A Soft-Reference Breast Ultrasound Image Quality Assessment Method That Considers the Local Lesion Area
Abstract
1. Introduction
- An integrated global–local breast ultrasound IQA framework is established, in which the images without lesions are evaluated globally, and the images with lesions are evaluated locally (i.e., with a focus on the lesion area).
- A soft-reference IQA method based on lesion segmentation is proposed, in which the segmented lesion is taken as a reference image and transforms the medical ultrasound image assessment problem, from no reference to reduced reference.
- A comprehensive evaluation index for medical ultrasound images with lesions is proposed, including pixel-level features MSE (Mean Squared Error), PSNR (Peak Signal-to-Noise Ratio) and semantic-level features CR (Contrast Ratio) and SSIM (Structural Similarity).
2. Materials and Methods
2.1. Data Collection
2.2. Data Labeling
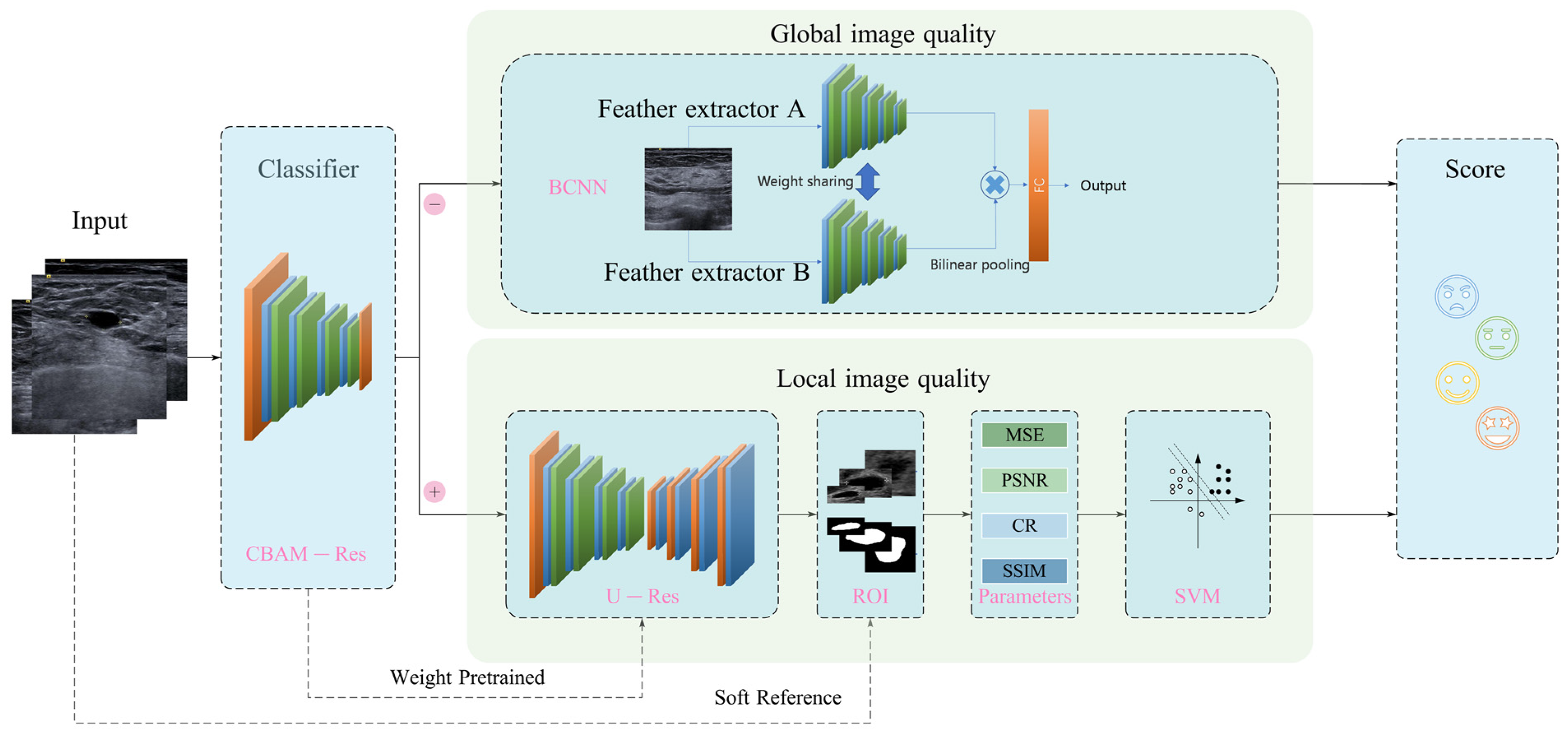
2.3. Proposed Framework for Breast Ultrasound Image Quality Assessment
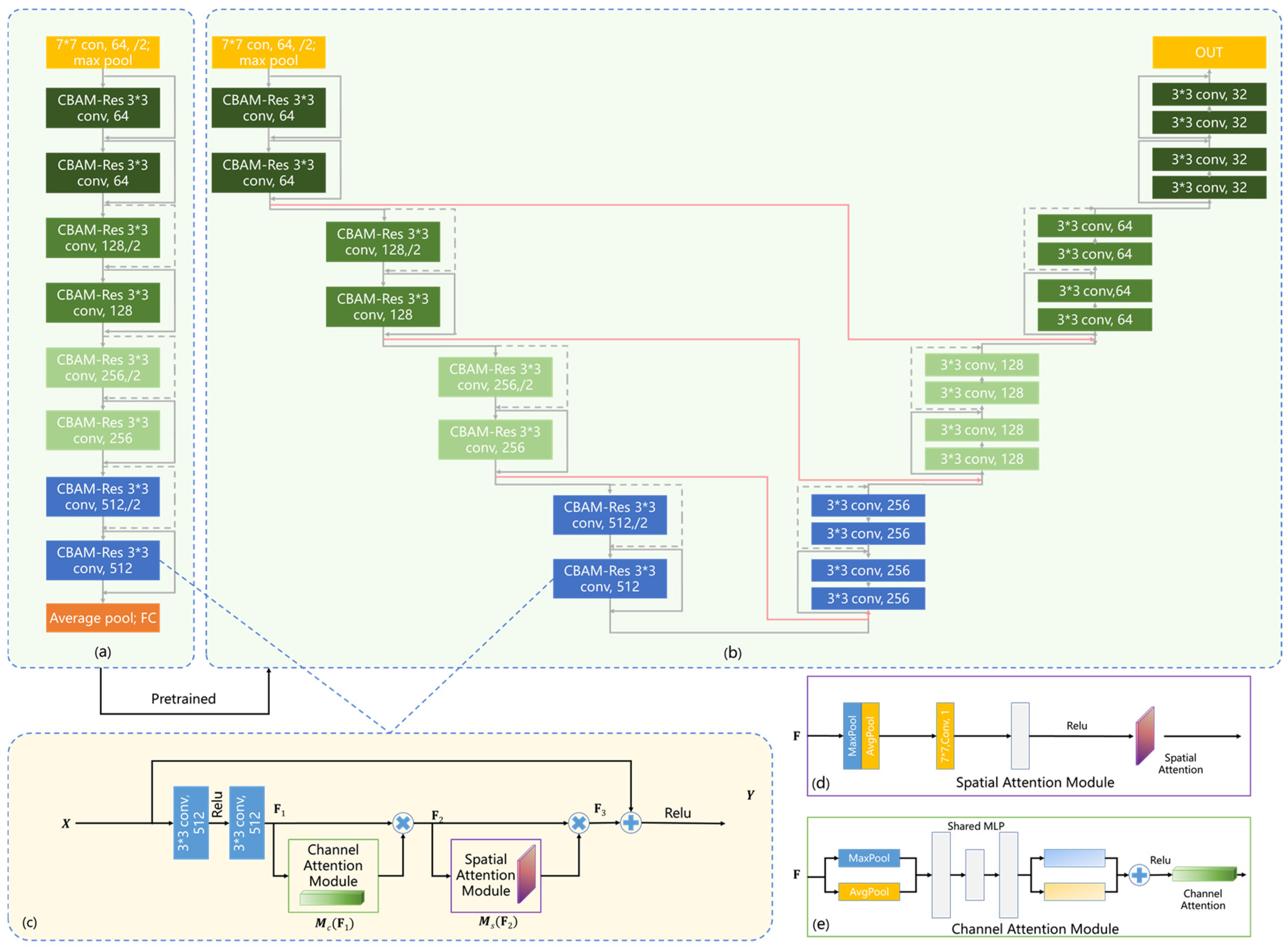
2.4. Classification Network
2.5. BCNN Global IQA Network
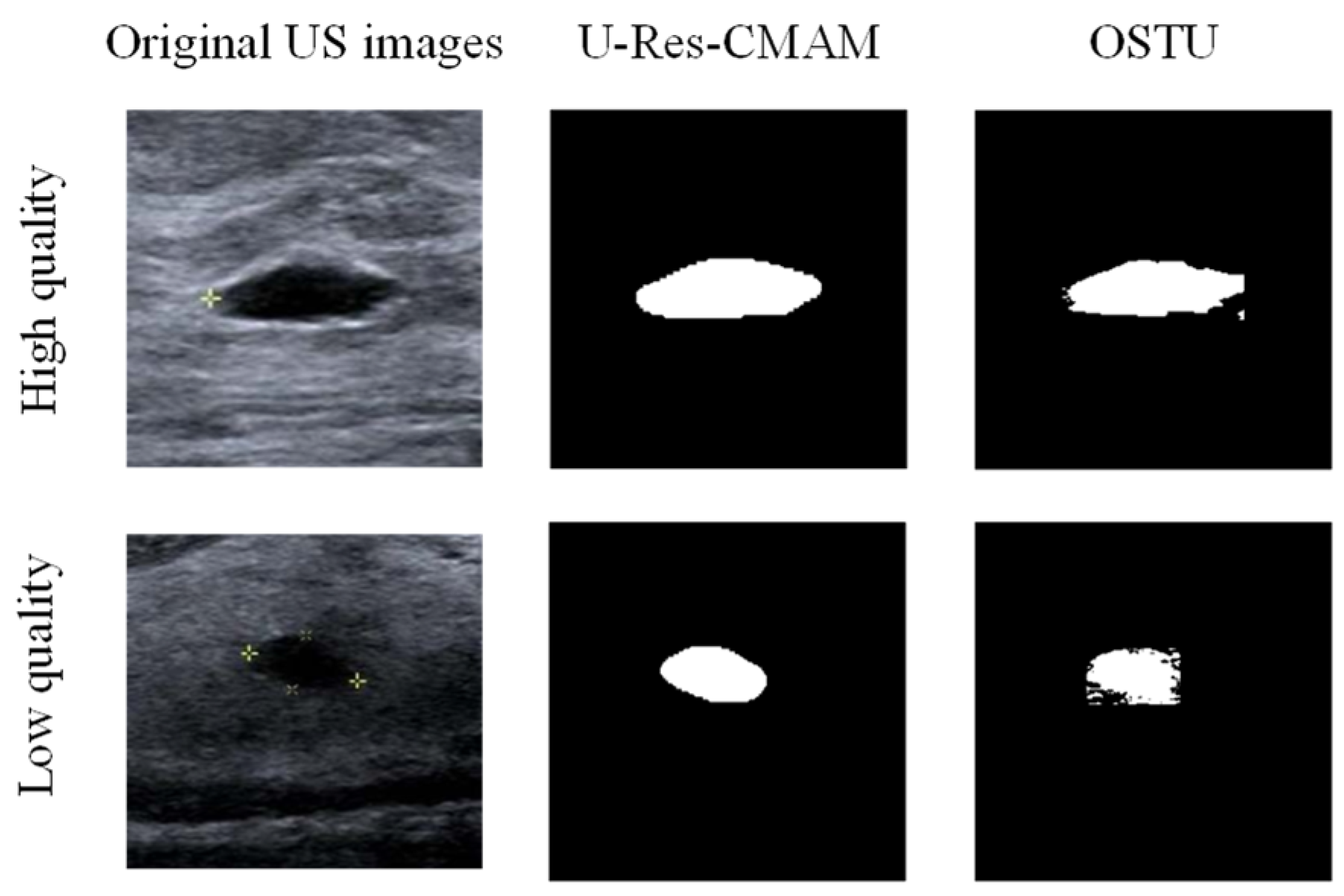
2.6. Lesion Segmentation Network
2.7. Parameters of Local Image Quality Assessment
2.7.1. Mean Squared Error (MSE)
2.7.2. Peak Signal-to-Noise Ratio (PSNR)
2.7.3. Contrast Ratio (CR)
2.7.4. Structural Similarity (SSIM)
2.8. Support Vector Machine (SVM)
3. Results
3.1. Implementation Details
3.2. Quantitative Evaluation Metrics
3.2.1. Pearson Linear Correlation Coefficient (PLCC)
3.2.2. Spearman Rank-Order Correlation Coefficient (SRCC)
3.2.3. Accuracy
3.2.4. Root Mean Square Error (RMSE)
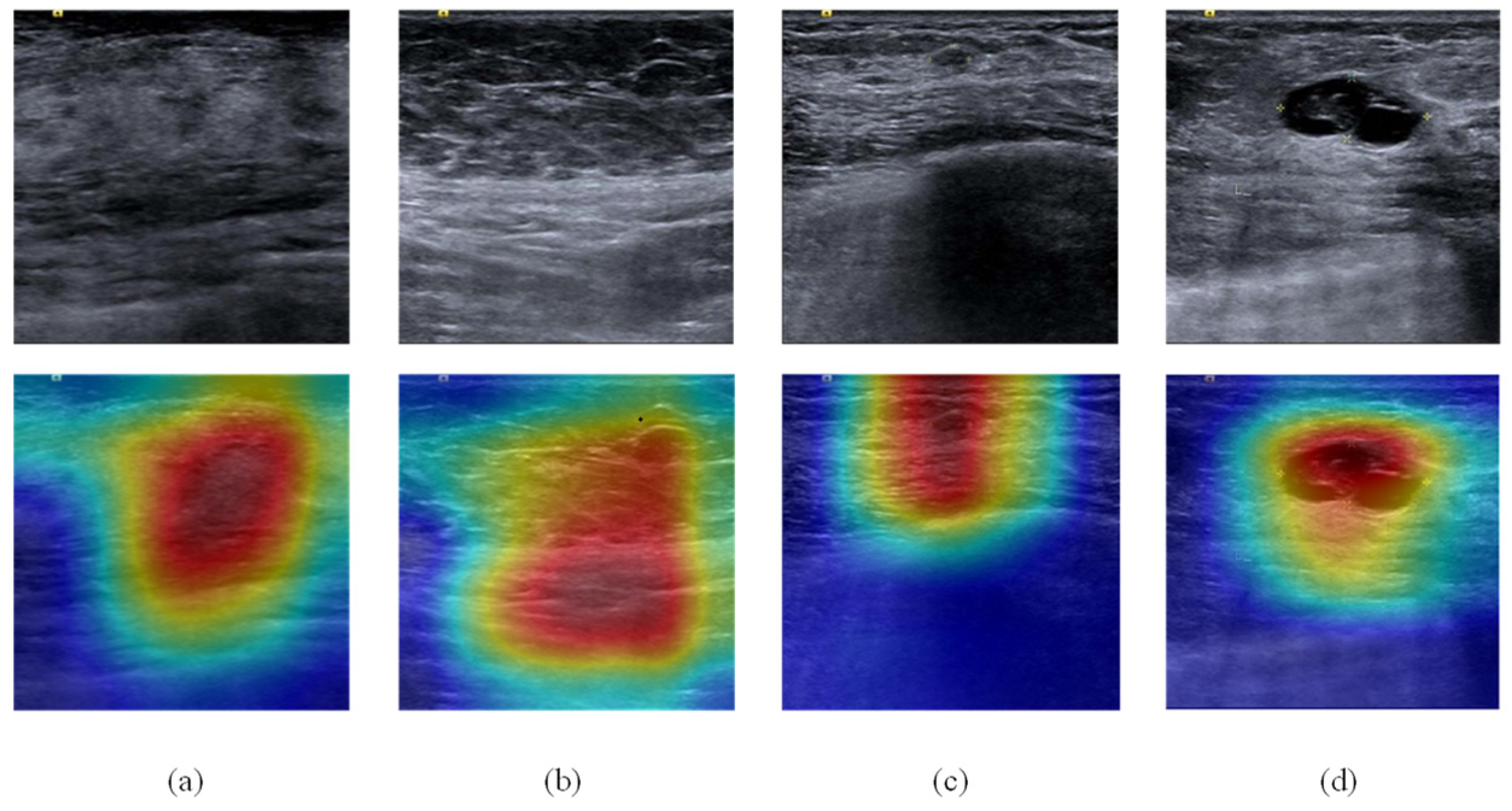
