Automated Prediction of Osteoarthritis Level in Human Osteochondral Tissue Using Histopathological Images
Abstract
1. Introduction
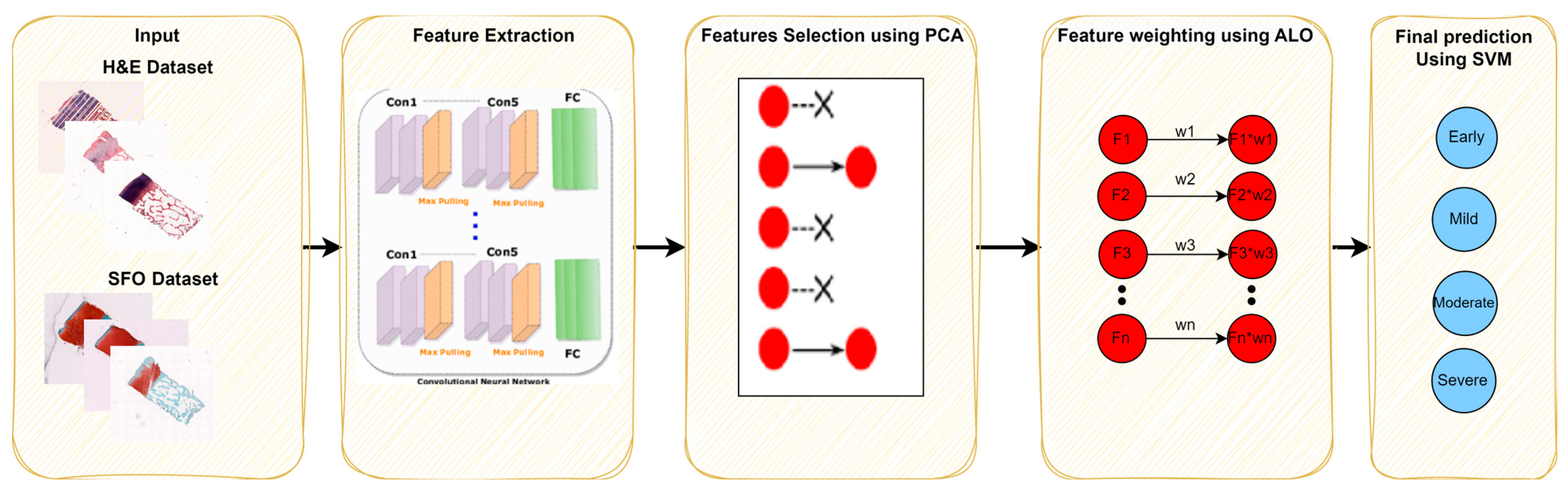
2. Materials and Methods
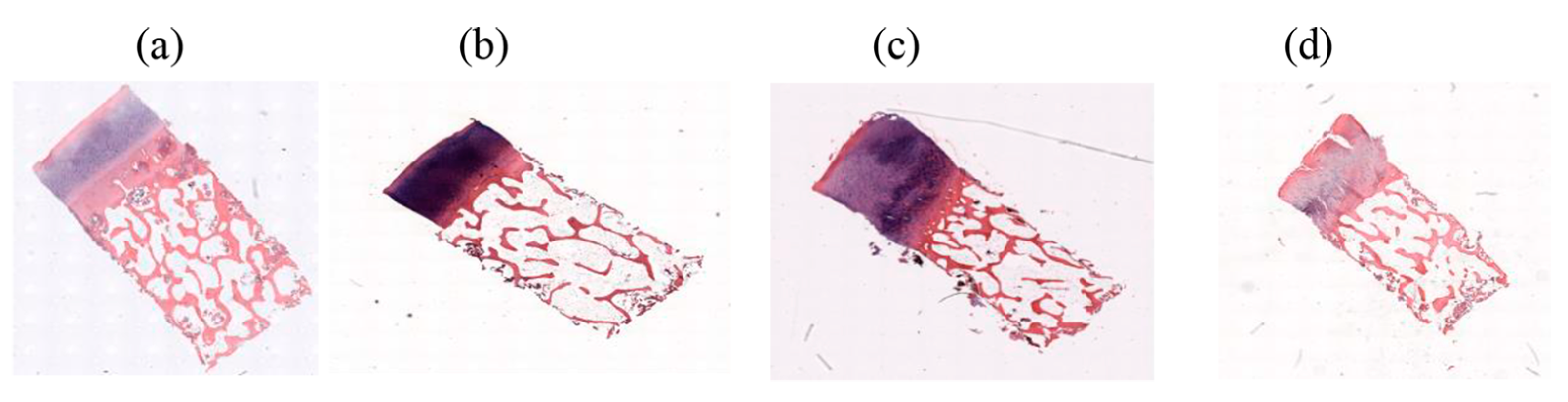
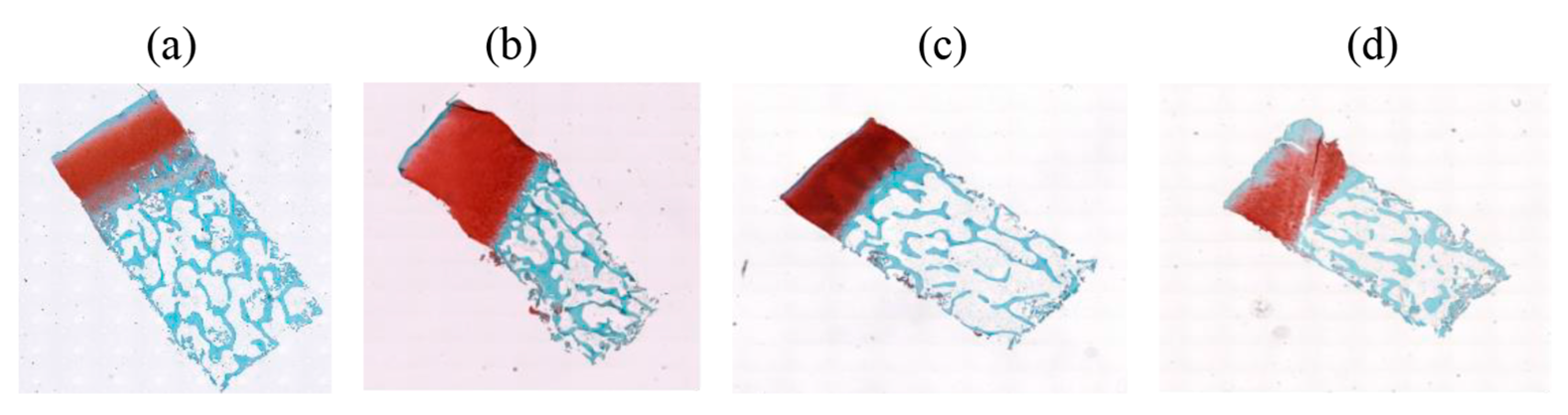
2.1. Database
2.2. Deep Learning Features
2.2.1. DarkNet-19
2.2.2. NasNet
2.2.3. ResNet-101
2.2.4. ShuffleNet
2.2.5. MobileNet
2.3. Features Engineering
2.3.1. Principal Component Analysis
- Standardization: this step is performed by standardizing each column feature that makes the mean for each feature zero, and the variance is unity.
- Covariance matrix: this step is performed by constructing the covariance matrix, which is a square matrix that reflects the variance between each pair of features; its diagonal represents the variance for each feature and the off-diagonal represents the covariance between each pair of features.
- Computation of the principal components: this step is performed by computing the eigenvector, which explains the direction of maxim variance, and the eigenvalue that quantifies the amount of maximum variance.
- Selection of the principal components: the principal components are selected based on 95% of the majority variance of the features.
- Mapping between the selected principal components and the features: this is performed by projecting the standardized features onto the best principal components.
2.3.2. Feature Weighting Using ALO
- Initialize the population of ant lions randomly.
- Evaluate the accuracy of each ant lion in the population based on both weight and k-value.
- Define the king ant lion based on the highest accuracy.
- Move the ant lions towards the king ant lion using a certain formula that simulates the hunting behavior of the ant lions.
- Calculate the accuracy for the new position.
- Repeat steps 3–5 until the stopping criterion is met.
- The results are the optimized weights.
2.4. Support Vector Machine
3. Results
3.1. Pre-Trained Model Classification
3.2. Deep Features with SVM
3.3. Principal Component Analysis (PCA)
- Four features from MobileNet.
- Three features from ShuffleNet.
- Two features from NasNet.
- One feature from ResNet-101.
- Three features from MobileNet.
- Three features from ShuffleNet.
