Self-Supervised Learning Application on COVID-19 Chest X-ray Image Classification Using Masked AutoEncoder
Abstract
1. Introduction
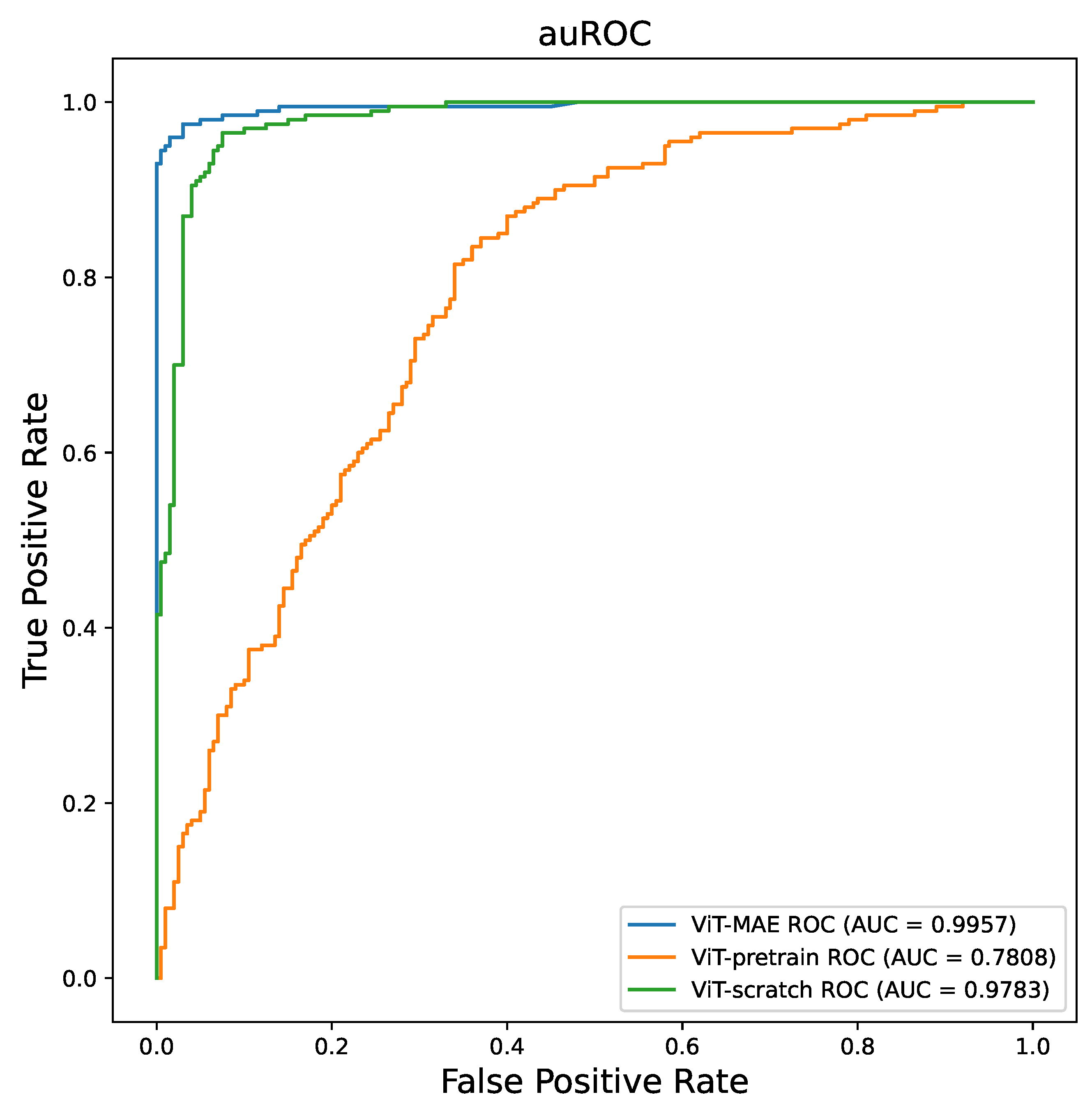
- We conducted a comparative analysis of various training strategies using the same public COVID-19 dataset and observed that the MAE model outperformed other approaches, demonstrating superior performance.