3.3. Classification Experiment
3.4. Global IQA Experiment
3.5. Segmentation Experiment
3.6. Local IQA Experiment
3.7. Global–Local Integrated IQA Experiment
3.8. Real-Time Performance
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, D.; Haybittle, J.L. The Curability of Breast Cancer. Lancet 1975, 306, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, D.; McConnell, B.; Royse, C. A review of diagnostic accuracy and clinical impact from the focused use of perioperative ultrasound. Can. J. Anesth. 2018, 65, 371–380. [Google Scholar] [CrossRef]
- Sencha, A.N.; Evseeva, E.V.; Mogutov, M.S.; Patrunov, Y.N. Breast Ultrasound; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Wang, Z.W.; Zhao, B.L.; Zhang, P.; Yao, L.; Wang, Q.; Li, B.; Meng, M.Q.H.; Hu, Y. Full-Coverage Path Planning and Stable Interaction Control for Automated Robotic Breast Ultrasound Scanning. IEEE Trans. Ind. Electron. 2023, 70, 7051–7061. [Google Scholar] [CrossRef]
- Liu, S.Y.; Thung, K.H.; Lin, W.L.; Shen, D.G.; Yap, P.T. Hierarchical Nonlocal Residual Networks for Image Quality Assessment of Pediatric Diffusion MRI With Limited and Noisy Annotations. IEEE Trans. Med. Imaging 2020, 39, 3691–3702. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.S.; Paramesran, R. Review of medical image quality assessment. Biomed. Signal Process. Control 2016, 27, 145–154. [Google Scholar] [CrossRef]
- Shiao, Y.H.; Chen, T.J.; Chuang, K.S.; Lin, C.H.; Chuang, C.C. Quality of compressed medical images. J. Digit. Imaging 2007, 20, 149–159. [Google Scholar] [CrossRef]
- Choong, M.K.; Logeswaran, R.; Bister, M. Improving diagnostic quality of MR images through controlled lossy compression using SPIHT. J. Med. Syst. 2006, 30, 139–143. [Google Scholar] [CrossRef]
