Identification of Cell-Attachment Factors Derived from Green Algal Cells Disrupted by Sonication in Fabrication of Cell Plastics
Abstract
1. Introduction
2. Materials and Methods
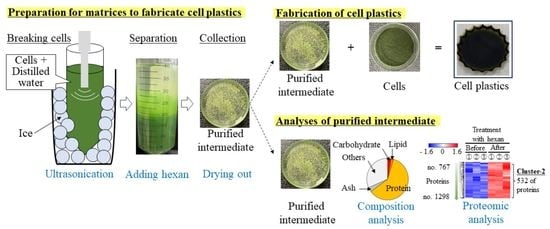
2.1. Use of Cells as Resources of Cell Plastics
2.2. Supply of Matrices
2.3. Fabrication of the Cell Plastics
2.4. Tensile Test
2.5. Evaluation of Composition in Matrix
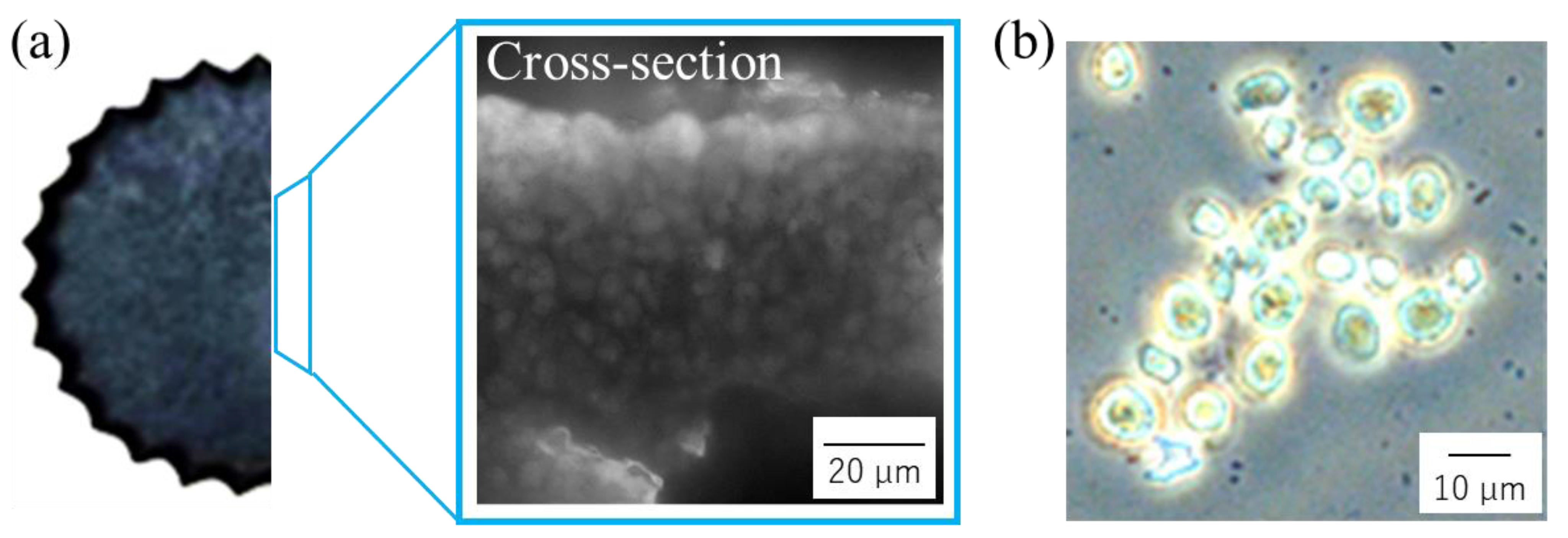
2.6. Observation by Microscopy
2.7. Evaluation of Remained Cell Numbers as Structures of Cell Plastics
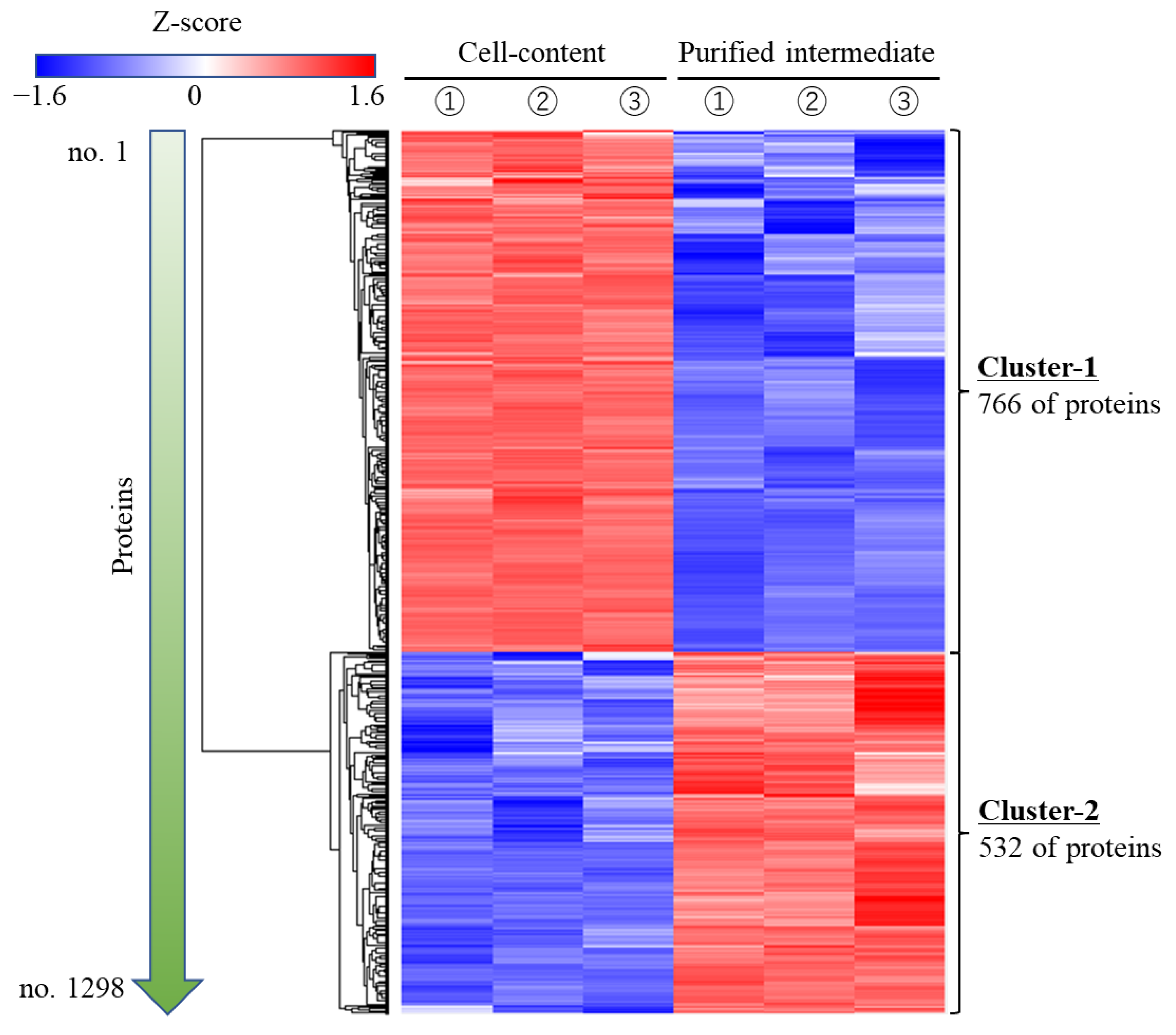
2.8. Analysis of Proteomics
3. Results
3.1. Evaluation of the Composition of Cells Prepared by Sonication
3.2. Tensile Test of Cell Plastics Composed of Intermediate as a Matrix
