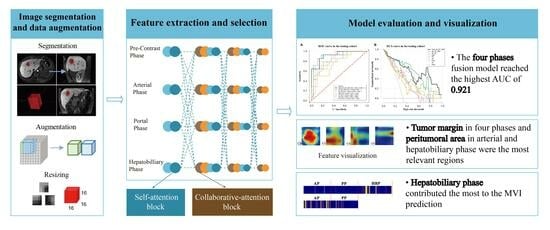
Clinical Interpretability of Deep Learning for Predicting Microvascular Invasion in Hepatocellular Carcinoma by Using Attention Mechanism
Abstract
1. Introduction
2. Material and Methods
2.1. Patients
2.2. Pathological Records
2.3. MRI Examination
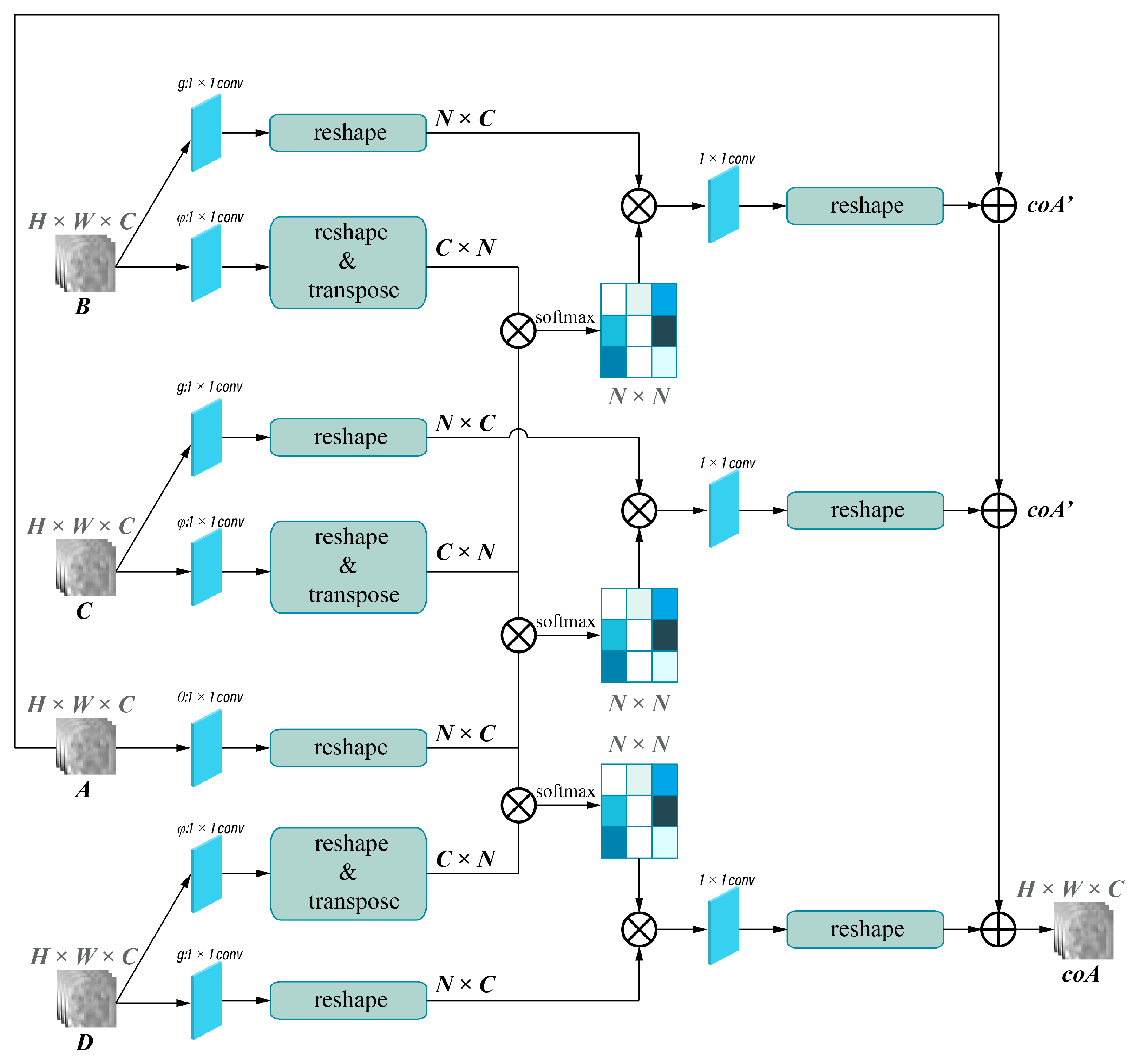
2.4. Deep Learning Models’ Development
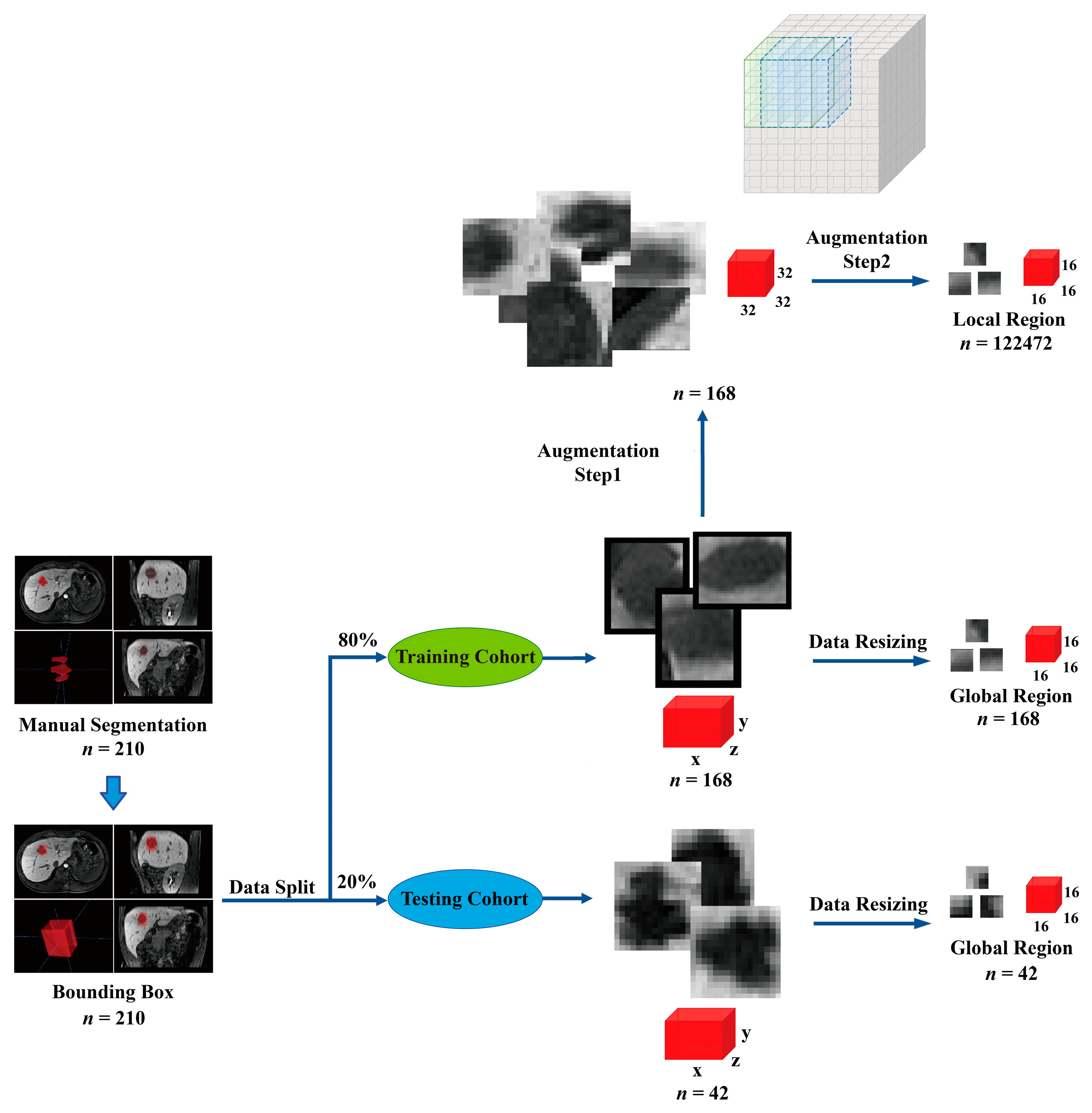
2.4.1. Image Preprocessing and Data Augmentation