- Two features from NasNet
- Two features from DarkNet
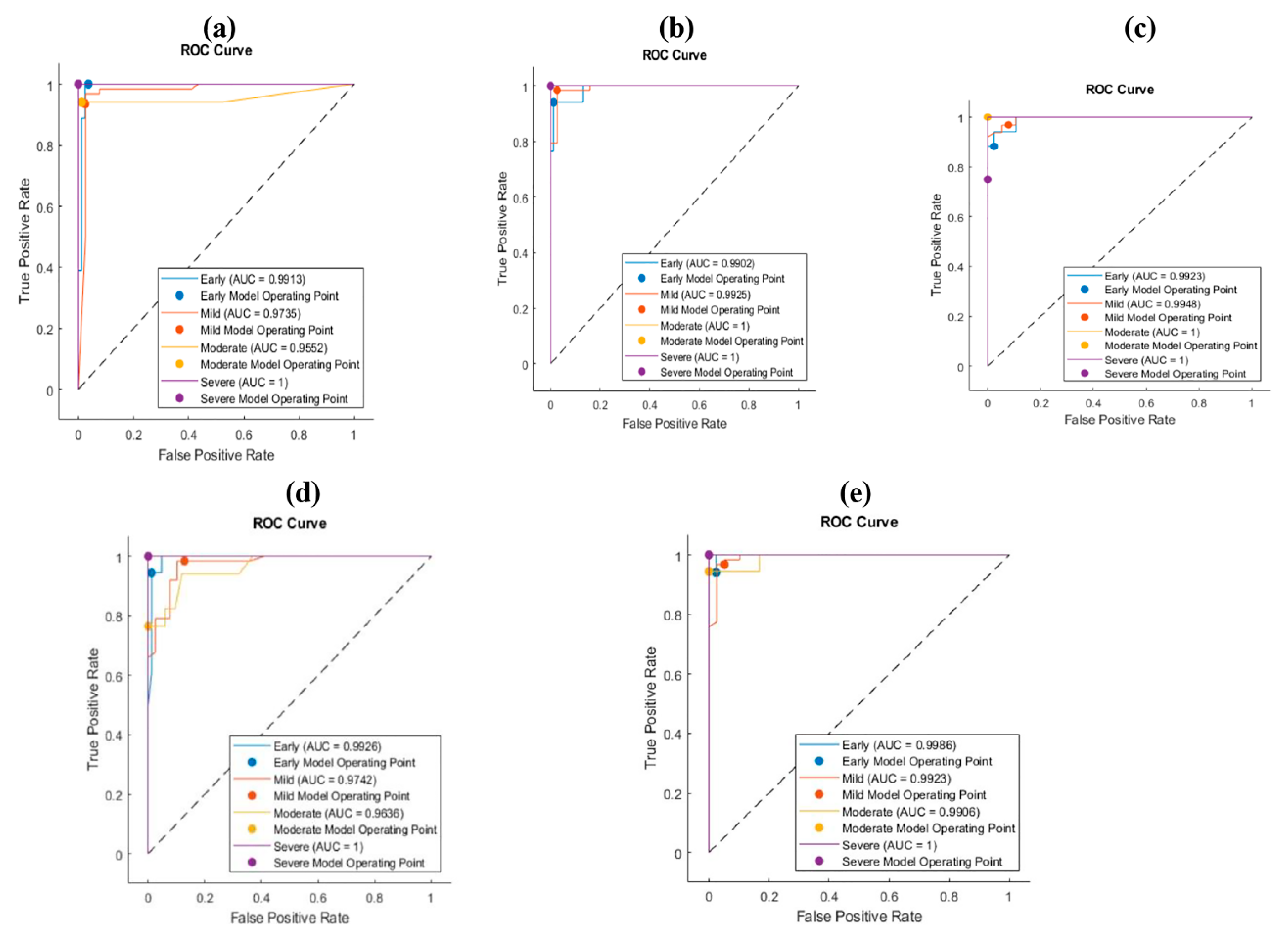
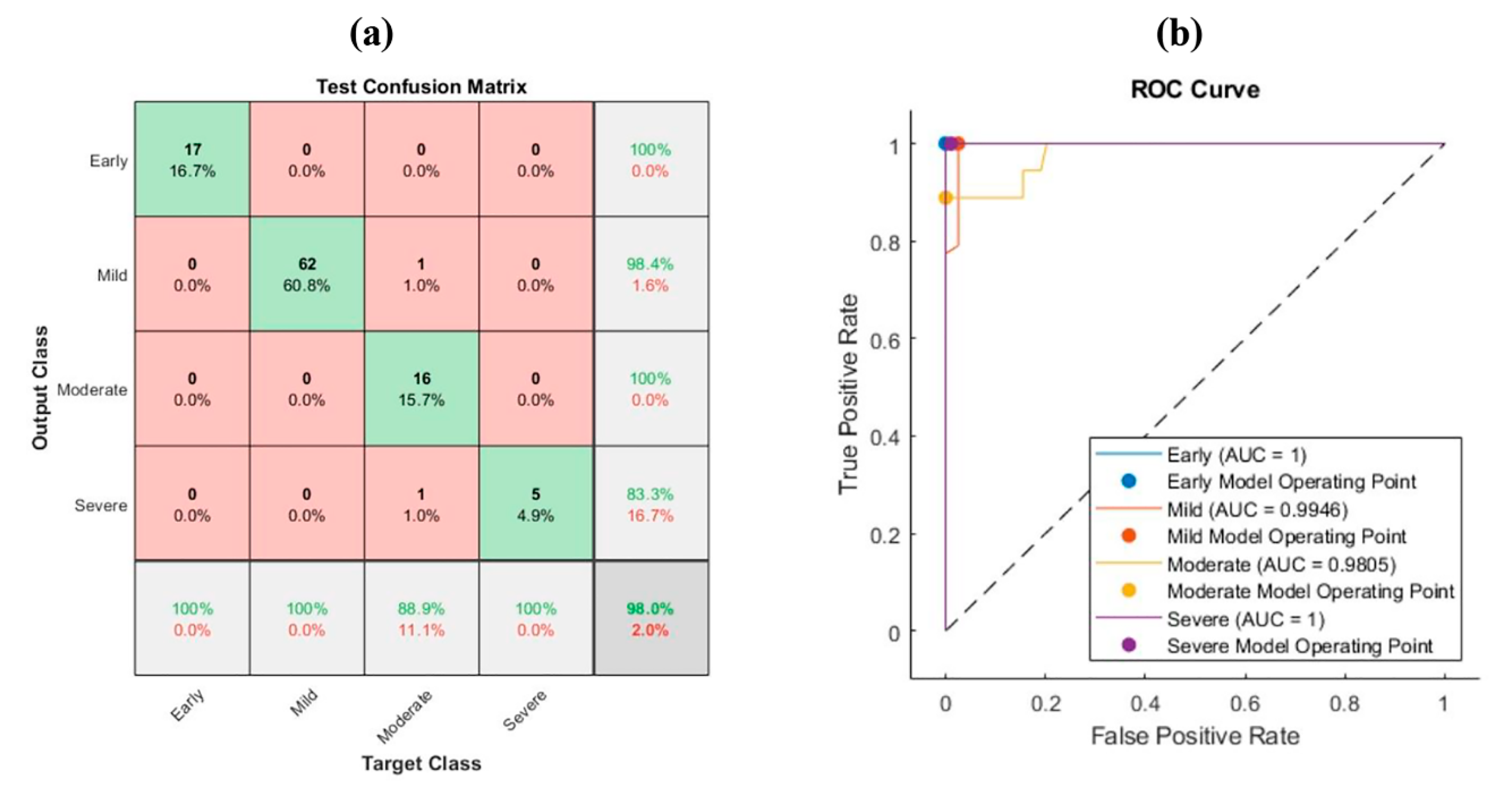
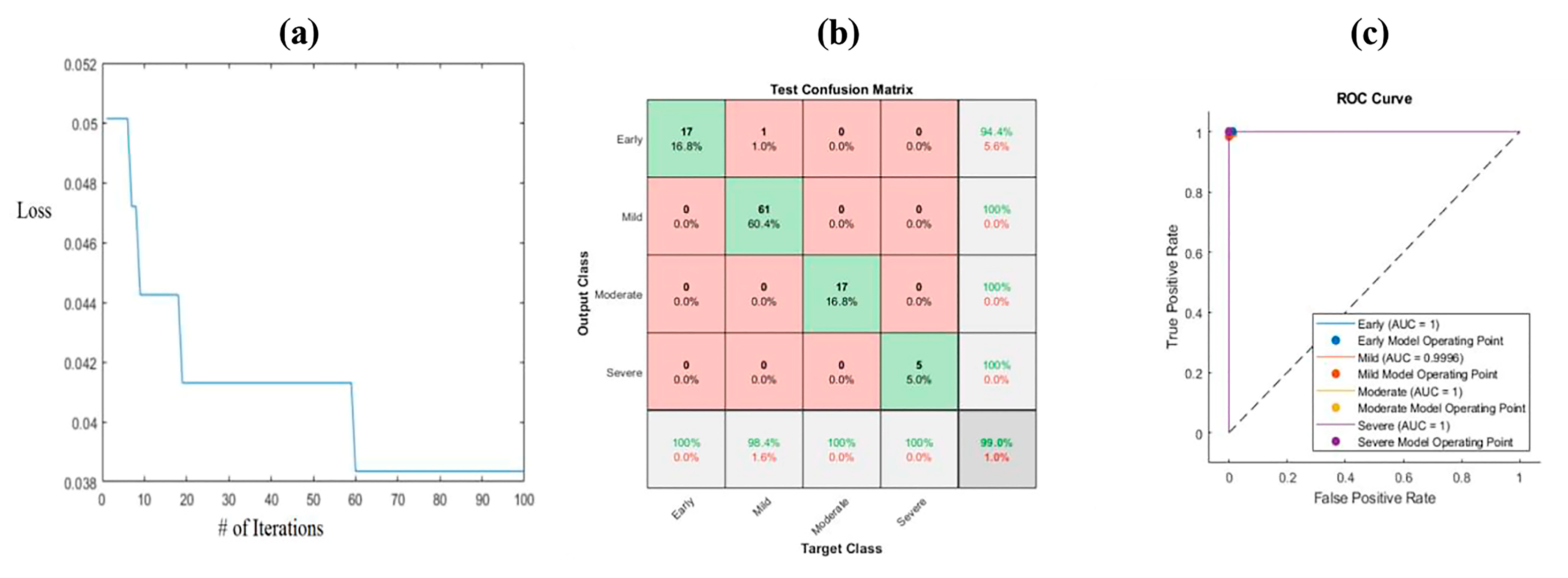
3.4. Ant Lion Optimization (ALO)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hubertsson, J.; Petersson, I.F.; Thorstensson, C.A.; Englund, M. Risk of sick leave and disability pension in working-age women and men with knee osteoarthritis. Ann. Rheum. Dis. 2013, 72, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.C.; Felson, D.T.; Helmick, C.G.; Arnold, L.M.; Choi, H.; Deyo, R.A.; Gabriel, S.; Hirsch, R.; Hochberg, M.C.; Hunder, G.G.; et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheumatol. 2008, 58, 26–35. [Google Scholar] [CrossRef]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheumatol. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y. Osteoarthritis year in review 2021: Biology. Osteoarthr. Cartil. 2022, 30, 207–215. [Google Scholar] [CrossRef]
- Fujii, Y.; Liu, L.; Yagasaki, L.; Inotsume, M.; Chiba, T.; Asahara, H. Cartilage homeostasis and osteoarthritis. Int. J. Mol. Sci. 2022, 23, 6316. [Google Scholar] [CrossRef] [PubMed]
- Pritzker, K.P.H.; Gay, S.; Jimenez, S.A.; Ostergaard, K.; Pelletier, J.P.; Revell, P.A.; Salter, D.; van den Berg, W.B. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthr. Cartil. 2006, 14, 13–29. [Google Scholar] [CrossRef]
- Rutgers, M.; van Pelt, M.J.; Dhert, W.J.; Creemers, L.B.; Saris, D.B. Evaluation of histological scoring systems for tissue-engineered, repaired and osteoarthritic cartilage. Osteoarthr. Cartil. 2010, 18, 12–23. [Google Scholar] [CrossRef]
- Custers, R.J.; Creemers, L.B.; Verbout, A.J.; van Rijen, M.H.; Dhert, W.J.; Saris, D.B. Reliability, reproducibility and variability of the traditional Histologic/Histochemical Grading System vs. the new OARSI Osteoarthritis Cartilage Histopathology Assessment System. Osteoarthr. Cartil. 2007, 15, 1241–1248. [Google Scholar] [CrossRef]
- Pollard, T.C.; Gwilym, S.E.; Carr, A.J. The assessment of early osteoarthritis. J. Bone Jt. Surg. Br. 2008, 90, 411–421. [Google Scholar] [CrossRef]
- Favero, M.; Ramonda, R.; Goldring, M.B.; Goldring, S.R.; Punzi, L. Early knee osteoarthritis. RMD Open 2015, 1, e000062. [Google Scholar] [CrossRef]
- Pauli, C.; Whiteside, R.; Heras, F.L.; Nesic, D.; Koziol, J.; Grogan, S.P.; Matyas, J.; Pritzker, K.P.; D’Lima, D.D.; Lotz, M.K. Comparison of cartilage histopathology assessment systems on human knee joints at all stages of osteoarthritis development. Osteoarthr. Cartil. 2012, 20, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Serey, J.; Alfaro, M.; Fuertes, G.; Vargas, M.; Durán, C.; Ternero, R.; Rivera, R.; Sabattin, J. Pattern Recognition and Deep Learning Technologies, Enablers of Industry 4.0, and Their Role in Engineering Research. Symmetry 2023, 15, 535. [Google Scholar] [CrossRef]