3.3. Evaluation of Cells as the Structural Components of Cell Plastics
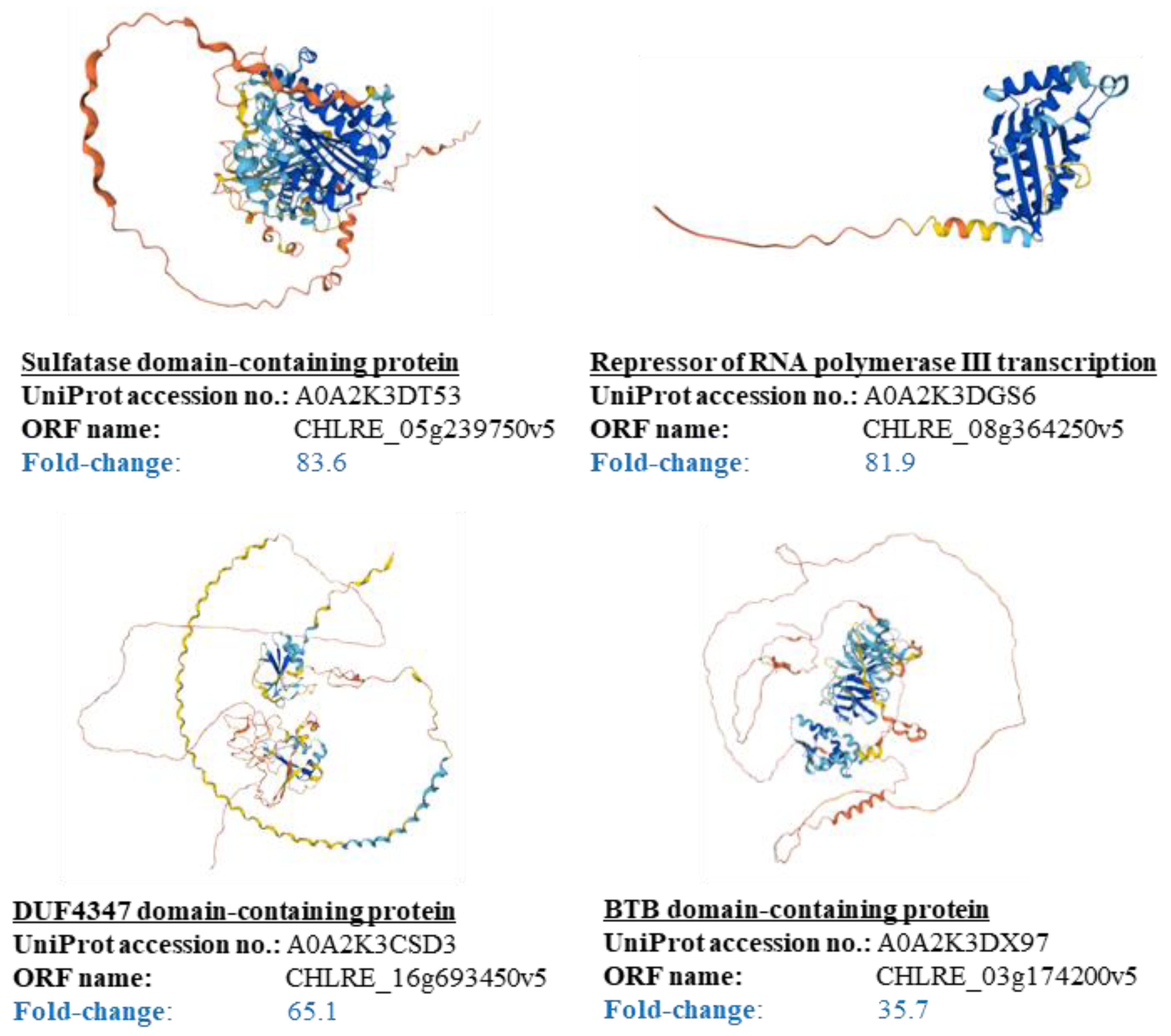
3.4. Suggestion of Attachment Factors to Cells in Cell Plastics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Department of Economic and Social Affairs. Available online: https://sdgs.un.org/2030agenda (accessed on 29 May 2023).