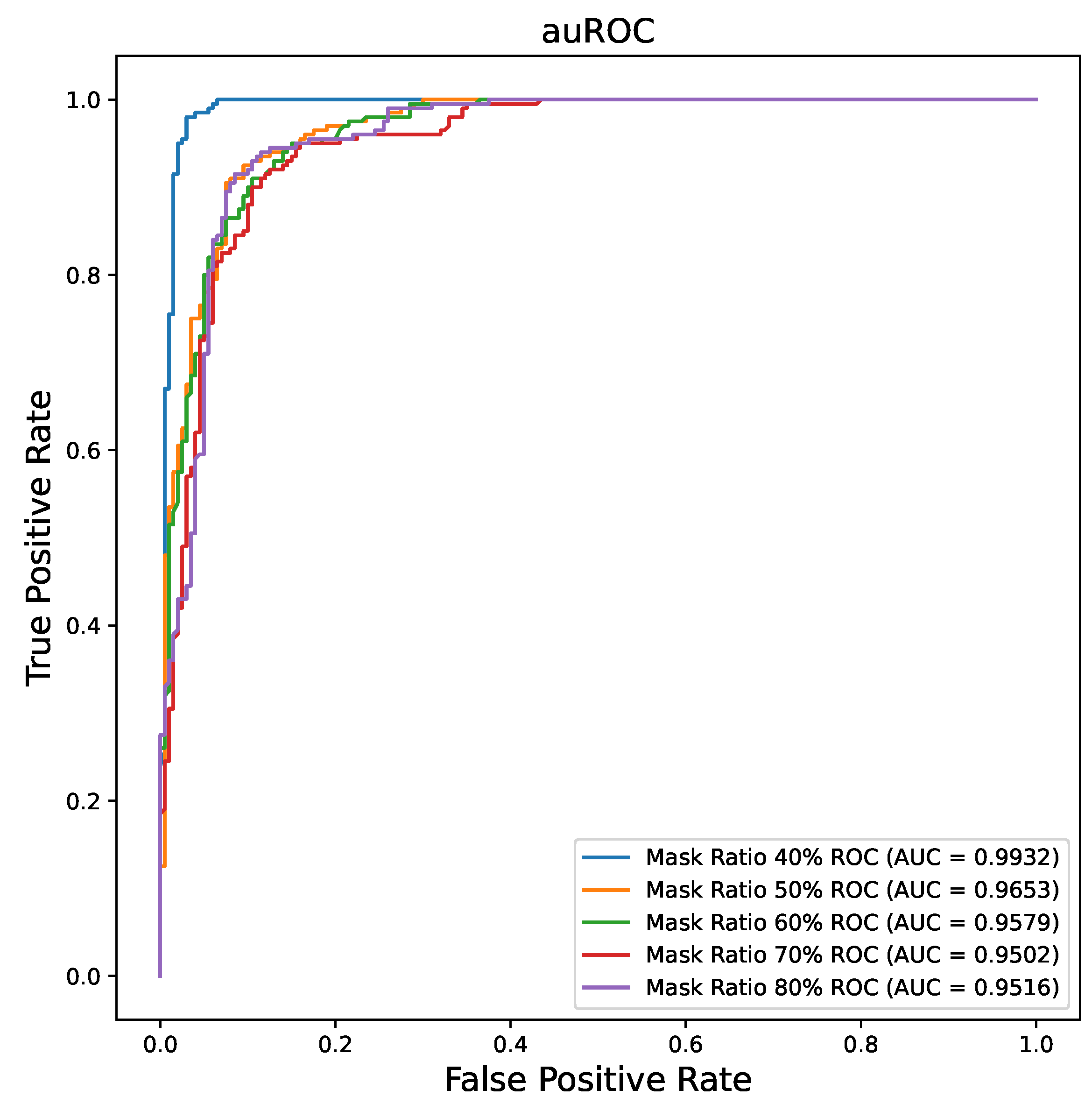
- To further investigate the impact of different mask ratios on the MAE model’s performance, we examined how varying mask ratios affected the effectiveness of the model. Our experiments revealed that the model achieved its best performance with a mask ratio of 0.4.
