Rationally Improving Doramectin Production in Industrial Streptomyces avermitilis Strains
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains, Plasmids, and Cultivation Conditions
2.2. DNA Cloning and Sequence Analysis
2.3. Vector Construction and Genetic Manipulation
2.4. Analysis of Doramectin Production with High-Performance Liquid Chromatography (HPLC)
2.5. Statistical Methods
3. Results
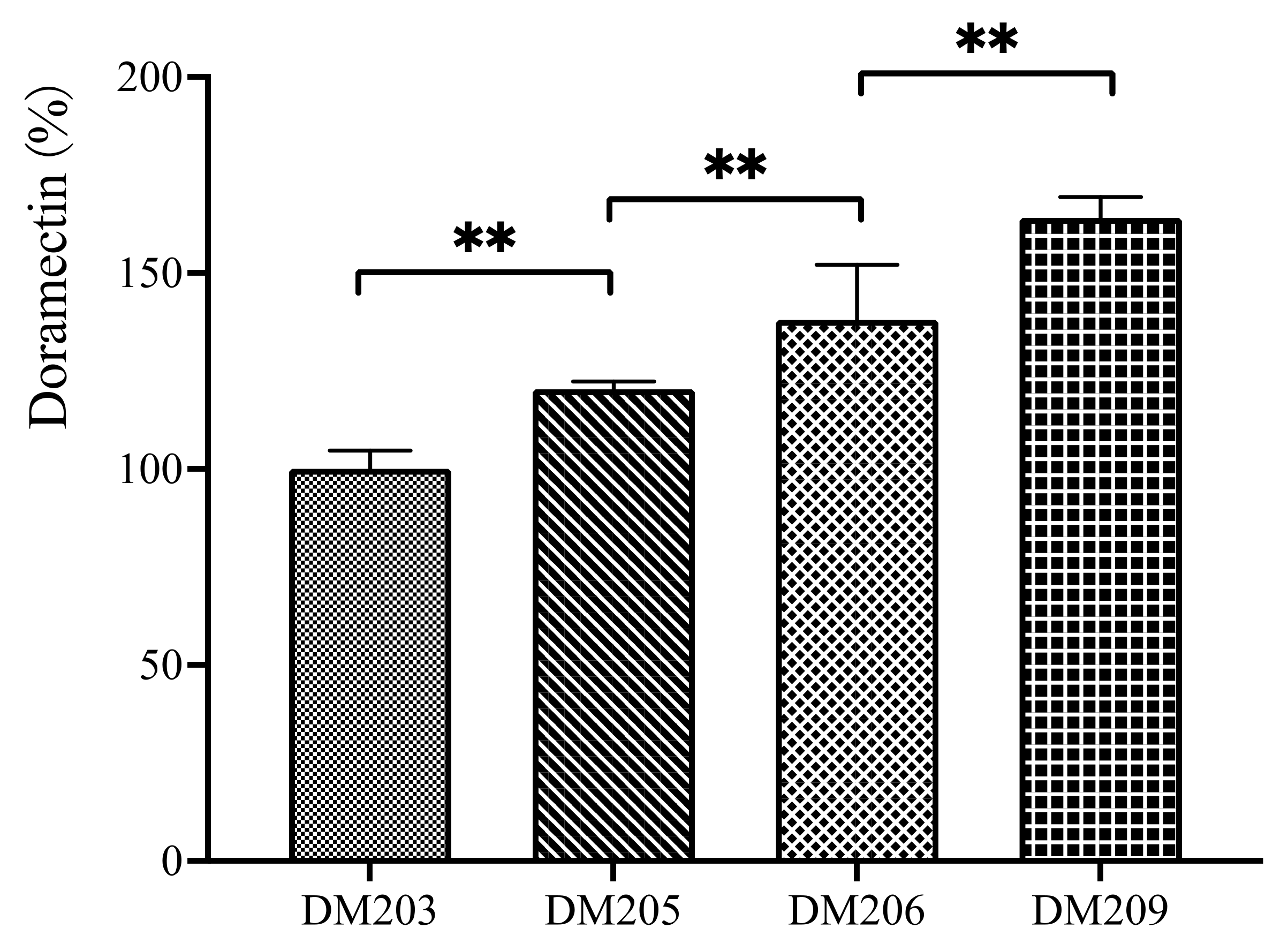
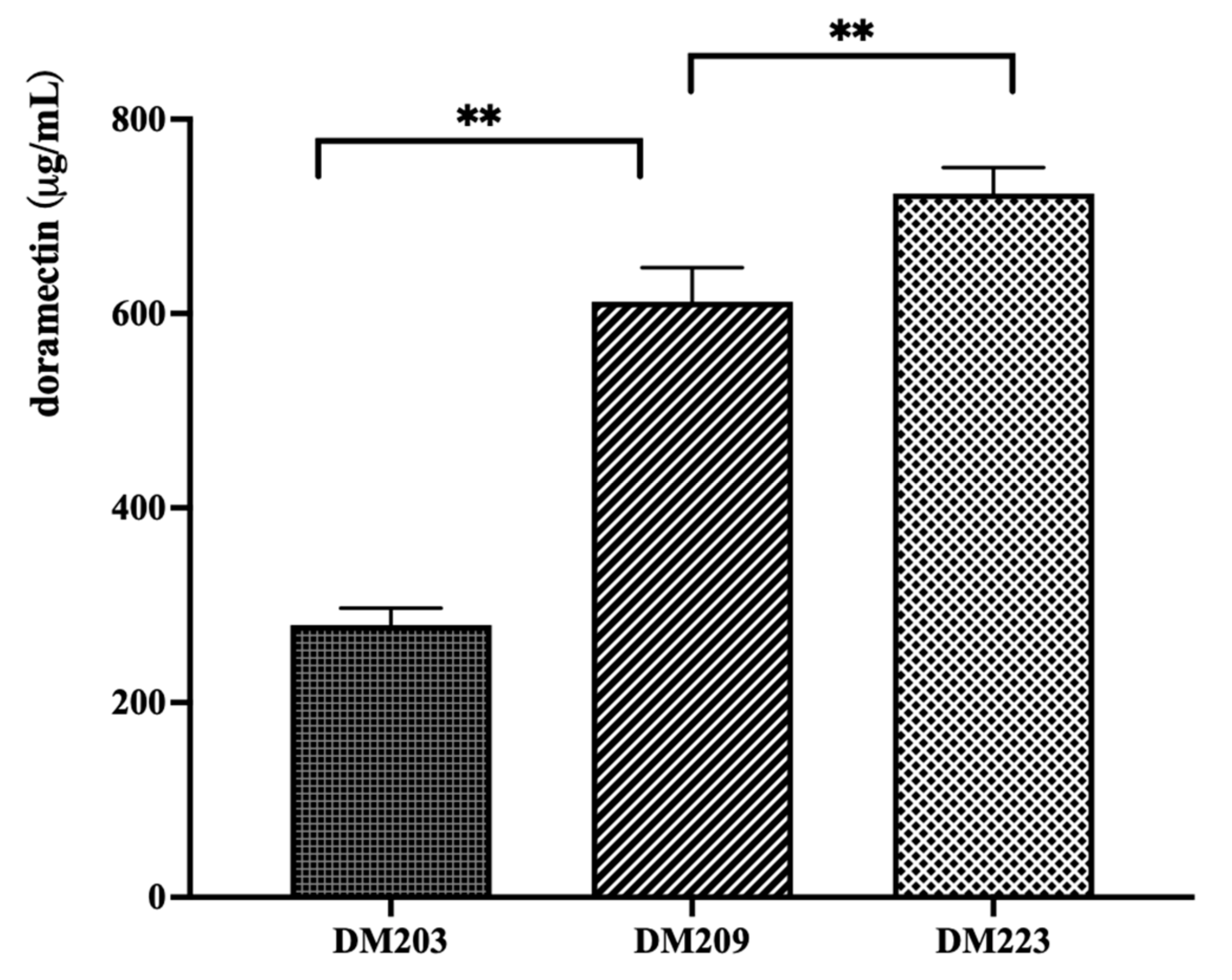
3.1. Sequential Deletion of the PKS Clusters from the Doramectin-Producing Strain
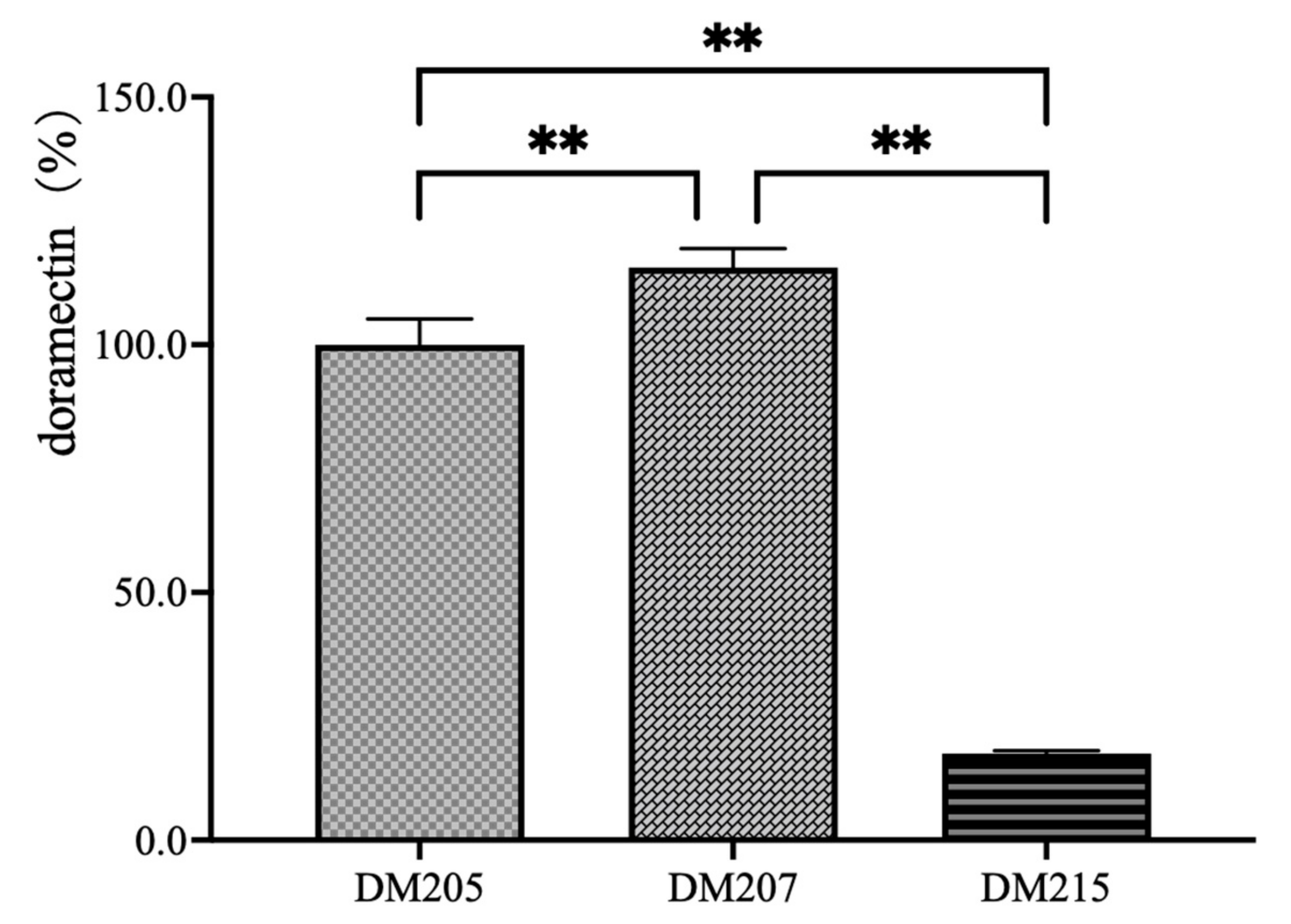
3.2. Verification of the CHC Activation Protein-Encoding Gene
3.3. Construction of a Doramectin Hyperproduction Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burg, R.W.; Miller, B.M.; Baker, E.E.; Birnbaum, J.; Currie, S.A.; Hartman, R.; Kong, Y.L.; Monaghan, R.L.; Olson, G.; Putter, I.; et al. Avermectins, new family of potent anthelmintic agents: Producing organism and fermentation. Antimicrob. Agents Chemother. 1979, 15, 361–367. [Google Scholar] [CrossRef]
- Thuan, N.H.; Pandey, R.P.; Sohng, J.K. Recent advances in biochemistry and biotechnological synthesis of avermectins and their derivatives. Appl. Microbiol. Biotechnol. 2014, 98, 7747–7759. [Google Scholar] [CrossRef]
- Chen, J.; Liu, M.; Zhang, L. Avermectin, from winning the Nobel Prize to “innovation in China”. Wei Sheng Wu Xue Bao 2016, 56, 543–558. [Google Scholar]
- Ikeda, H. Natural products discovery from micro-organisms in the post-genome era. Biosci. Biotechnol. Biochem. 2017, 81, 13–22. [Google Scholar] [CrossRef]