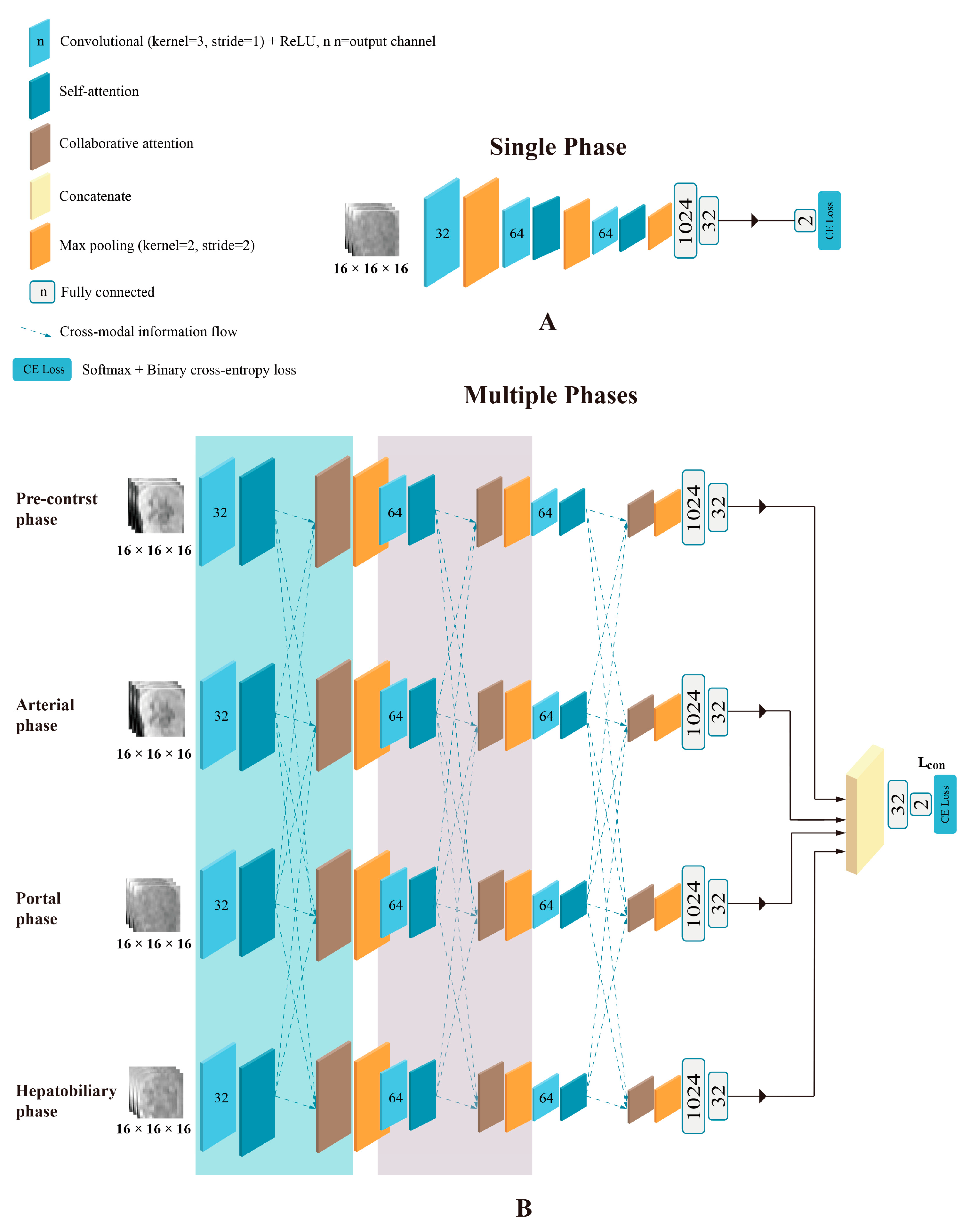
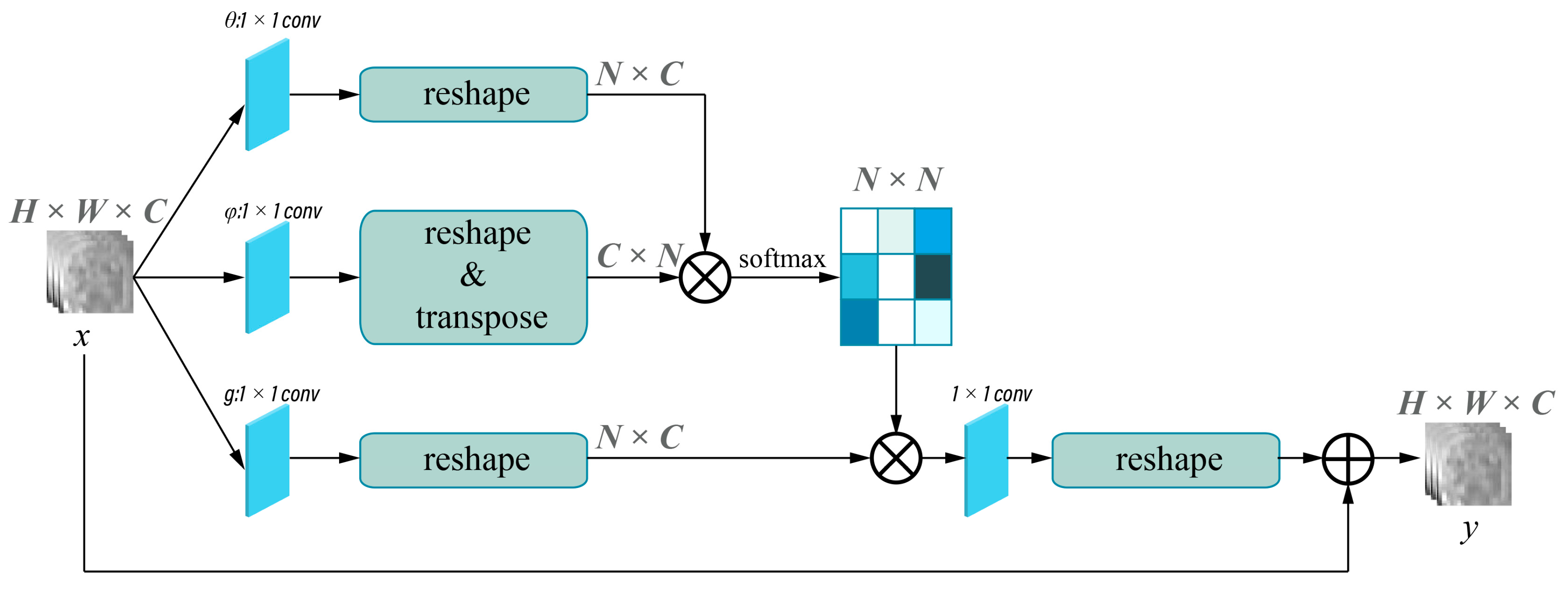
2.4.2. Model Development
- Data augmentation: We augmented the 168 original training samples to 122,640, allowing the model to have sufficient numbers of validation data to help the model converge in the 5-fold cross-validation session.
- Dropout: Dropout randomly removes a fraction of the neurons during the training process, forcing the model to be less reliant on specific neurons and preventing potential overfitting.
- Early stopping: We employed early stopping during the model training process. By outputting the learning curve in real time during the training process, we could monitor the model’s performance on a validation set and stop the training process once the performance started to deteriorate, which can prevent the model from excessively fitting to the training data.