- Abolmaesumi, P.; Salcudean, S.E.; Zhu, W.H.; Sirouspour, M.R.; DiMaio, S.P. Image-guided control of a robot for medical ultrasound. IEEE Trans. Robot. Autom. 2002, 18, 11–23. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Fantini, I.; Yasuda, C.; Bento, M.; Rittner, L.; Cendes, F.; Lotufo, R. Automatic MR image quality evaluation using a Deep CNN: A reference-free method to rate motion artifacts in neuroimaging. Comput. Med. Imaging Graph. 2021, 90, 101897. [Google Scholar] [CrossRef]
- Oszust, M.; Bielecka, M.; Bielecki, A.; Stepien, I.; Obuchowicz, R.; Piorkowski, A. Blind image quality assessment of magnetic resonance images with statistics of local intensity extrema. Inf. Sci. 2022, 606, 112–125. [Google Scholar] [CrossRef]
- Shen, Y.X.; Sheng, B.; Fang, R.G.; Li, H.T.; Dai, L.; Stolte, S.; Qin, J.; Jia, W.P.; Shen, D.G. Domain-invariant interpretable fundus image quality assessment. Med. Image Anal. 2020, 61, 101654. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Beghdadi, A.; Kaaniche, M.; Alaya-Cheikh, F.; Gharbi, O. A neural network based framework for effective laparoscopic video quality assessment. Comput. Med. Imaging Graph. 2022, 101, 102121. [Google Scholar] [CrossRef]
- Ali, S.; Zhou, F.; Bailey, A.; Braden, B.; East, J.E.; Lu, X.; Rittscher, J. A deep learning framework for quality assessment and restoration in video endoscopy. Med. Image Anal. 2021, 68, 101900. [Google Scholar] [CrossRef] [PubMed]
- Khanin, A.; Anton, M.; Reginatto, M.; Elster, C. Assessment of CT Image Quality Using a Bayesian Framework. IEEE Trans. Med. Imaging 2018, 37, 2687–2694. [Google Scholar] [CrossRef] [PubMed]