- Denoya, C.D.; Fedechko, R.W.; Hafner, E.W.; McArthur, H.A.; Morgenstern, M.R.; Skinner, D.D.; Stutzman-Engwall, K.; Wax, R.G.; Wernau, W.C. A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: Its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J. Bacteriol. 1995, 177, 3504–3511. [Google Scholar] [CrossRef] [PubMed]
- Dutton, C.J.; Gibson, S.P.; Goudie, A.C.; Holdom, K.S.; Pacey, M.S.; Ruddock, J.C.; Bu’Lock, J.D.; Richards, M.K. Novel avermectins produced by mutational biosynthesis. J. Antibiot. 1991, 44, 357–365. [Google Scholar] [CrossRef]
- Stutzman-Engwall, K.; Conlon, S.; Fedechko, R.; Kaczmarek, F.; McArthur, H.; Krebber, A.; Chen, Y.; Minshull, J.; Raillard, S.; Gustafsson, C. Engineering the aveC gene to enhance the ratio of doramectin to its CHC-B2 analogue produced in Streptomyces avermitilis. Biotechnol. Bioeng. 2003, 82, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Stutzman-Engwall, K.; Conlon, S.; Fedechko, R.; McArthur, H.; Pekrun, K.; Chen, Y.; Jenne, S.; La, C.; Trinh, N.; Kim, S.; et al. Semi-synthetic DNA shuffling of aveC leads to improved industrial scale production of doramectin by Streptomyces avermitilis. Metab. Eng. 2005, 7, 27–37. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Li, Z.; Zhang, J.; Fan, K.; Tan, G.; Ai, G.; Lam, S.M.; Shui, G.; Yang, Z.; et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat. Biotechnol. 2020, 38, 76–83. [Google Scholar] [CrossRef]
- Ikeda, H.; Kazuo, S.Y.; Omura, S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 2014, 41, 233–250. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Uchiyama, T.; Omura, S.; Cane, D.E.; Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 2646–2651. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Komatsu, K.; Koiwai, H.; Yamada, Y.; Kozone, I.; Izumikawa, M.; Hashimoto, J.; Takagi, M.; Omura, S.; Shin-ya, K.; et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 2013, 2, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Jing, X.; Xie, P.; Chen, W.; Wang, T.; Xia, H.; Qin, Z. Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol. Lett. 2012, 333, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cai, J. Improved fermentation yield of doramectin from Streptomyces avermitilis N72 by strain selection and glucose supplementation strategies. Fermentation 2023, 9, 121. [Google Scholar] [CrossRef]
- Wang, J.; Pan, H.; Tang, G. Production of doramectin by rational engineering of the avermectin biosynthetic pathway. Bioorg. Med. Chem. Lett. 2011, 21, 3320–3323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Wang, S.; Chen, Z.; Wen, Y.; Song, Y. Construction of a doramectin producer mutant from an avermectin-overproducing industrial strain of Streptomyces avermitilis. Can. J. Microbiol. 2009, 55, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Dang, F.; Xia, H.; Qin, Z. Increasing doramectin content of industrial strain by introduction of a synthetic multiple-mutated aveC* gene. Chin. J. Antibiot. 2014, 39, 574–578. [Google Scholar] [CrossRef]
- Xia, H.; Huang, J.; Hu, M.; Shen, M.; Xie, P.; Zhang, L.; Wang, H.; Qin, Z. Construction of an ordered cosmid library of Streptomyces avermitilis for genetic modification of the industrial strains. Chin. J. Antibiot. 2009, 34, 340–343. [Google Scholar]
- Hu, M.; Xia, H.; Qin, Z. Effects on productivity of avermectin by knockout of 10 polyketide biosynthetic gene clusters in Streptomyces avermitilis genome. Microbiol. China 2014, 41, 1471–1476. [Google Scholar] [CrossRef]