2.4.3. Model Evaluation and Features Visualization
2.5. Statistical Analysis
3. Results
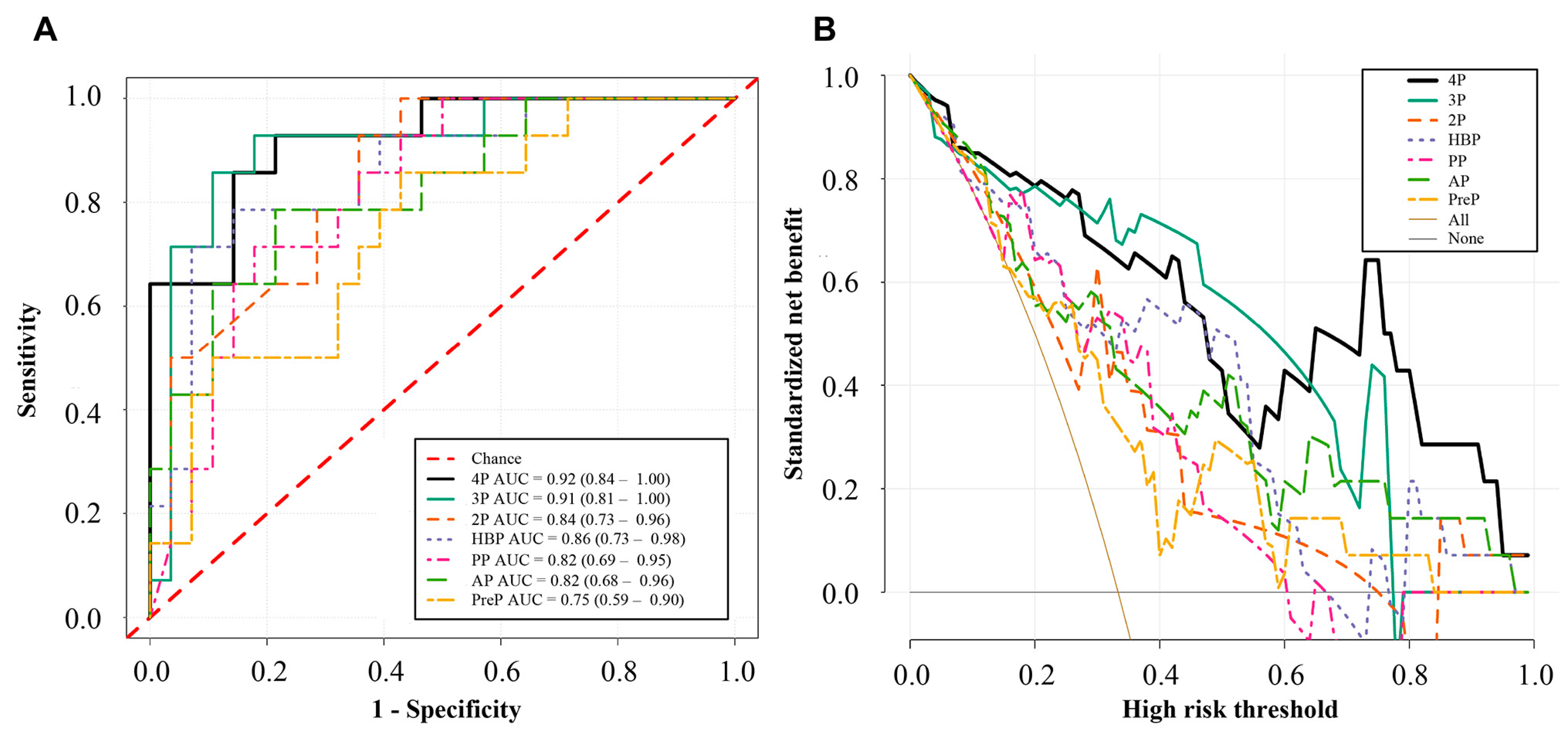
3.1. MVI Prediction Performance of the Deep Learning Models
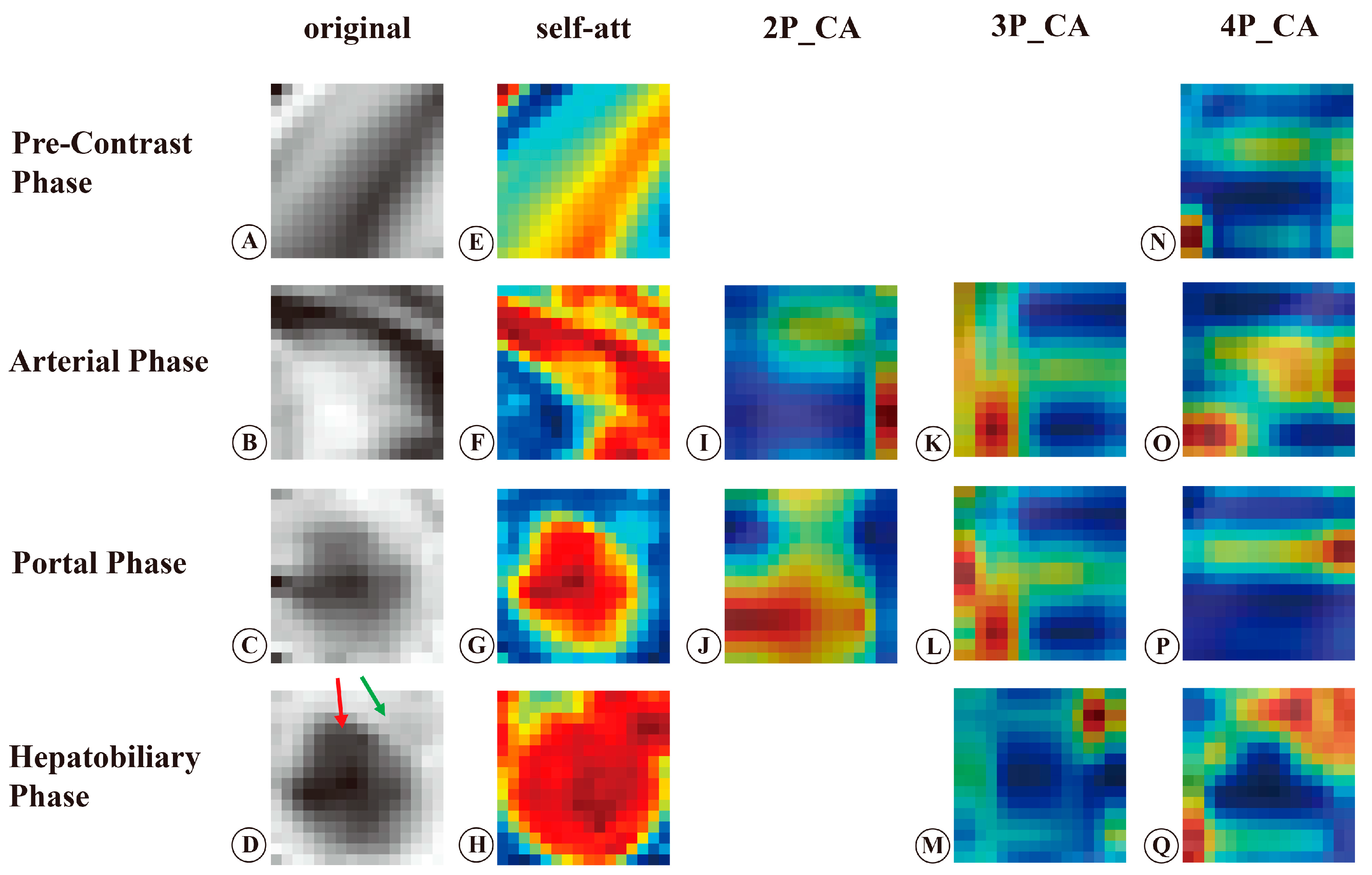
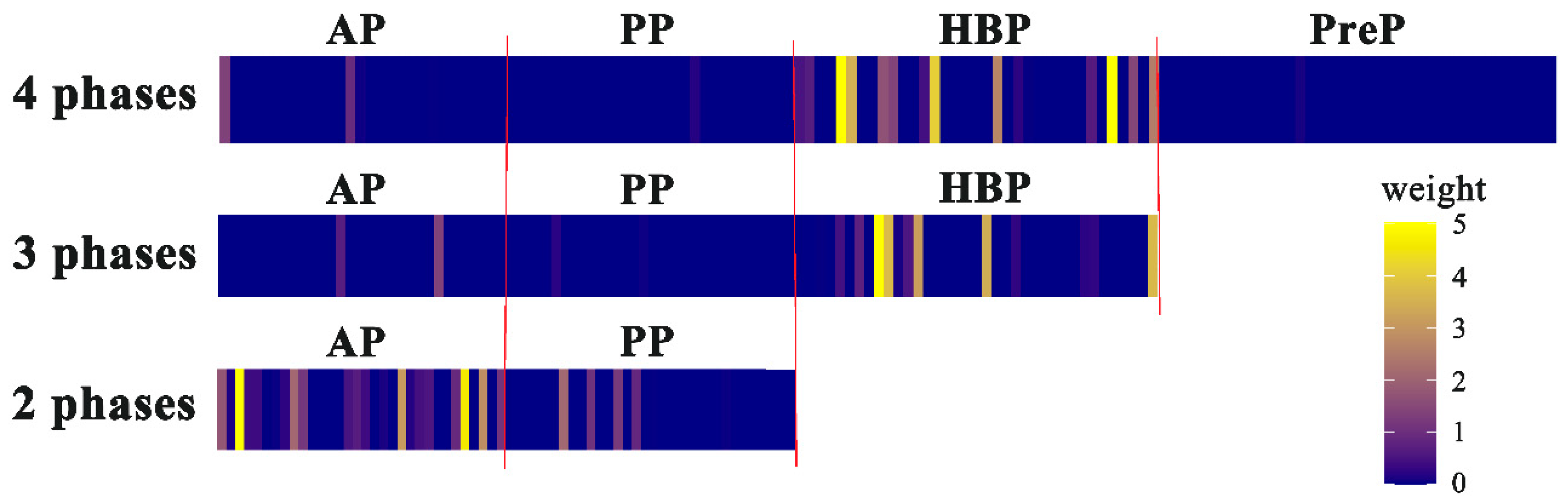
3.2. Visualization and Interpretation of the Deep Learning Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | hepatocellular carcinoma |
| MVI | microvascular invasion |
| HBP | hepatobiliary phase |
| AUC | area under the curve |
| ROC | receiver operating characteristic |
| DCA | decision curve analysis |
| CI | confidence interval |
| Grad-CAM | gradient-weighted class activation mapping |
| 3D | three-dimensional |
| CNN | convolutional neural network |
References
- Beaufrère, A.; Caruso, S.; Calderaro, J.; Poté, N.; Bijot, J.-C.; Couchy, G.; Cauchy, F.; Vilgrain, V.; Zucman-Rossi, J.; Paradis, V. Gene expression signature as a surrogate marker of microvascular invasion on routine hepatocellular carcinoma biopsies. J. Hepatol. 2022, 76, 343–352. [Google Scholar] [CrossRef]
- Yang, L.; Gu, D.; Wei, J.; Yang, C.; Rao, S.; Wang, W.; Chen, C.; Ding, Y.; Tian, J.; Zeng, M. A Radiomics Nomogram for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Liver Cancer 2019, 8, 373–386. [Google Scholar] [CrossRef]
- Toh, M.R.; Wong, E.Y.T.; Wong, S.H.; Ting, E.W.Y.; Loo, L.-H.; Chow, P.K.-H.; Ngeow, J. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology 2023, 164, 766–782. [Google Scholar] [CrossRef]
- Chan, A.W.H.; Zhong, J.; Berhane, S.; Toyoda, H.; Cucchetti, A.; Shi, K.; Tada, T.; Chong, C.C.; Xiang, B.-D.; Li, L.-Q.; et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J. Hepatol. 2018, 69, 1284–1293. [Google Scholar] [CrossRef]
- Renzulli, M.; Brocchi, S.; Cucchetti, A.; Mazzotti, F.; Mosconi, C.; Sportoletti, C.; Brandi, G.; Pinna, A.D.; Golfieri, R. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology 2016, 279, 432–442. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.H.; Lee, J.E.; Sinn, D.H.; Park, C.K. Preoperative gadoxetic acid–enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J. Hepatol. 2017, 67, 526–534. [Google Scholar] [CrossRef]
- Lei, Z.; Li, J.; Wu, D.; Xia, Y.; Wang, Q.; Si, A.; Wang, K.; Wan, X.; Lau, W.Y.; Wu, M.; et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus–Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg. 2016, 151, 356–363. [Google Scholar] [CrossRef]
- Chen, Q.; Xiao, H.; Gu, Y.; Weng, Z.; Wei, L.; Li, B.; Liao, B.; Li, J.; Lin, J.; Hei, M.; et al. Deep learning for evaluation of microvascular invasion in hepatocellular carcinoma from tumor areas of histology images. Hepatol. Int. 2022, 16, 590–602. [Google Scholar] [CrossRef]