- Dou, T.; Zhou, W. 2D and 3D convolutional neural network fusion for predicting the histological grade of hepatocellular carcinoma. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3832–3837. [Google Scholar]
- Zhang, L.; Ren, Z. Comparison of CT and MRI images for the prediction of soft-tissue sarcoma grading and lung metastasis via a convolutional neural networks model. Clin. Radiol. 2020, 75, 64–69. [Google Scholar] [CrossRef]
- Lee, J.H.; Shih, Y.T.; Wei, M.L.; Sun, C.K.; Chiang, B.L. Classification of established atopic dermatitis in children with the in vivo imaging methods. J. Biophotonics 2019, 12, e201800148. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Nir, G.; Kwak, J.T.; Xu, S.; Wilson, S.; Iczkowski, K.A.; Lucia, M.S.; et al. Detection and grading of prostate cancer using temporal enhanced ultrasound: Combining deep neural networks and tissue mimicking simulations. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1293–1305. [Google Scholar] [CrossRef]
- Ashinsky, B.G.; Bouhrara, M.; Coletta, C.E.; Lehallier, B.; Urish, K.L.; Lin, P.C.; Goldberg, I.G.; Spencer, R.G. Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J. Orthop. Res. 2017, 35, 2243–2250. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Almajalid, R.; Shan, J.; Zhang, M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans. NanoBiosci. 2018, 17, 228–236. [Google Scholar] [CrossRef]
- Leung, K.; Zhang, B.; Tan, J.; Shen, Y.; Geras, K.J.; Babb, J.S.; Cho, K.; Chang, G.; Deniz, C.M. Prediction of Total Knee Replacement and Diagnosis of Osteoarthritis by Using Deep Learning on Knee Radiographs: Data from the Osteoarthritis Initiative. Radiology 2020, 296, 584–593. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, R.; Deng, Y.; Chen, K.; Jiang, T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS ONE 2017, 12, e0178992. [Google Scholar] [CrossRef] [PubMed]
- Panfilov, E.; Tiulpin, A.; Nieminen, M.T.; Saarakkala, S.; Casula, V. Deep learning-based segmentation of knee MRI for fully automatic subregional morphological assessment of cartilage tissues: Data from the Osteoarthritis Initiative. J. Orthop. Res. 2022, 40, 1113–1124. [Google Scholar] [CrossRef]
- Tiulpin, A.; Saarakkala, S. Automatic Grading of Individual Knee Osteoarthritis Features in Plain Radiographs Using Deep Convolutional Neural Networks. Diagnostics 2020, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Rytky, S.J.O.; Tiulpin, A.; Frondelius, T.; Finnilä, M.A.J.; Karhula, S.S.; Leino, J.; Pritzker, K.P.H.; Valkealahti, M.; Lehenkari, P.; Joukainen, A.; et al. Automating three-dimensional osteoarthritis histopathological grading of human osteochondral tissue using machine learning on contrast-enhanced micro-computed tomography. Osteoarthr. Cartil. 