- Meng, L.; Xiong, Z.; Chu, J.; Wang, Y. Enhanced production of avermectin by deletion of type III polyketide synthases biosynthetic cluster rpp in Streptomyces avermitilis. Lett. Appl. Microbiol. 2016, 63, 384–390. [Google Scholar] [CrossRef]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Bierman, M.; Logan, R.; O’Brien, K.; Seno, E.; Rao, R.; Schoner, B. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Xia, H.; Qin, Z. Construction of the industrial strains for nemadectin production with less byproducts by genetically engineering. Chin. J. Antibiot. 2014, 39, 579–583. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K.; Hopwood, D. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Cropp, T.; Wilson, D.; Reynolds, K. Identification of a cyclohexylcarbonyl CoA biosynthetic gene cluster and application in the production of doramectin. Nat. Biotechnol. 2000, 18, 980–983. [Google Scholar] [CrossRef]
- Tan, L.L.; Heng, E.; Zulkarnain, N.; Hsiao, W.C.; Wong, F.T.; Zhang, M.M. CRISPR/Cas-mediated genome editing of Streptomyces. Methods Mol. Biol. 2022, 2479, 207–225. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, L.; Zheng, G.; Jiang, W.; Deng, Z.; Wang, Z.; Lu, Y. CRISPR/dCas9-mediated multiplex gene repression in Streptomyces. Biotechnol. J. 2018, 13, e1800121. [Google Scholar] [CrossRef]
- Li, L.; Wei, K.; Zheng, G.; Liu, X.; Chen, S.; Jiang, W.; Lu, Y. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl. Environ. Microbiol. 2018, 84, e00827-18. [Google Scholar] [CrossRef]
- Tian, J.; Yang, G.; Gu, Y.; Sun, X.; Lu, Y.; Jiang, W. Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res. 2020, 48, 8188–8202. [Google Scholar] [CrossRef]

| Strain or Plasmids | Genotype/Description | Sources/Reference |