- Hu, H.; Zheng, Q.; Huang, Y.; Huang, X.W.; Lai, Z.C.; Liu, J.; Xie, X.; Feng, S.T.; Wang, W.; De Lu, M. A non-smooth tumor margin on preoperative imaging assesses microvascular invasion of hepatocellular carcinoma: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 15375. [Google Scholar] [CrossRef]
- Min, J.H.; Lee, M.W.; Park, H.S.; Lee, D.H.; Park, H.J.; Lim, S.; Choi, S.-Y.; Lee, J.; Lee, J.E.; Ha, S.Y.; et al. Interobserver Variability and Diagnostic Performance of Gadoxetic Acid–enhanced MRI for Predicting Microvascular Invasion in Hepatocellular Carcinoma. Radiology 2020, 297, 573–581. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.-L.; Liu, Q.-P.; Sun, S.-W.; Zhang, J.; Zhu, F.-P.; Yang, G.; Yan, X.; Zhang, Y.-D.; Liu, X.-S. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J. Hepatol. 2019, 70, 1133–1144. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Zhou, C.; Ma, X. Deep learning radiomics based on contrast enhanced computed tomography predicts microvascular invasion and survival outcome in early stage hepatocellular carcinoma. Eur. J. Surg. Oncol. 2022, 48, 1068–1077. [Google Scholar] [CrossRef]
- Zhou, W.; Jian, W.; Cen, X.; Zhang, L.; Guo, H.; Liu, Z.; Liang, C.; Wang, G. Prediction of Microvascular Invasion of Hepatocellular Carcinoma Based on Contrast-Enhanced MR and 3D Convolutional Neural Networks. Front. Oncol. 2021, 11, 588010. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, X.; Qiu, J.; Zhang, B.; Zhang, L.; Fang, J.; Li, M.; Chen, L.; Wang, F.; Liu, S.; et al. Deep Learning With 3D Convolutional Neural Network for Noninvasive Prediction of Microvascular Invasion in Hepatocellular Carcinoma. J. Magn. Reson. Imaging 2021, 54, 134–143. [Google Scholar] [CrossRef]
- Song, D.; Wang, Y.; Wang, W.; Wang, Y.; Cai, J.; Zhu, K.; Lv, M.; Gao, Q.; Zhou, J.; Fan, J.; et al. Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J. Cancer Res. Clin. Oncol. 2021, 147, 3757–3767. [Google Scholar] [CrossRef]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Li, J.; Jin, K.; Zhou, D.; Kubota, N.; Ju, Z. Attention mechanism-based CNN for facial expression recognition. Neurocomputing 2020, 411, 340–350. [Google Scholar] [CrossRef]
- Sheng, C.; Wang, L.; Huang, Z.; Wang, T.; Guo, Y.; Hou, W.; Xu, L.; Wang, J.; Yan, X. Transformer-Based Deep Learning Network for Tooth Segmentation on Panoramic Radiographs. J. Syst. Sci. Complex. 2022, 36, 257–272. [Google Scholar] [CrossRef]
- Yang, H.; Wang, L.; Xu, Y.; Liu, X. CovidViT: A novel neural network with self-attention mechanism to detect COVID-19 through X-ray images. Int. J. Mach. Learn. Cybern. 2023, 14, 973–987. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Y.; Zhang, L.; Zhou, J.T.; Liu, Y.; Goh, R.S.M.; Zhen, L. Adversarial multimodal fusion with attention mechanism for skin lesion classification using clinical and dermoscopic images. Med. Image Anal. 2022, 81, 102535. [Google Scholar] [CrossRef]