2020, 28, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Power, L.; Acevedo, L.; Yamashita, R.; Rubin, D.; Martin, I.; Barbero, A. Deep learning enables the automation of grading histological tissue engineered cartilage images for quality control standardization. Osteoarthr. Cartil. 2021, 29, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Mantripragada, V.P.; Piuzzi, N.S.; Muschler, G.F.; Erdemir, A.; Midura, R.J. A comprehensive dataset of histopathology images, grades and patient demographics for human Osteoarthritis Cartilage. Data Brief 2021, 37, 107129. [Google Scholar] [CrossRef]
- Schmitz, N.; Laverty, S.; Kraus, V.B.; Aigner, T. Basic methods in histopathology of joint tissues. Osteoarthr. Cartil. 2010, 18, S113–S116. [Google Scholar] [CrossRef]
- Cooper, J.A.; Mintz, B.R.; Palumbo, S.L.; Li, W.J. 4—Assays for determining cell differentiation in biomaterials. In Characterization of Biomaterials; Jaffe, M., Hammond, W., Tolias, P., Arinzeh, T., Eds.; Woodhead Publishing: Sawston, UK, 2013; pp. 101–137. [Google Scholar]
- Mantripragada, V.P.; Piuzzi, N.S.; Zachos, T.; Obuchowski, N.A.; Muschler, G.F.; Midura, R.J. High occurrence of osteoarthritic histopathological features unaccounted for by traditional scoring systems in lateral femoral condyles from total knee arthroplasty patients with varus alignment. Acta Orthop. 2018, 89, 197–203. [Google Scholar] [CrossRef]
- Alquran, H.; Al-Issa, Y.; Alsalatie, M.; Mustafa, W.A.; Qasmieh, I.A.; Zyout, A. Intelligent Diagnosis and Classification of Keratitis. Diagnostics 2022, 12, 1344. [Google Scholar] [CrossRef] [PubMed]
- Alquran, H.; Mustafa, W.-A.; Qasmieh, I.-A.; Yacob, Y.-M.; Alsalatie, M.; Al-Issa, Y.; Alqudah, A.-M. Cervical Cancer Classification Using Combined Machine Learning and Deep Learning Approach. Comput. Mater. Contin. 2022, 72, 5117–5134. [Google Scholar] [CrossRef]
- Tawalbeh, S.; Alquran, H.; Alsalatie, M. Deep Feature Engineering in Colposcopy Image Recognition: A Comparative Study. Bioengineering 2023, 10, 105. [Google Scholar] [CrossRef]
- Alquran, H.; Alsalatie, M.; Mustafa, W.A.; Abdi, R.A.; Ismail, A.R. Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion. Bioengineering 2022, 9, 578. [Google Scholar] [CrossRef]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Hybridization of feature selection and feature weighting for high dimensional data. Appl. Intell. 2019, 49, 1580–1596. [Google Scholar] [CrossRef]
- Alquran, H.; Qasmieh, I.A.; Alqudah, A.M.; Alhammouri, S.; Alawneh, E.; Abughazaleh, A.; Hasayen, F. The melanoma skin cancer detection and classification using support vector machine. In Proceedings of the 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT), Aqaba, Jordan, 11–13 October 2017; pp. 1–5. [Google Scholar]