- Chatelain, P.; Krupa, A.; Navab, N. Confidence-Driven Control of an Ultrasound Probe. IEEE Trans. Robot. 2017, 33, 1410–1424. [Google Scholar] [CrossRef]
- Karamalis, A.; Wein, W.; Klein, T.; Navab, N. Ultrasound confidence maps using random walks. Med. Image Anal. 2012, 16, 1101–1112. [Google Scholar] [CrossRef]
- Wu, L.Y.; Cheng, J.Z.; Li, S.L.; Lei, B.Y.; Wang, T.F.; Ni, D. FUIQA: Fetal Ultrasound Image Quality Assessment with Deep Convolutional Networks. IEEE Trans. Cybern. 2017, 47, 1336–1349. [Google Scholar] [CrossRef]
- Lin, Z.H.; Li, S.L.; Ni, D.; Liao, Y.M.; Wen, H.X.; Du, J.; Chen, S.P.; Wang, T.F.; Lei, B.Y. Multi-task learning for quality assessment of fetal head ultrasound images. Med. Image Anal. 2019, 58, 101548. [Google Scholar] [CrossRef]
- Camps, S.M.; Houben, T.; Carneiro, G.; Edwards, C.; Antico, M.; Dunnhofer, M.; Martens, E.; Baeza, J.A.; Vanneste, B.G.L.; van Limbergen, E.J.; et al. Automatic quality assessment of transperineal ultrasound images of the male pelvic region, using deep learning. Ultrasound Med. Biol. 2020, 46, 445–454. [Google Scholar] [CrossRef]
- Antico, M.; Vukovic, D.; Camps, S.M.; Sasazawa, F.; Takeda, Y.; Le, A.T.H.; Jaiprakash, A.T.; Roberts, J.; Crawford, R.; Fontanarosa, D.; et al. Deep Learning for US Image Quality Assessment Based on Femoral Cartilage Boundary Detection in Autonomous Knee Arthroscopy. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 2543–2552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Wang, Y.F.; Jiang, J.Y.; Dong, J.X.; Yi, W.W.; Hou, W.G. CNN-Based Medical Ultrasound Image Quality Assessment. Complexity 2021, 2021, 9938367. [Google Scholar] [CrossRef]