- Fu, X.; Bi, L.; Kumar, A.; Fulham, M.; Kim, J. An attention-enhanced cross-task network to analyse lung nodule attributes in CT images. Pattern Recognit. 2022, 126, 108576. [Google Scholar] [CrossRef]
- Ou, C.; Zhou, S.; Yang, R.; Jiang, W.; He, H.; Gan, W.; Chen, W.; Qin, X.; Luo, W.; Pi, X.; et al. A deep learning based multimodal fusion model for skin lesion diagnosis using smartphone collected clinical images and metadata. Front. Surg. 2022, 9, 1029991. [Google Scholar] [CrossRef]
- Li, S.; Xie, Y.; Wang, G.; Zhang, L.; Zhou, W. Attention guided discriminative feature learning and adaptive fusion for grading hepatocellular carcinoma with Contrast-enhanced MR. Comput. Med. Imaging Graph. 2022, 97, 102050. [Google Scholar] [CrossRef]
- Li, S.; Xie, Y.; Wang, G.; Zhang, L.; Zhou, W. Adaptive Multimodal Fusion With Attention Guided Deep Supervision Net for Grading Hepatocellular Carcinoma. IEEE J. Biomed. Health Inform. 2022, 26, 4123–4131. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, Y.; Wu, J.; Pan, J.; Liao, C.; Su, H. The utility of Vision Transformer in preoperatively predicting microvascular invasion status of hepatocellular carcinoma. HPB 2023, 25, 533–542. [Google Scholar] [CrossRef]
- Cong, W.-M.; Bu, H.; Chen, J.; Dong, H.; Zhu, Y.-Y.; Feng, L.-H.; Chen, J. Practice Guideline Committee Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J. Gastroenterol. 2016, 22, 9279–9287. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Liu, J.; Yao, J.; Seff, A.; Cherry, K.; Kim, L.; Summers, R.M. Improving Computer-Aided Detection Using Convolutional Neural Networks and Random View Aggregation. IEEE Trans. Med. Imaging 2016, 35, 1170–1181. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Jiang, Y.Q.; Cao, S.E.; Cao, S.; Chen, J.N.; Wang, G.Y.; Shi, W.Q.; Deng, Y.N.; Cheng, N.; Ma, K.; Zeng, K.N.; et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J. Cancer Res. Clin. Oncol. 2020, 147, 821–833. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Zhu, Y.; Xu, J.; Wei, J.; Xie, J.; Zhang, J. Modality-based attention and dual-stream multiple instance convolutional neural network for predicting microvascular invasion of hepatocellular carcinoma. Front. Oncol. 2023, 13, 1195110. [Google Scholar] [CrossRef]
- Dou, T.; Zhang, L.; Zhou, W. 3D Deep Feature Fusion In Contrast-Enhanced Mr For Malignancy Characterization of Hepatocellular Carcinoma. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 29–33. [Google Scholar]