- Through extensive evaluations, we examined the applicability of the MAE model across different proportions of available training data. Remarkably, the MAE model achieved comparable performance even when trained with only 30% of the available data.
2. Materials and Methods
2.1. Data
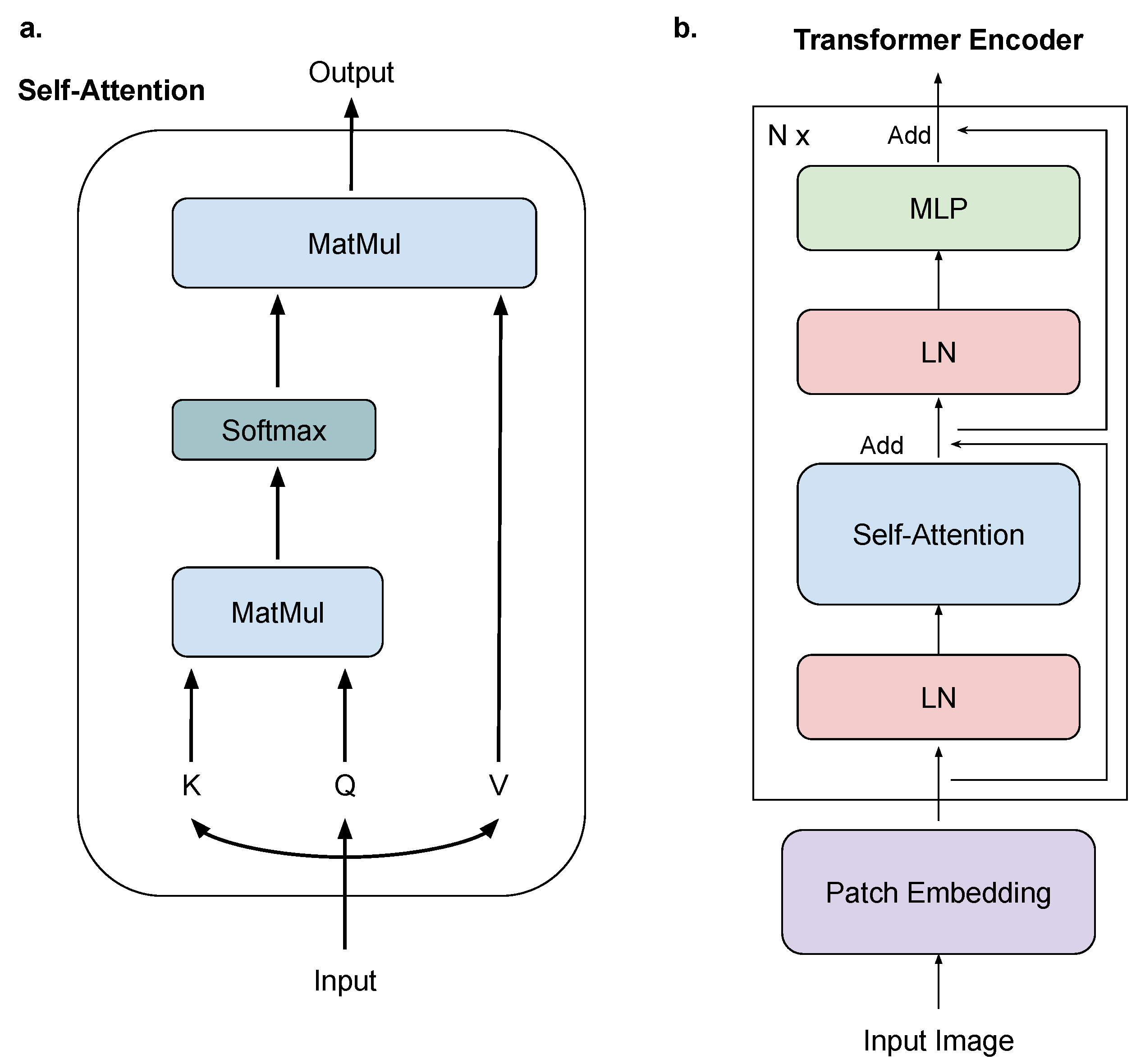
2.2. Vision Transformer
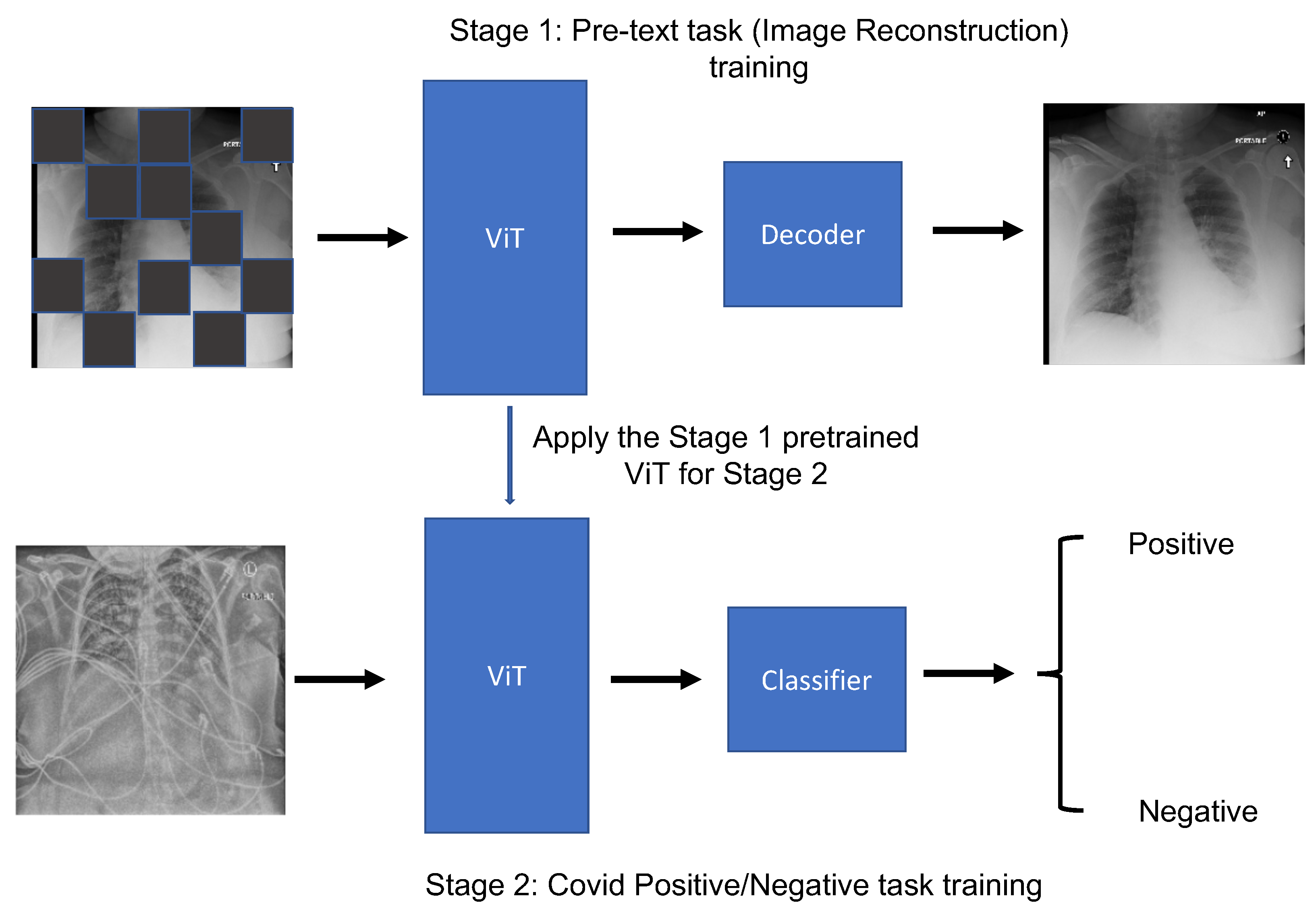
2.3. MAE
2.4. Loss Function
2.5. Implementation and Metrics
3. Results
3.1. Model Performance Increasing by MAE
3.2. Mask Ratio Influence on MAE Performance
3.3. MAE Performance on the Limited Training Dataset
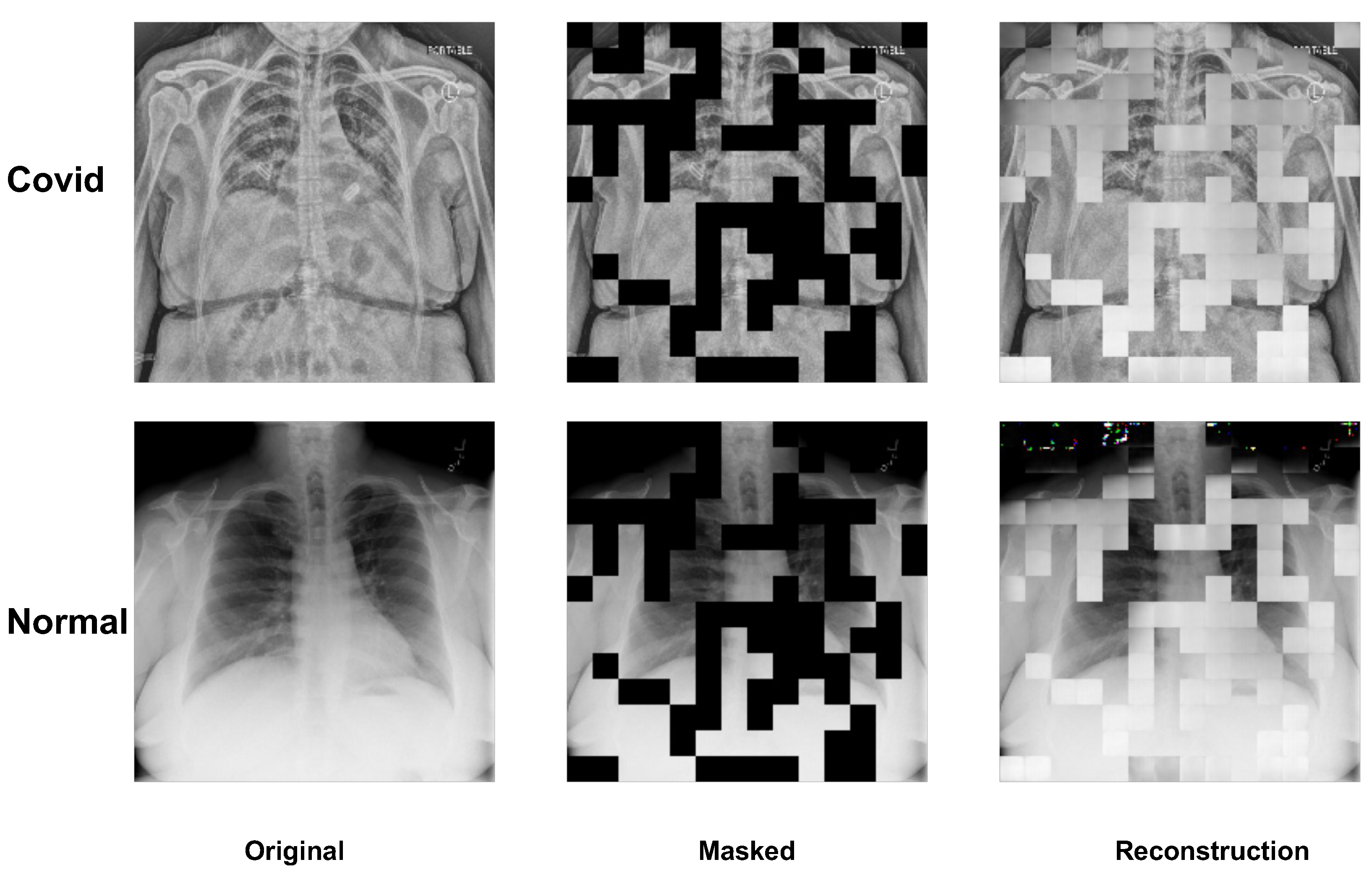
3.4. Visualization of MAE on Image Reconstruction
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Xing, X.; Peng, C.; Zhang, Y.; Lin, A.L.; Jacobs, N. AssocFormer: Association Transformer for Multi-label Classification. In Proceedings of the 33rd British Machine Vision Conference, London, UK, 21–24 November 2022. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3146–3154. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Ranjbarzadeh, R.; Jafarzadeh Ghoushchi, S.; Anari, S.; Safavi, S.; Tataei Sarshar, N.; Babaee Tirkolaee, E.; Bendechache, M. A deep learning approach for robust, multi-oriented, and curved text detection. Cogn. Comput. 2022, 1–13. [Google Scholar] [CrossRef]
- Anari, S.; Tataei Sarshar, N.; Mahjoori, N.; Dorosti, S.; Rezaie, A. Review of deep learning approaches for thyroid cancer diagnosis. Math. Probl. Eng. 2022, 2022, 5052435. [Google Scholar] [CrossRef]