|---|---|---|
| Streptomyces avermitilis | ||
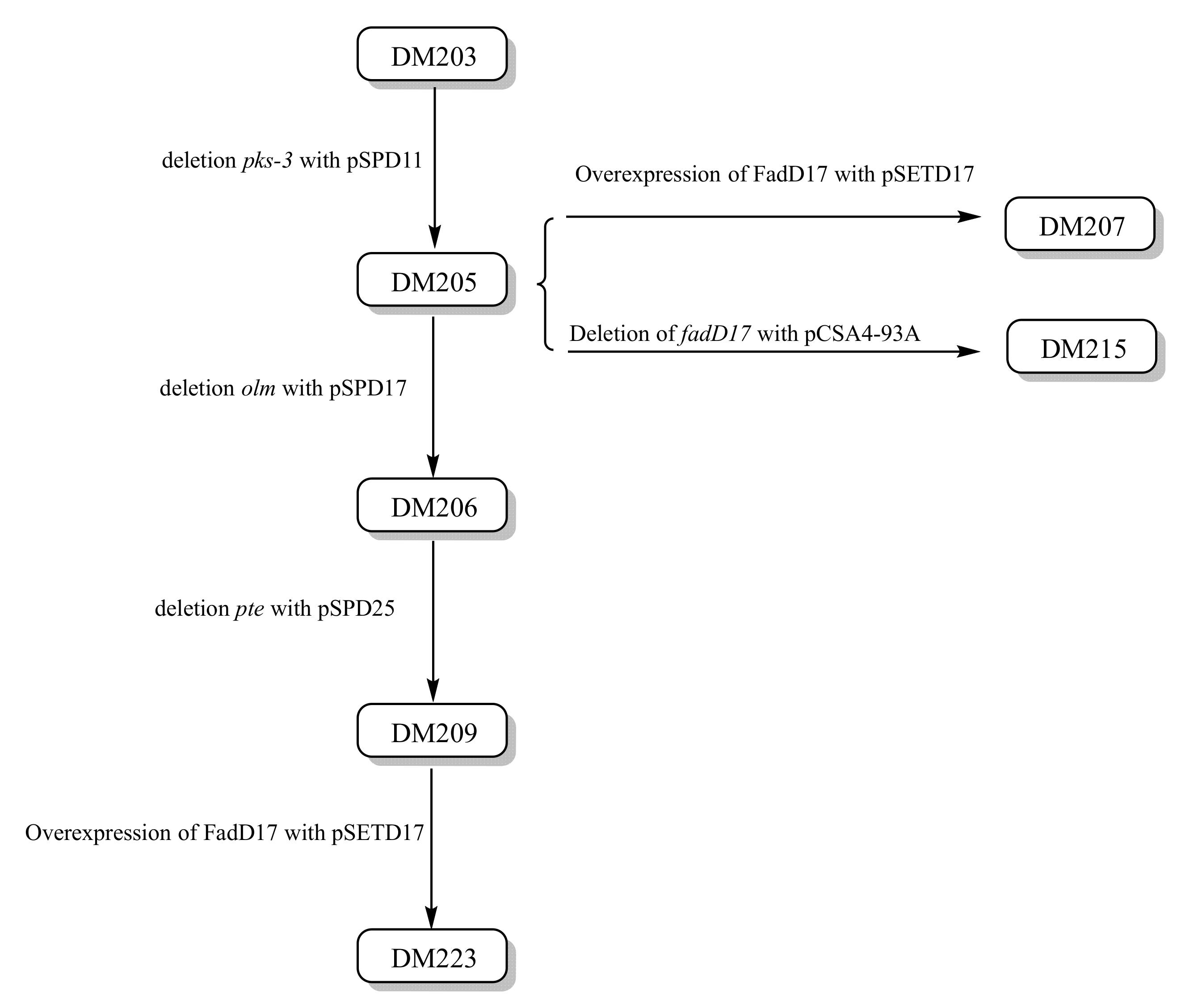
| DM203 | Doramectin-producing strain | [17] |
| DM205 | ∆pks3 mutant of DM203 | This study |
| DM206 | ∆pks3-∆olm mutant of DM205 | This study |
| DM209 | ∆pks3-∆olm-∆pte mutant of DM206 | This study |
| DM207 | DM205 with a copy of PermE*-fadD17 | This study |
| DM215 | ∆fadD17 mutant of DM205 | This study |
| DM223 | DM209 with a copy of PermE*-fadD17 | This study |
| Escherichia coli | ||
| DH5α | Host for general cloning | Invitrogen |
| BW25113 (pIJ790) | Host containing L-arabinose-inducible lambda Red recombination system | [21] |
| BT340 | Host containing inducible FLP recombination system | [21] |
| ET12567 (pUZ8002) | dam dcm hsdM cm kan, containing a nontransmissible RP4-derived plasmid pUZ8002 | [22] |
| Plasmids | ||
| pIJ773 | bla colE1-ori FRT-aac(3)IV-oriT-FRT cassette | [21] |
| pHY773 | bla colE1-ori FRT-aac(3)IV-FRT cassette | [23] |
| pHY642 | Bla colE1-oritsr oriT | [23] |
| pSET152 | colE1 ori aac(3)IV oriT phiC31int attP lacZα | [22] |
| pCSA4-65 | Cosmid contains a c. 40-kb insert (includes the pks3 gene cluster) of S. avermitilis | [19] |
| pCSA11-69 | Cosmid contains a c. 40-kb insert (includes the pteA1 gene) of S. avermitilis | [19] |
| pCSA4-93 | Cosmid contains a c. 40-kb insert (includes the fadD17 gene) of S. avermitilis | [19] |
| pCSA4-65A | pks3 gene cluster deletion of pCSA4-65 (PCR-targeting) | This study |
| pCSA11-69A | pte gene cluster deletion of pCSA11-69 (PCR-targeting) | This study |
| pSPD11 | pks3 gene cluster markerless deletion plasmid | This study |
| pSPD17 | olm gene cluster markerless deletion plasmid | This study |