- Song, Y.X.; Zhong, Z.M.; Zhao, B.L.; Zhang, P.; Wang, Q.; Wang, Z.W.; Yao, L.; Lv, F.Q.; Hu, Y. Medical Ultrasound Image Quality Assessment for Autonomous Robotic Screening. IEEE Robot. Autom. Lett. 2022, 7, 6290–6296. [Google Scholar] [CrossRef]
- He, F.X.; Liu, T.L.; Tao, D.C. Why ResNet Works? Residuals Generalize. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 5349–5362. [Google Scholar] [CrossRef]
- Woo, S.H.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Lin, T.Y.; RoyChowdhury, A.; Maji, S. Bilinear CNN Models for Fine-Grained Visual Recognition. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 11–18 December 2015; pp. 1449–1457. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Bindu, C.H.; Prasad, K.S. An efficient medical image segmentation using conventional OTSU method. Int. J. Adv. Sci. Technol. 2012, 38, 67–74. [Google Scholar]
- Malkomes, G.; Schaff, C.; Garnett, R. Bayesian optimization for automated model selection. In Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS), Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Zhao, H.S.; Shi, J.P.; Qi, X.J.; Wang, X.G.; Jia, J.Y. Pyramid Scene Parsing Network. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar]
- Chen, L.C.E.; Zhu, Y.K.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 833–851. [Google Scholar]
- Zhang, Q.; Cui, Z.P.; Niu, X.G.; Geng, S.J.; Qiao, Y. Image Segmentation with Pyramid Dilated Convolution Based on ResNet and U-Net. In Proceedings of the 24th International Conference on Neural Information Processing (ICONIP), Guangzhou, China, 14–18 November 2017; pp. 364–372. [Google Scholar]
- Matsoukas, C.; Haslum, J.F.; Söderberg, M.; Smith, K. Is It Time to Replace CNNS with Transformers for Medical Images? arXiv 2021, arXiv:2108.09038. [Google Scholar]