- Xing, X.; Liang, G.; Zhang, Y.; Khanal, S.; Lin, A.L.; Jacobs, N. Advit: Vision transformer on multi-modality pet images for alzheimer disease diagnosis. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–4. [Google Scholar]
- Xing, X.; Rafique, M.U.; Liang, G.; Blanton, H.; Zhang, Y.; Wang, C.; Jacobs, N.; Lin, A.L. Efficient Training on Alzheimer’s Disease Diagnosis with Learnable Weighted Pooling for 3D PET Brain Image Classification. Electronics 2023, 12, 467. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Xing, X.; Liu, L.; Zhang, Y.; Ying, Q.; Lin, A.L.; Jacobs, N. Alzheimer’s disease classification using 2d convolutional neural networks. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 3008–3012. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the CVPR09, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Ying, X. An overview of overfitting and its solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Wang, X.; Liang, G.; Zhang, Y.; Blanton, H.; Bessinger, Z.; Jacobs, N. Inconsistent performance of deep learning models on mammogram classification. J. Am. Coll. Radiol. 2020, 17, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Xing, X.; Liang, G.; Blanton, H.; Rafique, M.U.; Wang, C.; Lin, A.L.; Jacobs, N. Dynamic image for 3d mri image alzheimer’s disease classification. In Proceedings of the Computer Vision—ECCV 2020 Workshops, Glasgow, UK, 23–28 August 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 355–364. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Durand, T.; Mordan, T.; Thome, N.; Cord, M. Wildcat: Weakly supervised learning of deep convnets for image classification, pointwise localization and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 642–651. [Google Scholar]