- Ralston, J.; Cooper, K.; Powis, J. Obesity, SDGs and ROOTS: A framework for impact. Curr. Obes. Rep. 2021, 10, 54–60. [Google Scholar] [CrossRef]
- Plastic Production Worldwide 2021|Statista. Available online: https://www.statista.com/statistics/282732/global-production-of-plastics-since-1950/ (accessed on 29 May 2023).
- Karaagac, E.; Jones, M.P.; Koch, T.; Archodoulaki, V.M. Polypropylene contamination in post-consumer polyolefin waste: Characterisation, consequences and compatibilisation. Polymers 2021, 13, 2618. [Google Scholar] [CrossRef] [PubMed]
- Center for International Environment Law. Available online: https://www.ciel.org/wp-content/uploads/2017/09/Fueling-Plastics-Fossils-Plastics-Petrochemical-Feedstocks.pdf. (accessed on 25 November 2021).
- Adhikari, D.; Mukai, M.; Kubota, K.; Kai, T.; Kaneko, N.; Araki, K.S.; Kubo, T. Degradation of bioplastics in soil and their degradation effects on environmental microorganisms. J. Agric. Chem. Environ. 2016, 5, 23–34. [Google Scholar]
- Lambert, S.; Wagner, M. Environmental performance of bio-based and biodegradable plastics: The road ahead. Chem. Soc. Rev. 2017, 46, 6855–6871. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Iritani, K.; Sakihama, Y. Developing neo-bioplastics for the realization of carbon sustainable society. J. Nanotechnol. Nanomater. 2020, 1, 72–85. [Google Scholar]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic pollution in the world’s oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef]
- Ferreira-Filipe, D.A.; Paço, A.; Duarte, A.C.; Rocha-Santos, T.; Patrício Silva, A.T. Are biobased plastics green alternatives?—A critical review. Int. J. Environ. Res. Public Health 2021, 18, 7729. [Google Scholar] [CrossRef]
- Garavand, F.; Rouhi, M.; Razavi, S.H.; Cacciotti, I.; Mohammadi, R. Improving the integrity of natural biopolymer films used in food packaging by crosslinking approach: A review. Int. J. Biol. Macromol. 2017, 104, 687–707. [Google Scholar] [CrossRef]
- Hottle, T.A.; Bilec, M.M.; Landis, A.E. Sustainability assessments of bio-based polymers. Polym. Degrad. Stab. 2013, 98, 1898–1907. [Google Scholar] [CrossRef]
- Arraiza, M.P.; López, J.V.; Fernando, A. Aerobic degradation of bioplastic materials. In Environmental Security and Solid Waste Management, Proceedings of the 1st International Workshop on Environmental Security, Geological Hazards and Management, San Cristobal de La Laguna, Spain, 10–12 April 2013; Santamarta-Cerezal, J.C., Gutiérrez, L.E.H., Eds.; Universidad de La Laguna: San Cristóbal de La Laguna, Spain; pp. 10–12.