- Yacob, Y.M.; Alquran, H.; Mustafa, W.A.; Alsalatie, M.; Sakim, H.A.M.; Lola, M.S.H. pylori Related Atrophic Gastritis Detection Using Enhanced Convolution Neural Network (CNN) Learner. Diagnostics 2023, 13, 336. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, K.; Alquran, H.; Alsalatie, M.; Mustafa, W.A.; Al-Issa, Y.; Alqudah, A.; Badarneh, A. LiverNet: Diagnosis of Liver Tumors in Human CT Images. Appl. Sci. 2022, 12, 5501. [Google Scholar] [CrossRef]
- Sulzbacher, I. Osteoarthritis: Histology and pathogenesis. Wien. Med. Wochenschr. 2013, 163, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shi, R.; Wei, D.; Liu, Z.; Zhao, L.; Ke, B.; Pfister, H.; Ni, B. MedMNIST v2—A large-scale lightweight benchmark for 2D and 3D biomedical image classification. Sci. Data 2023, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Qi, S.; Zhou, K.; Lu, T.; Ning, H.; Xiao, R. Pairwise attention-enhanced adversarial model for automatic bone segmentation in CT images. Phys. Med. Biol. 2023, 68, 035019. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, B.; Zhou, K.; He, W.; Yan, F.; Wang, Z.; Xiao, R. CSR-Net: Cross-Scale Residual Network for multi-objective scaphoid fracture segmentation. Comput. Biol. Med. 2022, 137, 104776. [Google Scholar] [CrossRef]
- Das, A.; Mohapatra, S.K.; Mohanty, M.N. Design of deep ensemble classifier with fuzzy decision method for biomedical image classification. Appl. Soft Comput. 2022, 115, 108178. [Google Scholar] [CrossRef]
- Al-Badarneh, I.; Habib, M.; Aljarah, I.; Faris, H. Neuro-evolutionary models for imbalanced classification problems. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 2787–2797. [Google Scholar] [CrossRef]
- Jamshidi, A.; Leclercq, M.; Labbe, A.; Pelletier, J.-P.; Abram, F.; Droit, A.; Martel-Pelletier, J. Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods. Ther. Adv. Musculoskelet. Dis. 2020, 12, 1759720X20933468. [Google Scholar] [CrossRef]
- Pedoia, V.; Lee, J.; Norman, B.; Link, T.M.; Majumdar, S. Diagnosing osteoarthritis from T2 maps using deep learning: An analysis of the entire Osteoarthritis Initiative baseline cohort. Osteoarthr. Cartil. 2019, 27, 1002–1010. [Google Scholar] [CrossRef]
- Hayashi, D.; Roemer, F.W.; Guermazi, A. Magnetic resonance imaging assessment of knee osteoarthritis: Current and developing new concepts and techniques. Clin. Exp. Rheumatol. 2019, 37, 88–95. [Google Scholar]
- Martel-Pelletier, J.; Paiement, P.; Pelletier, J.-P. Magnetic resonance imaging assessments for knee segmentation and their use in combination with machine/deep learning as predictors of early osteoarthritis diagnosis and prognosis. Ther. Adv. Musculoskelet. Dis. 2023, 15, 1759720X231165560. [Google Scholar] [CrossRef]
