 129, 94–106. [Google Scholar] [CrossRef]
- Gupta, V.; Chopda, M.D.; Pachori, R.B. Cross-subject emotion recognition using flexible analytic wavelet transform from EEG signals. IEEE Sens. J. 2018, 19, 2266–2274. [Google Scholar] [CrossRef]
- Galvão, F.; Alarcão, S.M.; Fonseca, M.J. Predicting exact valence and arousal values from EEG. Sensors 2021, 21, 3414. [Google Scholar] [CrossRef]
- Seal, A.; Reddy, P.P.N.; Chaithanya, P.; Meghana, A.; Jahnavi, K.; Krejcar, O.; Hudak, R. An EEG database and its initial benchmark emotion classification performance. Comput. Math. Methods Med. 2020, 2020, 8303465. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; He, H. Implementation of EEG emotion recognition system based on hierarchical convolutional neural networks. In Proceedings of the Advances in Brain Inspired Cognitive Systems: 8th International Conference (BICS), Beijing, China, 28–30 November 2016; pp. 22–33. [Google Scholar] [CrossRef]
- Li, X.; Song, D.; Zhang, P.; Yu, G.; Hou, Y.; Hu, B. Emotion recognition from multi-channel EEG data through convolutional recurrent neural network. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 352–359. [Google Scholar] [CrossRef]
- Chen, J.X.; Jiang, D.M.; Zhang, Y.N. A hierarchical bidirectional GRU model with attention for EEG-based emotion classification. IEEE Access 2019, 7, 118530–118540. [Google Scholar] [CrossRef]
- Etkin, A.; Egner, T.; Kalisch, R. Emotional processing in anterior cingulate and medial prefrontal cortex. Trends Cogn. Sci. 2011, 15, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Lotze, M.; Erb, M.; Grodd, W.; Birbaumer, N. Brain activity underlying emotional valence and arousal: A response-related fMRI study. Hum. Brain Mapp. 2004, 23, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Heller, W.; Nitscke, J.B. Regional brain activity in emotion: A framework for understanding cognition in depresion. Cogn. Emot. 1997, 11, 637–661. [Google Scholar] [CrossRef]
- Davidson, R.J. Affective style, psychopathology, and resilience: Brain mechanisms and plasticity. Am. Psychol. 2000, 55, 1196. [Google Scholar] [CrossRef]
- Lindquist, K.A.; Wager, T.D.; Kober, H.; Bliss-Moreau, E.; Barrett, L.F. The brain basis of emotion: A meta-analytic review. Behav. Brain Sci. 2012, 35, 121–143. [Google Scholar] [CrossRef]
- Zhang, P.; Min, C.; Zhang, K.; Xue, W.; Chen, J. Hierarchical spatiotemporal electroencephalogram feature learning and emotion recognition with attention-based antagonism neural network. Front. Neurosci. 2021, 15, 738167. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Hu, C.; Yin, Z.; Song, Y. Transformers for EEG-based emotion recognition: A hierarchical spatial information learning model. IEEE Sens. J. 2022, 22, 4359–4368. [Google Scholar] [CrossRef]
- Ribas, G.C. The cerebral sulci and gyri. Neurosurg. Focus 2010, 28, E2. [Google Scholar] [CrossRef]
- Cohen, I.; Berdugo, B. Speech enhancement for non-stationary noise environments. Signal Process. 2001, 81, 2403–2418. [Google Scholar] [CrossRef]