- Meixner, K.; Kovalcik, A.; Sykacek, E.; Gruber-Brunhumerabe, M.; Zeilinger, W.; Markl, K.; Haas, C.; Fritz, I.; Mundigler, N.; Stelzer, F.; et al. Cyanobacteria biorefinery—Production of poly (3-hydroxybutyrate) with Synechocystis salina and utilization of residual biomass. J. Biotechnol. 2018, 265, 46–53. [Google Scholar] [CrossRef]
- Nakanishi, A.; Iritani, K.; Sakihama, Y.; Ozawa, N.; Mochizuki, A.; Watanabe, M. Construction of cell-plastics as neo-plastics consisted of cell-layer provided green alga Chlamydomonas reinhardtii covered by two-dimensional polymer. AMB Expr. 2020, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Iritani, K.; Sakihama, Y.; Watanabe, M. Investigation of the mechanical strength of cell-plastics fabricated using unicellular green algal cells and varying weight ratios of biodegradable polybutylene succinate. Int. J. Microbiol. Biotechnol. 2020, 5, 159–164. [Google Scholar] [CrossRef]
- Nakanishi, A.; Iritani, K.; Sakihama, Y.; Watanabe, M.; Mochizuki, A.; Tsuruta, A.; Sakamoto, S.; Ota, A. Fabrication and biodegradability of starch cell-plastics as recyclable resources. Appl. Sci. 2021, 11, 847. [Google Scholar] [CrossRef]
- Iritani, K.; Nakanishi, A.; Ota, A.; Yamashita, T. Fabrication of novel functional cell-plastic using polyvinyl alcohol: Effects of Nakanishi, A cross-linking structure and mixing ratio of components on the mechanical and thermal properties. Glob. Chall. 2021, 5, 2100026. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Iritani, K.; Tsuruta, A.; Yamamoto, N.; Watanabe, M.; Ozawa, N.; Watanabe, M.; Zhang, K.; Tokudome, A. Fabrication of cell-plastics composed only of unicellular green alga Chlamydomonas reinhardtii as a raw material. Appl. Microbiol. Biotechnol. 2022, 106, 4459–4468. [Google Scholar] [CrossRef]
- Schreiber, C.; Schiedung, H.; Harrison, L.; Briese, C.; Ackermann, B.; Kant, J.; Schrey, S.D.; Hofmann, D.; Singh, D.; Ebenhöh, O.; et al. Evaluating potential of green alga Chlorella vulgaris to accumulate phosphorus and to fertilize nutrient-poor soil substrates for crop plants. J. Appl. Phycol. 2018, 30, 2827–2836. [Google Scholar] [CrossRef]
- Alvarez, A.L.; Weyers, S.L.; Goemann, H.M.; Peyton, B.M.; Gardner, R.D. Microalgae, soil and plants: A critical review of microalgae as renewable resources for agriculture. Algal Res. 2021, 54, 102200. [Google Scholar] [CrossRef]
- Dementyeva, P.; Freudenberg, R.A.; Baier, T.; Rojek, K.; Wobbe, L.; Weisshaar, B.; Kruse, O. A novel, robust and mating-competent Chlamydomonas reinhardtii strain with an enhanced transgene expression capacity for algal biotechnology. Biotechnol. Rep. 2021, 31, e00644. [Google Scholar] [CrossRef]
- Sathe, S.; Durand, P.M. Cellular aggregation in Chlamydomonas (Chlorophyceae) is chimaeric and depends on traits like cell size and motility. Eur. J. Phycol. 2016, 51, 129–138. [Google Scholar] [CrossRef]
- Williams, J.G.K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Meth. Enzymol. 1988, 167, 766–778. [Google Scholar]