| Lesion | Samples | ||||
|---|---|---|---|---|---|
| Score | 1 | 2 | 3 | 4 | |
| Negative | Image | 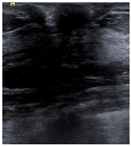 |  | 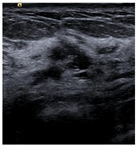 | 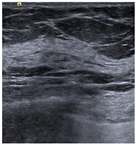 |
| Number | 98 | 112 | 345 | 289 | |
| Positive | Image | 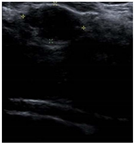 |  |  |  |
| Number | 47 | 49 | 161 | 184 | |
| Model | Accuracy of Validation Set | Accuracy of Test Set | Best Validation Epoch |
|---|---|---|---|
| ResNet18 without pre-training | 0.966 | 0.953 | 15 |
| ResNet18 with pre-training | 0.970 | 0.965 | 11 |
| CBAM-ResNet18 with pre-training | 0.989 | 0.992 | 5 |
| Dataset | PLCC | SRCC | Accuracy | RMSE |
|---|---|---|---|---|
| Negative (A.3) | 0.8964 | 0.8765 | 0.8402 | 0.4615 |
| Model | Pretrained | Val-Dice | Test-Dice |
|---|---|---|---|
| VGG16 | ImageNet | 0.8346 ± 0.0839 | 0.8633 ± 0.0936 |
| PSPNet | ImageNet | 0.8700 ± 0.1174 | 0.8789 ± 0.1024 |
| DeepLab V3+ | ImageNet | 0.8968 ± 0.0560 | 0.8824 ± 0.0865 |
| U-Res | ImageNet | 0.9056 ± 0.1676 | 0.8932 ± 1.1895 |
| ViT | ImageNet | 0.9162 ± 0.0964 | 0.9065 ± 0.6352 |
| U-Res-CBAM | ImageNet | 0.9272 ± 0.1155 | 0.9126 ± 0.0971 |
| Model | ValPLCC | ValSRCC | ValAccuracy | ValRMSE | TestPLCC | TestSRCC | TestAccuracy | TestRMSE |
|---|---|---|---|---|---|---|---|---|
| Vgg16 | 0.5587 | 0.5219 | 0.6232 | 0.7161 | 0.6381 | 0.6011 | 0.6250 | 0.6657 |
| PSPNet | 0.6546 | 0.6307 | 0.6289 | 0.6028 | 0.6830 | 0.7055 | 0.6364 | 0.6307 |
| Deeplab v3+ | 0.6500 | 0.6516 | 0.6601 | 0.6584 | 0.7254 | 0.7118 | 0.6932 | 0.5839 |
| U-Res | 0.7735 | 0.7800 | 0.7507 | 0.5160 | 0.8031 | 0.7945 | 0.7500 | 0.5000 |
