Named Entity Recognition of Diabetes Online Health Community Data Using Multiple Machine Learning Models
Abstract
1. Introduction
2. Method
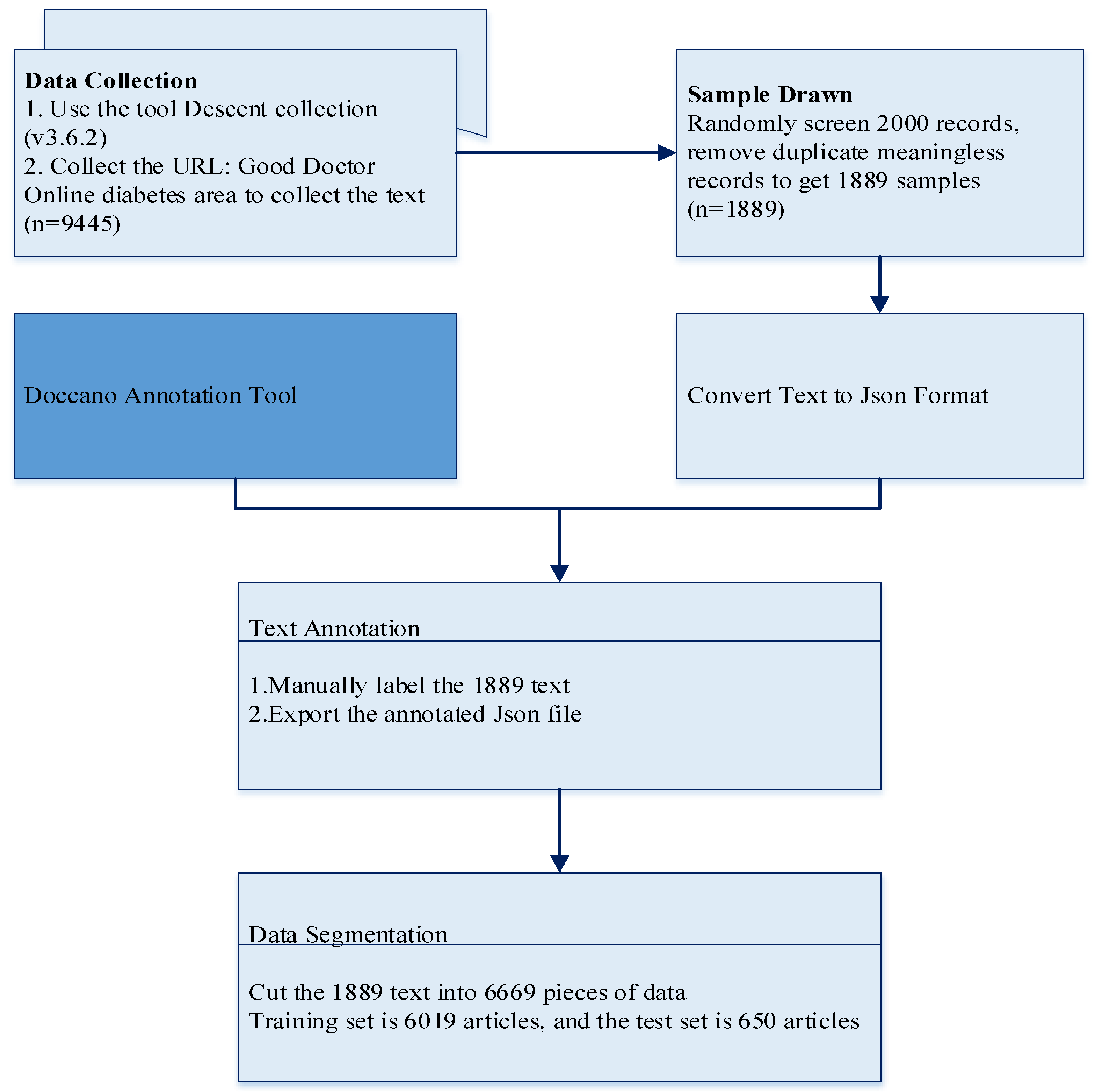
2.1. Data Collection and Preprocessing
2.2. RoBERTa-BiLSTM-CRF Model Construction
2.2.1. RoBERTa Pre-Training Layer to Construct Word Vectors
2.2.2. Layer of BiLSTM for Semantic Encoding
2.2.3. CRF Optimized Tag Sequence
3. Result
3.1. Text Annotation
3.2. Experimental Setup