- Ma, N.; Liu, D.; Liu, Y.; Sui, G. Extraction and characterization of nanocellulose from Xanthoceras sorbifolia husks. Int. J. Nanosci. Nanotechnol. 2015, 2, 43–50. [Google Scholar]
- Dreywood, R. Qualitative test for carbohydrate material. Ind. Eng. Chem. Anal. Ed. 1946, 18, 499. [Google Scholar] [CrossRef]
- Morris, D.L. Quantitative determination of carbohydrates with dreywood’s anthrone reagent. Science 1948, 107, 254–255. [Google Scholar] [CrossRef]
- Ho, S.H.; Nakanishi, A.; Ye, X.; Chang, J.S.; Chen, C.Y.; Hasunuma, T.; Kondo, A. Dynamic metabolic profiling of the marine microalga Chlamydomonas sp. JSC4 and enhancing its oil production by optimizing light intensity. Biotechnol. Biofuels 2015, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Ho, S.H.; Kato, Y.; Yamasaki, H.; Chang, J.S.; Misawa, N.; Hirose, Y.; Minagawa, J.; Hasunuma, T.; Kondo, A. Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci. Rep. 2017, 7, 45471. [Google Scholar]
- Wang, M.; Yuan, W.; Jiang, X.; Jing, Y.; Wang, Z. Disruption of microalgal cells using high-frequency focused ultrasound. Bioresour. Technol. 2014, 153, 315–321. [Google Scholar] [CrossRef]
- Duan, Z.; Tan, X.; Guo, J.; Kahehu, C.W.; Yang, H.; Zheng, X.; Zhu, F. Effects of biological and physical properties of microalgae on disruption induced by a low-frequency ultrasound. J. Appl. Phycol. 2017, 29, 2937–2946. [Google Scholar] [CrossRef]
- Prewo, K.M. Tension and flexural strength of silicon carbide fibre-reinforced glass ceramics. J. Mater. Sci. 1986, 21, 3590–3600. [Google Scholar] [CrossRef]
- Callister, W.D. Characteristics, applications, and processing of polymers. In Materials Science and Engineering, 7th ed.; Hayton, J., Ed.; John Wiley and Sons, Inc.: New York, NY, USA, 2007; pp. 523–576. ISBN 978-0-471-73696-7. [Google Scholar]
- Beer, F.P.; Johnston, E.R.; Dewolf, J.T.; Mazurek, D.F. Mechanics of Materials, 6th ed.; McGraw-Hill Companies: New York, NY, USA, 2012; pp. 52–139. ISBN 978-0-07-338028-5. [Google Scholar]
- Ridwan, R.; Prabowo, A.R.; Muhayat, N.; Putranto, T.; Sohn, J.M. Tensile analysis and assessment of carbon and alloy steels using FE approach as an idealization of material fractures under collision and grounding. Curved Layer Struct. 2020, 7, 188–198. [Google Scholar] [CrossRef]
- Boey, J.Y.; Lee, C.K.; Tay, G.S. Factors affecting mechanical properties of reinforced bioplastics. Polymers 2022, 14, 3737. [Google Scholar] [CrossRef]
- Stauber, E.J.; Fink, A.; Markert, C.; Kruse, O.; Johanningmeier, U.; Hippler, M. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell. 2003, 2, 978–994. [Google Scholar] [CrossRef] [PubMed]