- Xiao, H.; Guo, Y.; Zhou, Q.; Chen, Q.; Du, Q.; Chen, S.; Fu, S.; Lin, J.; Li, D.; Song, X.; et al. Prediction of microvascular invasion in hepatocellular carcinoma with expert-inspiration and skeleton sharing deep learning. Liver Int. 2022, 42, 1423–1431. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, B.; Xu, Y.; Zhou, W. An attention-based deep learning model for predicting microvascular invasion of hepatocellular carcinoma using an intra-voxel incoherent motion model of diffusion-weighted magnetic resonance imaging. Phys. Med. Biol. 2021, 66, 185019. [Google Scholar] [CrossRef]
- Hong, S.B.; Choi, S.H.; Kim, S.Y.; Shim, J.H.; Lee, S.S.; Byun, J.H.; Park, S.H.; Kim, K.W.; Kim, S.; Lee, N.K. MRI Features for Predicting Microvascular Invasion of Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. Liver Cancer 2021, 10, 94–106. [Google Scholar] [CrossRef]
- Wang, W.-T.; Yang, L.; Yang, Z.-X.; Hu, X.-X.; Ding, Y.; Yan, X.; Fu, C.-X.; Grimm, R.; Zeng, M.-S.; Rao, S.-X. Assessment of Microvascular Invasion of Hepatocellular Carcinoma with Diffusion Kurtosis Imaging. Radiology 2018, 286, 571–580. [Google Scholar] [CrossRef]
- Jiang, H.; Wei, J.; Fu, F.; Wei, H.; Qin, Y.; Duan, T.; Chen, W.; Xie, K.; Lee, J.M.; Bashir, M.R.; et al. Predicting microvascular invasion in hepatocellular carcinoma: A dual-institution study on gadoxetate disodium-enhanced MRI. Liver Int. 2022, 42, 1158–1172. [Google Scholar] [CrossRef]
- Rhee, H.; An, C.; Kim, H.Y.; Yoo, J.E.; Park, Y.N.; Kim, M.J. Hepatocellular Carcinoma with Irregular Rim-Like Arterial Phase Hyperenhancement: More Aggressive Pathologic Features. Liver Cancer 2019, 8, 24–40. [Google Scholar] [CrossRef]
- Kim, H.; Park, M.-S.; Choi, J.Y.; Park, Y.N.; Kim, M.-J.; Kim, K.S.; Han, K.-H.; Kim, E.; Kim, K.W. Can microvessel invasion of hepatocellular carcinoma be predicted by pre-operative MRI? Eur. Radiol. 2009, 19, 1744–1751. [Google Scholar] [CrossRef]
- Miyata, R.; Tanimoto, A.; Wakabayashi, G.; Shimazu, M.; Nakatsuka, S.; Mukai, M.; Kitajima, M. Accuracy of preoperative prediction of microinvasion of portal vein in hepatocellular carcinoma using superparamagnetic iron oxide-enhanced magnetic resonance imaging and computed tomography during hepatic angiography. J. Gastroenterol. 2006, 41, 987–995. [Google Scholar] [CrossRef]

| Parameters | MVI-Negative (n = 140) | MVI-Positive (n = 70) | p-Value |
|---|---|---|---|
| Age (years) * | 55 ± 11 | 54 ± 13 | 0.818 |
| Sex | >0.99 | ||
| Men | 124 (88.6) | 62 (88.6) | |
| Women | 16 (11.4) | 8 (11.4) | |
| Aspartate aminotransferase (U/L) † | 32.0 (24.2–48.0) | 33.0 (24.0–60.0) | 0.378 |
| Alanine aminotransferase (U/L) † | 33.5 (23.0–53.7) | 32.0 (20.7–51.2) | 0.682 |
| Alpha-fetoprotein | <0.001 | ||
| ≥400 µg/L | 28 (20.0) | 30 (42.9) | |
| <400 µg/L | 112 (80.0) | 40 (57.1) | |
| Tumor diameter (mm) † | 42.5 (30.0–65.0) | 57.0 (41.5–91.2) | <0.001 |