- Zhou, Z.H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018, 5, 44–53. [Google Scholar] [CrossRef]
- Liang, G.; Wang, X.; Zhang, Y.; Jacobs, N. Weakly-supervised self-training for breast cancer localization. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1124–1127. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wu, H.; Xiao, B.; Codella, N.; Liu, M.; Dai, X.; Yuan, L.; Zhang, L. Cvt: Introducing convolutions to vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; pp. 22–31. [Google Scholar]
- Goyal, P.; Caron, M.; Lefaudeux, B.; Xu, M.; Wang, P.; Pai, V.; Singh, M.; Liptchinsky, V.; Misra, I.; Joulin, A.; et al. Self-supervised pretraining of visual features in the wild. arXiv 2021, arXiv:2103.01988. [Google Scholar]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised representation learning by predicting image rotations. arXiv 2018, arXiv:1803.07728. [Google Scholar]
- Doersch, C.; Gupta, A.; Efros, A.A. Unsupervised visual representation learning by context prediction. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1422–1430. [Google Scholar]
- Grill, J.B.; Strub, F.; Altché, F.; Tallec, C.; Richemond, P.; Buchatskaya, E.; Doersch, C.; Avila Pires, B.; Guo, Z.; Gheshlaghi Azar, M.; et al. Bootstrap your own latent-a new approach to self-supervised learning. Adv. Neural Inf. Process. Syst. 2020, 33, 21271–21284. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 image data collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Figure 1-COVID-19 Chest X-ray Dataset Initiative. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset (accessed on 8 May 2020).