- Tolpadi, A.A.; Lee, J.J.; Pedoia, V.; Majumdar, S. Deep learning predicts total knee replacement from magnetic resonance images. Sci. Rep. 2020, 10, 6371. [Google Scholar] [CrossRef]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The Basic Science of Articular Cartilage: Structure, Composition, and Function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Mantripragada, V.P.; Gao, W.; Piuzzi, N.S.; Hoemann, C.D.; Muschler, G.F.; Midura, R.J. Comparative Assessment of Primary Osteoarthritis Progression Using Conventional Histopathology, Polarized Light Microscopy, and Immunohistochemistry. Cartilage 2021, 13, 1494s–1510s. [Google Scholar] [CrossRef]
- Mantripragada, V.P.; Piuzzi, N.S.; Zachos, T.; Obuchowski, N.A.; Muschler, G.F.; Midura, R.J. Histopathological assessment of primary osteoarthritic knees in large patient cohort reveal the possibility of several potential patterns of osteoarthritis initiation. Curr. Res. Transl. Med. 2017, 65, 133–139. [Google Scholar] [CrossRef]
- Saarakkala, S.; Julkunen, P.; Kiviranta, P.; Mäkitalo, J.; Jurvelin, J.S.; Korhonen, R.K. Depth-wise progression of osteoarthritis in human articular cartilage: Investigation of composition, structure and biomechanics. Osteoarthr. Cartil. 2010, 18, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Nagira, K.; Ikuta, Y.; Shinohara, M.; Sanada, Y.; Omoto, T.; Kanaya, H.; Nakasa, T.; Ishikawa, M.; Adachi, N.; Miyaki, S.; et al. Histological scoring system for subchondral bone changes in murine models of joint aging and osteoarthritis. Sci. Rep. 2020, 10, 10077. [Google Scholar] [CrossRef] [PubMed]
- Namhong, S.; Wongdee, K.; Suntornsaratoon, P.; Teerapornpuntakit, J.; Hemstapat, R.; Charoenphandhu, N. Knee osteoarthritis in young growing rats is associated with widespread osteopenia and impaired bone mineralization. Sci. Rep. 2020, 10, 15079. [Google Scholar] [CrossRef] [PubMed]
- Radakovich, L.B.; Marolf, A.J.; Shannon, J.P.; Pannone, S.C.; Sherk, V.D.; Santangelo, K.S. Development of a microcomputed tomography scoring system to characterize disease progression in the Hartley guinea pig model of spontaneous osteoarthritis. Connect. Tissue Res. 2018, 59, 523–533. [Google Scholar] [CrossRef] [PubMed]



| CNN | DarkNet-19 | MobileNet | NasNet | ResNet-101 | ShuffleNet | |
|---|---|---|---|---|---|---|
| Images | ||||||
| HE | 69.6% | 61.8% | 70.6% | 69.6% | 64.7% | |
| SafO | 80.2% | 77.2% | 73.3% | 76.2% | 74.3% | |
| CNN | DarkNet-19 | MobileNet | NasNet | ResNet-101 | ShuffleNet | |
|---|---|---|---|---|---|---|
| Images | ||||||
| HE | 60.8% | 99% | 95.1% | 94.1% | 96.1% | |
| SafO | 95% | 98% | 95% | 94.1% | 95% | |
| HE Images | SafO Images | ||||
|---|---|---|---|---|---|
| Class | Sensitivity | Precision | Sensitivity | Precision | |