| pSPD25 | pte gene cluster markerless deletion plasmid | This study |
| pSETD17 | pSET152 derivative for PermE*-driven expression of fadD17 | This study |
| pCSA4-93A | fadD17 gene deletion of pCSA4-93 (PCR-targeting) | This study |
| Primer | Sequences (5′-3′) | Used for |
|---|---|---|
| pks3F | GCAGATCTTGCGCAGGGAGACGAACGCGTCGGGCCCGGCTGTAGGCTGGAGCTGCTTC | Amplification of the FRT-aac(3)IV-FRT cassette to target the pks3 cluster in pCSA4-65 via lambda Red recombination |
| pks3R | AGGCCCCGGCCGCGGCGCCCTTGGGCCGTCAGCGGGGTGATTCCGGGGATCCGTCGACC | |
| pks3DF | GACCACCTTCACCTCGTTGC | To verify the deletion and replacement of pks3 in S. avermitilis DM205 and pCSA4-65A, respectively. |
| pks3DR | CATCAGCCTTTCCGACTTCC | |
| olmLF | CGGAAGCTTTCCTCGAGGGACTCACCGAC | Amplification of the upstream and downstream homologous arms to construct pSPD17. |
| olmLR | GAGGGATCCTGAGAGCGCCTCCAGTTCCC | |
| olmRF | TGCGGATCCCCATAGAACCTTTCGTGCTA | |
| olmRR | TACGAATTCAACTCGGCGTCCATGTGGAT | |
| olmDF | ATGACGGGAAGGGCGGAGGTGT | To verify the deletion of the olm cluster in S. avermitilis DM206 |
| olmDR | GTCGTAGAGGAGGAAGAGCGGTGC | |
| pteF | CGACGAACGTGGTCACGCCCAGCTCGTGCAGGGTGTGCAATTCCGGGGATCCGTCGACC | Amplification of the FRT-aac(3)IV-FRT cassette to target the pte cluster in pCSA11-69 via lambda Red recombination |
| pteR | GCCCACACCCGCGTCGCCAGAAACTCCCGCACCTGCGCCTGTAGGCTGGAGCTGCTTC | |
| pteDF | AGGGATGCGTCCTGAGTGAGA | To verify the deletion and replacement of pte in S. avermitilis DM209 and pCSA11-69A, respectively |
| pteDR | CGGTGAACATTGCGACTGCTT | |
| fadD17F | GGAATTCCATATGGTGAACGACACCGCACACGCGCTCA | Construction of pSETD17 and pHX85D17 |
| fadD17R | GGAATTCCATATG TCACCGCGCATAGCGCTCCCTCAGC | |