| U-Res-CBAM | 0.8307 | 0.8430 | 0.8130 | 0.4516 | 0.8418 | 0.8462 | 0.8068 | 0.4395 |
| Ground-truth | 0.9255 | 0.9394 | 0.9065 | 0.3058 | 0.8996 | 0.9251 | 0.8636 | 0.3693 |
| Dataset | Model | PLCC | SRCC | Accuracy | RMSE | 95% CI of Acc |
|---|---|---|---|---|---|---|
| Positive (B.3) | SR-IQA | 0.8418 | 0.8462 | 0.8068 | 0.4395 | (0.7243, 0.8893) |
| Positive (B.3) | BCNN | 0.6606 | 0.7062 | 0.7159 | 0.6946 | (0.6217, 0.8101) |
| Mixed (A.3 + B.3) | Global-local | 0.8851 | 0.8834 | 0.8288 | 0.4541 | (0.7501, 0.9075) |
| Mixed (A.3 + B.3) | BCNN | 0.8306 | 0.8338 | 0.8016 | 0.5649 | (0.7183, 0.8849) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Song, Y.; Zhao, B.; Zhong, Z.; Yao, L.; Lv, F.; Li, B.; Hu, Y. A Soft-Reference Breast Ultrasound Image Quality Assessment Method That Considers the Local Lesion Area. Bioengineering 2023, 10, 940. https://doi.org/10.3390/bioengineering10080940
Wang Z, Song Y, Zhao B, Zhong Z, Yao L, Lv F, Li B, Hu Y. A Soft-Reference Breast Ultrasound Image Quality Assessment Method That Considers the Local Lesion Area. Bioengineering. 2023; 10(8):940. https://doi.org/10.3390/bioengineering10080940
Chicago/Turabian StyleWang, Ziwen, Yuxin Song, Baoliang Zhao, Zhaoming Zhong, Liang Yao, Faqin Lv, Bing Li, and Ying Hu. 2023. "A Soft-Reference Breast Ultrasound Image Quality Assessment Method That Considers the Local Lesion Area" Bioengineering 10, no. 8: 940. https://doi.org/10.3390/bioengineering10080940
APA StyleWang, Z., Song, Y., Zhao, B., Zhong, Z., Yao, L., Lv, F., Li, B., & Hu, Y. (2023). A Soft-Reference Breast Ultrasound Image Quality Assessment Method That Considers the Local Lesion Area. Bioengineering, 10(8), 940. https://doi.org/10.3390/bioengineering10080940