- Actualmed COVID-19 Chest X-ray 71 Dataset Initiative. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset (accessed on 11 November 2020).
- COVID-19 Radiography Database. Available online: https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database (accessed on 31 March 2021).
- RSNA Pneumonia Detection Challenge. Available online: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge (accessed on 17 October 2018).
- RSNA International COVID-19 Open Radiology Database. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70230281 (accessed on 15 January 2021).
- BIMCV-COVID19+. Available online: https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/ (accessed on 20 October 2020).
- COVID-19-NY-SBU. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=89096912 (accessed on 11 August 2021).
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Hasan, N.; Bao, Y.; Shawon, A.; Huang, Y. DenseNet convolutional neural networks application for predicting COVID-19 using CT image. SN Comput. Sci. 2021, 2, 389. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.C.; Xing, X.; Wang, C.; Ma, D.; Nho, K.; Crane, P.K.; Elahi, F.; Ziegler, D.A.; Liang, G.; Cheng, Q.; et al. β-amyloid and tau drive early Alzheimer’s disease decline while glucose hypometabolism drives late decline. Commun. Biol. 2020, 3, 352. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.C.; Xing, X.; Yanckello, L.M.; Stromberg, A.; Chang, Y.H.; Nelson, P.T.; Lin, A.L. Human Gray and White Matter Metabolomics to Differentiate APOE and Stage Dependent Changes in Alzheimer’s Disease. J. Cell. Immunol. 2021, 3, 397. [Google Scholar] [PubMed]