- Diniz, M.C.; Pacheco, A.C.L.; Farias, K.M.; Oliveira, D.M.D. The eukaryotic flagellum makes the day: Novel and unforeseen roles uncovered after post-genomics and proteomics data. Curr. Protein Pept. Sci. 2012, 13, 524–546. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A.; Pancholi, V.; Schneewind, O. Common characteristics of the surface proteins from grampositive cocci. In Genetics and Molecular Biology of STREPTOCOCCI, Lactococci and Enterococcil; Dunny, G.M., Cleary, P.P., McKay, L.L., Eds.; American Society for Microbiology: Washington, DC, USA, 1991; pp. 290–294. ISBN 978-155-581-034-4. [Google Scholar]
- Roditi, I.; Schwarz, H.; Pearson, T.W.; Beecroft, R.P.; Liu, M.K.; Richardson, J.P.; Buhring, H.J.; Pleiss, J.; Bulow, R.; Williams, R.O.; et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma Brucei. J. Cell Biol. 1989, 108, 737–746. [Google Scholar] [CrossRef]
- Timoney, J.F.; Muktar, M.; Ding, J. M proteins of the equine group C streptococci. In Genetics and Molecular Biology of Streptococci, Lactococci and Enterococci; Dunny, G.M., Cleary, P.P., McKay, L.L., Eds.; American Society for Microbiology: Washington, DC, USA, 1991; pp. 160–164. ISBN 978-155-581-034-4. [Google Scholar]
- Hedén, L.O.; Frithz, E.; Lindahl, G. Molecular characterization of an IgA receptor from group B streptococci: Sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 1991, 21, 1481–1490. [Google Scholar] [CrossRef]
- Pancholi, V.; Fischetti, V.A. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J. Bacteriol. 1988, 170, 2618–2624. [Google Scholar] [CrossRef]
- Fahnestock, S.R.; Alexander, P.; Nagle, J.; Filpula, D. Gene for an immunoglobulin-binding protein from a group G Streptococcus. J. Bacteriol. 1986, 167, 870–880. [Google Scholar] [CrossRef]
- Li, L.J.; Dougan, G.; Novotny, P.; Charles, I.G.P. 70 pertactin, an outer-membrane protein from Bordetella parapertussis: Cloning, nucleotide sequence and surface expression in Escherichia coli. Mol. Microbiol. 1991, 5, 409–417. [Google Scholar] [CrossRef]
- Cronmiller, E.; Toor, D.; Shao, N.C.; Kariyawasam, T.; Wang, M.H.; Lee, J.H. Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 12204. [Google Scholar] [CrossRef]
- Mitaku, S.; Suzuki, K.; Odashima, S.; Ikuta, K.; Suwa, M.; Kukita, F.; Ishikawa, M.; Itoh, H. Interaction stabilizing tertiary structure of bacteriorhodopsin studied by denaturation experiments. Proteins 1995, 22, 350–362. [Google Scholar] [CrossRef]
- Pekkala, M.; Hieta, R.; Kursula, P.; Kivirikko, K.I.; Wierenga, R.K.; Myllyharju, J. Crystallization of the proline-rich-peptide binding domain of human type I collagen prolyl 4-hydroxylase. Acta. Crystallogr. D Biol. Crystallogr. 2003, 59, 940–942. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.D.; Wagner, W.L.; Bennett, R.D.; Ysasi, A.B.; Belle, J.M.; Molter, K.; Straub, B.K.; Wang, D.; Chen, Z.; Ackermann, M.; et al. Extracellular assembly of the elastin cable line element in the developing lung. Anat. Rec. 2017, 300, 1670–1679. [Google Scholar] [CrossRef] [PubMed]



| Hexane Purification | Matrix Contents (wt%) | Young’s Modulus (MPa) | Tensile Strength (MPa) | Strains (%) |
|---|---|---|---|---|
| w/ | 0.5 | 227.3 ± 29.5 | 4.6 ± 2.0 | 1.6 ± 0.4 |
| 1.0 | 358.3 ± 143.2 | 3.6 ± 1.0 | 1.3 ± 0.4 | |