| Early | DarkNet-19 | 11.8% | 16.7% | 100% | 85.7% |
| MobileNet | 100% | 100% | 94.1% | 94.1% | |
| NasNet | 89.9% | 94.1% | 88.2% | 88.2% | |
| ResNet-101 | 82.4% | 100% | 94.4% | 94.4% | |
| ShuffleNet | 94.1% | 94.1% | 94.1% | 88.9% | |
| Mild | DarkNet-19 | 85.7% | 69.2% | 93.5% | 98.3% |
| MobileNet | 98.4% | 100% | 98.4% | 98.4% | |
| NasNet | 95.2% | 96.8% | 96.8% | 95.3% | |
| ResNet-101 | 98.4% | 95.4% | 98.4% | 92.4% | |
| ShuffleNet | 100% | 95.4% | 96.8% | 96.8% | |
| Moderate | DarkNet-19 | 29.4% | 55.6% | 94.1% | 94.1% |
| MobileNet | 100% | 94.4% | 100% | 100% | |
| NasNet | 100% | 98.5% | 100% | 100% | |
| ResNet-101 | 88.2% | 88.2% | 76.5% | 100% | |
| ShuffleNet | 83.3% | 100% | 94.4% | 100% | |
| Severe | DarkNet-19 | 20% | 16.7% | 100% | 100% |
| MobileNet | 100% | 100% | 100% | 100% | |
| NasNet | 100% | 100% | 75% | 100% | |
| ResNet-101 | 100% | 83.3% | 100% | 100% | |
| ShuffleNet | 100% | 100% | 100% | 100% | |
| Class | Feature Engineering | Sensitivity | Precision | Specificity | F1 Score |
|---|---|---|---|---|---|
| Early | PCA | 100% | 100% | 100% | 1 |
| ALO | 100% | 98.4% | 98.8% | 0.97 | |
| Mild | PCA | 100% | 98.4% | 97.5% | 0.991 |
| ALO | 100% | 98.4% | 97.5% | 0.991 | |
| Moderate | PCA | 88.9% | 100% | 100% | 0.941 |
| ALO | 100% | 100% | 100% | 1 | |
| Severe | PCA | 100% | 83.3% | 99% | 0.909 |
| ALO | 100% | 100% | 100% | 1 |
| Class | Feature Engineering | Sensitivity | Precision | Specificity | F1 Score |
|---|---|---|---|---|---|
| Early | PCA | 94.1% | 94.1% | 98.8% | 1 |
| ALO | 100% | 100% | 100% | 1 | |
| Mild | PCA | 98.4% | 96.8% | 94.8% | 0.971 |
| ALO | 100% | 98.4% | 97.4% | 0.991 | |
| Moderate | PCA | 100% | 100% | 100% | 1 |
| ALO | 94.1% | 100% | 100% | 0.97 | |
| Severe | PCA | 80% | 100% | 100% | 0.889 |
| ALO | 100% | 100% | 100% | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khader, A.; Alquran, H. Automated Prediction of Osteoarthritis Level in Human Osteochondral Tissue Using Histopathological Images. Bioengineering 2023, 10, 764. https://doi.org/10.3390/bioengineering10070764
Khader A, Alquran H. Automated Prediction of Osteoarthritis Level in Human Osteochondral Tissue Using Histopathological Images. Bioengineering. 2023; 10(7):764. https://doi.org/10.3390/bioengineering10070764
Chicago/Turabian StyleKhader, Ateka, and Hiam Alquran. 2023. "Automated Prediction of Osteoarthritis Level in Human Osteochondral Tissue Using Histopathological Images" Bioengineering 10, no. 7: 764. https://doi.org/10.3390/bioengineering10070764
APA StyleKhader, A., & Alquran, H. (2023). Automated Prediction of Osteoarthritis Level in Human Osteochondral Tissue Using Histopathological Images. Bioengineering, 10(7), 764. https://doi.org/10.3390/bioengineering10070764