| Sequences | TR (ms) | TE (ms) | FA | Thickness (mm) | Matrix | FOV (mm) | Breath-Hold |
|---|---|---|---|---|---|---|---|
| T1-weighted imaging | 2.75 | 1.05 | 12.5 | 2 | 320 × 192 | 380 × 380 | Yes |
| T2-weighted imaging | 2000 | 77 | 103 | 5 | 384 × 288 | 380 × 380 | No |
| Arterial/Portal phase | 2.75 | 1.05 | 12.5 | 2 | 320 × 192 | 380 × 380 | Yes |
| Hepatobiliary phase | 3.84 | 1.45 | 25 | 2 | 288 × 186 | 380 × 306 | Yes |
| Parameters | Training Set (n = 168) | Test Set (n = 42) | p-Value |
|---|---|---|---|
| Age (years) * | 54 ± 11 | 56 ± 12 | 0.266 |
| Sex | 0.914 | ||
| Men | 149 | 37 | |
| Women | 19 | 5 | |
| Aspartate aminotransferase (U/L) † | 33.0 (24.0–50.8) | 31.0 (24.0–62.0) | 0.933 |
| Alanine aminotransferase (U/L) † | 33.0 (21.0–49.8) | 32.0 (23.0–77.0) | 0.469 |
| Alpha-fetoprotein | 0.354 | ||
| ≥400 µg/L | 124 | 28 | |
| <400 µg/L | 44 | 14 | |
| Tumor diameter (mm) † | 47.5 (34.0–70.0) | 52.1 (31.8–71.0) | 0.701 |
| Model | AUC (95%CI) | Accuracy | Sensitivity | Specificity | p-Value |
|---|---|---|---|---|---|
| PreP | 0.75 (0.59–0.90) | 0.64 | 0.86 | 0.57 | 0.010 |
| AP | 0.82 (0.68–0.96) | 0.74 | 0.79 | 0.79 | 0.001 |
| PP | 0.82 (0.69–0.95) | 0.79 | 0.71 | 0.82 | 0.001 |
| HBP | 0.86 (0.73–0.98) | 0.71 | 0.79 | 0.86 | <0.001 |
| 2P | 0.84 (0.73–0.96) | 0.81 | 0.93 | 0.64 | <0.001 |
| 3P | 0.91 (0.81–1.00) | 0.88 | 0.93 | 0.82 | <0.001 |
| 4P | 0.92 (0.84–1.00) | 0.81 | 0.93 | 0.79 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, H.; Wang, J.; Ma, R.; Chen, Y.; Li, L.; Song, C.; Dong, Z.; Feng, S.; Zhou, X. Clinical Interpretability of Deep Learning for Predicting Microvascular Invasion in Hepatocellular Carcinoma by Using Attention Mechanism. Bioengineering 2023, 10, 948. https://doi.org/10.3390/bioengineering10080948
You H, Wang J, Ma R, Chen Y, Li L, Song C, Dong Z, Feng S, Zhou X. Clinical Interpretability of Deep Learning for Predicting Microvascular Invasion in Hepatocellular Carcinoma by Using Attention Mechanism. Bioengineering. 2023; 10(8):948. https://doi.org/10.3390/bioengineering10080948
Chicago/Turabian StyleYou, Huayu, Jifei Wang, Ruixia Ma, Yuying Chen, Lujie Li, Chenyu Song, Zhi Dong, Shiting Feng, and Xiaoqi Zhou. 2023. "Clinical Interpretability of Deep Learning for Predicting Microvascular Invasion in Hepatocellular Carcinoma by Using Attention Mechanism" Bioengineering 10, no. 8: 948. https://doi.org/10.3390/bioengineering10080948
APA StyleYou, H., Wang, J., Ma, R., Chen, Y., Li, L., Song, C., Dong, Z., Feng, S., & Zhou, X. (2023). Clinical Interpretability of Deep Learning for Predicting Microvascular Invasion in Hepatocellular Carcinoma by Using Attention Mechanism. Bioengineering, 10(8), 948. https://doi.org/10.3390/bioengineering10080948