- Dorran, D. Audio Time-Scale Modification. Ph.D. Thesis, Dublin Institute of Technology, Dublin, Ireland, 2005. [Google Scholar] [CrossRef]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Patras, I. DEAP: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Soleymani, M.; Lichtenauer, J.; Pun, T.; Pantic, M. A multimodal database for affect recognition and implicit tagging. IEEE Trans. Affect. Comput. 2011, 3, 42–55. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans. Auton. Ment. Develop. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Personalizing EEG-based affective models with transfer learning. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence (IJCAI), New York, NY, USA, 9–15 July 2016; pp. 2732–2738. [Google Scholar]


| Rating Values (RVs) | Valence | Arousal | Dominance |
|---|---|---|---|
| Low | Low | Low | |
| High | High | High |
| Rating Values (RVs) | Valence | Arousal | Dominance |
|---|---|---|---|
| Negative | Activated | Controlled | |
| Neutral | Moderate | Moderate | |
| Positive | Deactivated | Overpowered |
| Methods | Two-Level CL | Three-Level CL | ||||
|---|---|---|---|---|---|---|
| VAL | ARO | DOM | VAL | ARO | DOM | |
| CNN | 69.7 (11.82) | 66.7 (9.50) | 70.1 (11.84) | 65.3 (10.09) | 64.7 (10.43) | 65.9 (8.94) |
| LSTM | 75.2 (11.56) | 72.3 (10.30) | 75.3 (9.11) | 71.1 (8.89) | 69.7 (11.13) | 71.6 (9.46) |
| BGRU | 76.8 (11.62) | 74.4 (11.45) | 77.2 (8.96) | 73.2 (9.02) | 71.5 (8.60) | 73.1 (8.81) |
| CRNN [24] | 81.1 (9.21) | 78.9 (11.31) | 81.4 (11.01) | 77.4 (11.04) | 76.1 (10.43) | 77.4 (10.58) |
| HA-BGRU [25] | 83.4 (10.26) | 81.5 (10.04) | 84.1 (9.42) | 80.2 (9.48) | 78.8 (10.12) | 80.1 (11.34) |
| R2G HA-BGRU | 87.6 (9.73) | 85.3 (9.46) | 88.1 (11.66) | 83.9 (10.54) | 82.9 (11.14) | 84.1 (10.92) |
| R2G TF-Encoder | 88.1 (10.05) | 86.4 (8.37) | 88.4 (9.55) | 84.5 (10.95) | 83.5 (11.29) | 84.5 (11.14) |
| R2G HSA-BGRU | 88.4 (8.95) | 87.1 (10.79) | 89.3 (10.59) | 85.1 (9.31) | 84.2 (10.43) | 85.2 (11.20) |
| HSCFLM | 92.1 (9.16) | 90.5 (9.81) | 92.3 (8.94) | 88.5 (8.52) | 87.4 (8.35) | 88.3 (9.76) |
| Methods | Two-Level CL | Three-Level CL | ||||
|---|---|---|---|---|---|---|
| VAL | ARO | DOM | VAL | ARO | DOM | |
| HA-BGRU [25] | 84.6 (10.90) | 82.7 (8.25) | 84.3 (9.67) | 80.2 (8.66) | 79.8 (9.60) | 80.7 (9.61) |
| R2G HA-BGRU | 88.6 (8.57) | 87.1 (8.64) | 88.3 (9.95) | 84.5 (11.30) | 84.4 (10.45) | 84.8 (9.48) |
| R2G TF-Encoder | 89.2 (8.07) | 87.8 (10.74) | 88.9 (8.86) | 85.1 (10.32) | 84.9 (11.27) | 85.4 (10.90) |
| R2G HSA-BGRU | 90.1 (10.22) | 88.5 (10.95) | 89.5 (9.65) | 85.8 (9.89) | 85.6 (8.42) | 86.1 (11.13) |
| HSCFLM | 93.3 (9.74) | 91.6 (10.71) | 92.8 (8.99) | 88.9 (10.62) | 89.1 (8.89) | 89.4 (10.38) |
| Methods | Four-Level CL (HAHV vs. LAHV vs. HALV vs. LALV) | Three-Level CL (VAL) | |
|---|---|---|---|
| DEAP | MAHNOB-HCI | SEED | |
| HA-BGRU [25] | 74.9 (10.86) | 75.2 (8.76) | 81.9 (11.59) |