| 1.5 | 422.1 ± 105.9 | 4.3 ± 2.6 | 1.1 ± 0.3 | |
| 2.0 | 332.5 ± 110.0 | 3.5 ± 1.8 | 1.2 ± 0.6 | |
| w/o | 0.5 | 562.9 ± 11.8 | 7.7 ± 3.3 | 1.3 ± 0.5 |
| 1.0 | 550.1 ± 37.9 | 5.1 ± 1.4 | 1.0 ± 0.3 | |
| 1.5 | 564.4 ± 26.8 | 6.8 ± 4.6 | 0.4 ± 0.6 | |
| 2.0 | 553.4 ± 65.8 | 7.0 ± 1.0 | 1.4 ± 0.1 |
| Fold-Change (B/A) | Protein Name | AA Residue- Number of Exposed Protein Parts | Composition of AA (%) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | D | E | F | G | H | I | K | L | M | N | P | Q | R | S | T | V | W | Y | ||||
| 83.62 | Sulfatase N- terminal domain- containing protein | 114 | 8.8 | 0.9 | 4.4 | 1.8 | 0.9 | 12.3 | 0.9 | 0.9 | 3.5 | 7.0 | 2.6 | 2.6 | 24.6 | 4.4 | 4.4 | 11.4 | 1.8 | 3.5 | 0.9 | 2.6 | |
| 81.90 | Repressor of RNA polymerase III transcription | 38 | 15.8 | 0.0 | 7.9 | 0.0 | 5.3 | 10.5 | 2.6 | 2.6 | 5.3 | 7.9 | 0.0 | 0.0 | 15.8 | 2.6 | 0.0 | 5.3 | 5.3 | 5.3 | 0.0 | 7.9 | |
| 81.51 | UBC core domain-containing protein | 163 | 6.1 | 3.1 | 5.5 | 6.1 | 2.5 | 7.4 | 1.8 | 4.9 | 3.1 | 8.0 | 3.7 | 3.7 | 8.6 | 3.1 | 7.4 | 9.2 | 3.1 | 7.4 | 2.5 | 3.1 | |
| 65.07 | DUF4347 domain-containing protein | 226 | 7.5 | 0.0 | 0.4 | 5.3 | 0.0 | 7.1 | 0.9 | 0.4 | 1.8 | 0.9 | 0.4 | 0.0 | 45.1 | 0.0 | 1.3 | 14.2 | 3.5 | 4.4 | 0.0 | 6.2 | |
| 35.74 | BTB domain-containing protein | 400 | 24.5 | 0.3 | 2.0 | 1.3 | 1.3 | 10.0 | 0.5 | 1.8 | 0.5 | 4.3 | 1.0 | 0.5 | 16.5 | 10.5 | 2.3 | 8.5 | 5.3 | 1.8 | 0.3 | 7.3 | |
| 32.51 | C3H1-type domain-containing protein | Arm A | 171 | 15.8 | 0.6 | 0.6 | 2.9 | 0.6 | 19.9 | 1.8 | 1.2 | 1.8 | 4.7 | 5.3 | 7.6 | 4.1 | 4.1 | 2.3 | 17.5 | 6.4 | 2.9 | 0.0 | 0.0 |
| Arm B | 227 | 26.9 | 0.0 | 0.4 | 0.4 | 0.4 | 15.0 | 4.8 | 1.8 | 0.0 | 7.9 | 4.8 | 1.8 | 4.4 | 13.7 | 1.3 | 5.7 | 2.2 | 5.3 | 0.0 | 2.6 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakanishi, A.; Nemoto, S.; Yamamoto, N.; Iritani, K.; Watanabe, M. Identification of Cell-Attachment Factors Derived from Green Algal Cells Disrupted by Sonication in Fabrication of Cell Plastics. Bioengineering 2023, 10, 893. https://doi.org/10.3390/bioengineering10080893
Nakanishi A, Nemoto S, Yamamoto N, Iritani K, Watanabe M. Identification of Cell-Attachment Factors Derived from Green Algal Cells Disrupted by Sonication in Fabrication of Cell Plastics. Bioengineering. 2023; 10(8):893. https://doi.org/10.3390/bioengineering10080893
Chicago/Turabian StyleNakanishi, Akihito, Shintaro Nemoto, Naotaka Yamamoto, Kohei Iritani, and Marina Watanabe. 2023. "Identification of Cell-Attachment Factors Derived from Green Algal Cells Disrupted by Sonication in Fabrication of Cell Plastics" Bioengineering 10, no. 8: 893. https://doi.org/10.3390/bioengineering10080893
APA StyleNakanishi, A., Nemoto, S., Yamamoto, N., Iritani, K., & Watanabe, M. (2023). Identification of Cell-Attachment Factors Derived from Green Algal Cells Disrupted by Sonication in Fabrication of Cell Plastics. Bioengineering, 10(8), 893. https://doi.org/10.3390/bioengineering10080893