- Ying, Q.; Xing, X.; Liu, L.; Lin, A.L.; Jacobs, N.; Liang, G. Multi-modal data analysis for alzheimer’s disease diagnosis: An ensemble model using imagery and genetic features. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 3586–3591. [Google Scholar]
- Zhao, Y.; Zeng, K.; Zhao, Y.; Bhatia, P.; Ranganath, M.; Kozhikkavil, M.L.; Li, C.; Hermosillo, G. Deep learning solution for medical image localization and orientation detection. Med. Image Anal. 2022, 81, 102529. [Google Scholar] [CrossRef] [PubMed]



| Type | Negative | Positive | Total |
|---|---|---|---|
| Images Distribution | |||
| Train | 13,992 | 15,994 | 29,986 |
| Test | 200 | 200 | 400 |
| Patients Distribution | |||
| Train | 13,850 | 2808 | 16,648 |
| Test | 200 | 178 | 378 |
| Type | Acc | AUC | F1 | Precision | Recall | AP |
|---|---|---|---|---|---|---|
| DenseNet121 | 0.9775 | 0.9970 | 0.9771 | 0.9948 | 0.96 | 0.9750 |
| ResNet50 | 0.9650 | 0.9969 | 0.9641 | 0.9894 | 0.94 | 0.9601 |
| ViT-scratch | 0.7075 | 0.7808 | 0.7082 | 0.7065 | 0.7100 | 0.6466 |
| ViT-pretrain | 0.9783 | 0.9340 | 0.9484 | 0.9200 | 0.9125 | |
| ViT-MAE | 0.9850 | 0.9957 | 0.9850 | 0.9950 | 0.9850 | 0.9859 |
| Ratio | Acc | AUC | F1 | Precision | Recall | AP |
|---|---|---|---|---|---|---|
| 0.4 | 0.9850 | 0.9957 | 0.9850 | 0.9850 | 0.9850 | 0.9559 |
| 0.5 | 0.9100 | 0.9653 | 0.9086 | 0.9277 | 0.8950 | 0.8783 |
| 0.6 | 0.8875 | 0.9579 | 0.8819 | 0.9282 | 0.8400 | 0.8597 |
| 0.7 | 0.8900 | 0.9502 | 0.8894 | 0.8939 | 0.8850 | 0.8486 |
| 0.8 | 0.8925 | 0.9516 | 0.8900 | 0.9110 | 0.8700 | 0.8576 |
| Percentage (%) | Acc | AUC | F1 | Precision | Recall | AP |