| fadD17-59F | GCGCTCAGCACCTCCGGCACGCTCTGGGAACTCGTCGTCATTCCGGGGATCCGTCGACC | Amplification of the FRT-aac(3)IV-oriT-FRT cassette to target fadD17 in pCSA4-93 via lambda Red recombination |
| fadD17-58R | GATCCTCGATCTCCTTCGCCGAGATGTTCTCGCCCTTGCTGTAGGCTGGAGCTGCTTC | |
| fadD17-PF | CTGGATACCAGCAGGCTAGTGCC | To verify the deletion and replacement of pks-3 in S. avermitilis DM207 and pCSA4-93A, respectively |
| fadD17-PR | CGCCCGCAGATACGAGGTCAT |
| Deleted Clusters | Annotated Region * | Deletion Region * | Length of Deletion Region (bp) | ||
|---|---|---|---|---|---|
| Start (nt) | End (nt) | Start (nt) | End (nt) | ||
| pks3 | 2,773,878 | 2,784,841 | 2,777,920 | 2,784,838 | 6918 |
| olm | 3,534,525 | 3,634,592 | 3,534,529 | 3,634,591 | 100,062 |
| pte | 487,415 | 567,017 | 546,803 | 568,027 | 21,224 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, F.; Xu, Q.; Qin, Z.; Xia, H. Rationally Improving Doramectin Production in Industrial Streptomyces avermitilis Strains. Bioengineering 2023, 10, 739. https://doi.org/10.3390/bioengineering10060739
Dang F, Xu Q, Qin Z, Xia H. Rationally Improving Doramectin Production in Industrial Streptomyces avermitilis Strains. Bioengineering. 2023; 10(6):739. https://doi.org/10.3390/bioengineering10060739
Chicago/Turabian StyleDang, Fujun, Qingyu Xu, Zhongjun Qin, and Haiyang Xia. 2023. "Rationally Improving Doramectin Production in Industrial Streptomyces avermitilis Strains" Bioengineering 10, no. 6: 739. https://doi.org/10.3390/bioengineering10060739
APA StyleDang, F., Xu, Q., Qin, Z., & Xia, H. (2023). Rationally Improving Doramectin Production in Industrial Streptomyces avermitilis Strains. Bioengineering, 10(6), 739. https://doi.org/10.3390/bioengineering10060739