| R2G HA-BGRU | 79.4 (11.03) | 79.0 (11.46) | 85.8 (12.10) |
| R2G TF-Encoder | 78.9 (8.11) | 79.8 (11.83) | 86.8 (9.89) |
| R2G HSA-BGRU | 79.5 (10.16) | 80.4 (10.34) | 87.3 (9.58) |
| HSCFLM | 83.2 (9.04) | 83.9 (9.86) | 90.9 (10.15) |
| Module | Number of Network Parameters | Number of Additions | Number of Multiplications | |
|---|---|---|---|---|
| Regional Brain-Level Encoding | Temporal Encoding | 350,976 | 124,489 | 13,568,576 |
| Spatial Encoding | 527,616 | 249,864 | 627,200 | |
| Global Brain-Level Encoding and Classification | 639,744 | 269,097 | 3,805,440 | |
| Total | 1,518,336 | 643,450 | 18,001,216 | |
| Model | Network Parameters | Training Time (h:m:s) | Test Time Per EEG Data (ms) | Network Size (MB) | Average Accuracy (%) |
|---|---|---|---|---|---|
| HSCFLM | 1,518,336 | 02:59:07 | 25 | 14.8 | 83.2 |
| Methods | Four-Level CL (HAHV vs. LAHV vs. HALV vs. LALV) | Three-Level CL (VAL) | ||||
|---|---|---|---|---|---|---|
| DEAP | MAHNOB-HCI | SEED | ||||
| ACC (STD) | p-Value | ACC (STD) | p-Value | ACC (STD) | p-Value | |
| HA-BGRU [25] | 83.6 (9.85) | 0.038 | 83.8 (11.56) | 0.031 | 88.3 (10.06) | 0.032 |
| R2G HA-BGRU | 88.1 (11.28) | 0.016 | 87.7 (10.37) | 0.015 | 91.9 (12.28) | 0.024 |
| R2G TF-Encoder | 87.8 (9.20) | 0.017 | 88.5 (9.43) | 0.018 | 92.9 (10.19) | 0.017 |
| R2G HSA-BGRU | 88.4 (10.79) | 0.015 | 89.3 (11.14) | 0.012 | 93.7 (9.56) | 0.014 |
| HSCFLM | 92.4 (9.06) | 93.1 (8.95) | 97.2 (10.11) | |||
| Brain Regions | DEAP Two-Level CL | MAHNOB-HCI Two-Level CL | SEED Three-Level CL | ||
|---|---|---|---|---|---|
| VAL | ARO | VAL | ARO | VAL | |
| R1 (Prefrontal) | 86.8 | 86.5 | 87.7 | 86.5 | 85.4 |
| R2 (Frontal) | 86.7 | 86.1 | 87.3 | 86.4 | 85.2 |
| R3 (Left Temporal) | 86.1 | 85.9 | 86.5 | 85.8 | 85.3 |
| R4 (Right Temporal) | 86.2 | 84.3 | 86.2 | 84.8 | 85.7 |
| R5 (Central) | 85.8 | 85.4 | 86.9 | 85.7 | 84.5 |
| R6 (Left Parietal) | 84.1 | 84.8 | 84.1 | 85.4 | 83.6 |
| R7 (Parietal) | 85.7 | 86.2 | 85.4 | 86.2 | 84.5 |
| R8 (Right Parietal) | 85.3 | 84.6 | 85.7 | 86.8 | 84.3 |
| R9 (Occipital) | 83.6 | 83.1 | 84.1 | 83.5 | 82.7 |
| All Regions | 92.1 | 90.5 | 93.3 | 91.6 | 90.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, D.-K.; Kim, H.-G.; Kim, J.-Y. Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions. Bioengineering 2023, 10, 1040. https://doi.org/10.3390/bioengineering10091040
Jeong D-K, Kim H-G, Kim J-Y. Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions. Bioengineering. 2023; 10(9):1040. https://doi.org/10.3390/bioengineering10091040
Chicago/Turabian StyleJeong, Dong-Ki, Hyoung-Gook Kim, and Jin-Young Kim. 2023. "Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions" Bioengineering 10, no. 9: 1040. https://doi.org/10.3390/bioengineering10091040
APA StyleJeong, D.-K., Kim, H.-G., & Kim, J.-Y. (2023). Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions. Bioengineering, 10(9), 1040. https://doi.org/10.3390/bioengineering10091040