|---|---|---|---|---|---|---|
| 10 | 0.8800 | 0.9394 | 0.8776 | 0.8958 | 0.8600 | 0.8404 |
| 20 | 0.8925 | 0.9588 | 0.8877 | 0.9290 | 0.8500 | 0.8646 |
| 30 | 0.9425 | 0.9772 | 0.9415 | 0.9585 | 0.9250 | 0.9242 |
| 40 | 0.9600 | 0.9866 | 0.9602 | 0.9554 | 0.9650 | 0.9395 |
| 50 | 0.9675 | 0.9879 | 0.9673 | 0.9746 | 0.9600 | 0.9556 |
| 60 | 0.9650 | 0.9876 | 0.9645 | 0.9794 | 0.9500 | 0.9554 |
| 70 | 0.9675 | 0.9896 | 0.9669 | 0.9845 | 0.9500 | 0.9602 |
| 80 | 0.9775 | 0.9978 | 0.9771 | 0.9948 | 0.9600 | 0.9750 |
| 90 | 0.9825 | 0.9944 | 0.9823 | 0.9949 | 0.9700 | 0.9800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, X.; Liang, G.; Wang, C.; Jacobs, N.; Lin, A.-L. Self-Supervised Learning Application on COVID-19 Chest X-ray Image Classification Using Masked AutoEncoder. Bioengineering 2023, 10, 901. https://doi.org/10.3390/bioengineering10080901
Xing X, Liang G, Wang C, Jacobs N, Lin A-L. Self-Supervised Learning Application on COVID-19 Chest X-ray Image Classification Using Masked AutoEncoder. Bioengineering. 2023; 10(8):901. https://doi.org/10.3390/bioengineering10080901
Chicago/Turabian StyleXing, Xin, Gongbo Liang, Chris Wang, Nathan Jacobs, and Ai-Ling Lin. 2023. "Self-Supervised Learning Application on COVID-19 Chest X-ray Image Classification Using Masked AutoEncoder" Bioengineering 10, no. 8: 901. https://doi.org/10.3390/bioengineering10080901
APA StyleXing, X., Liang, G., Wang, C., Jacobs, N., & Lin, A.-L. (2023). Self-Supervised Learning Application on COVID-19 Chest X-ray Image Classification Using Masked AutoEncoder. Bioengineering, 10(8), 901. https://doi.org/10.3390/bioengineering10080901