3.3. Evaluation
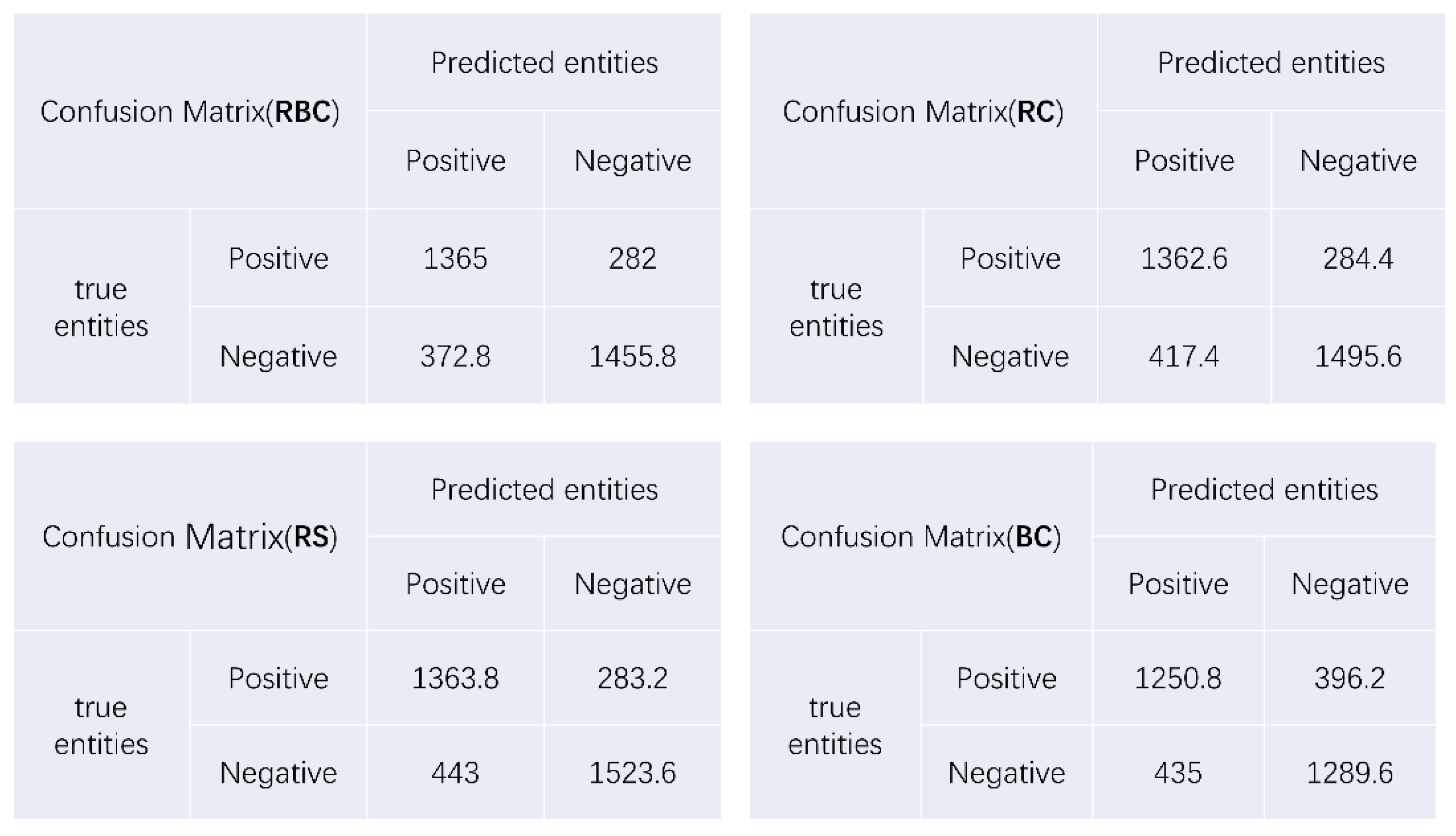
3.4. Model Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 13. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Hoti, K.; Hughes, J.D.; Emmerton, L.M. Consumer Use of “Dr Google”: A Survey on Health Information-Seeking Behaviors and Navigational Needs. J. Med. Internet Res. 2015, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Nakikj, D.; Mamykina, L. A park or a highway: Overcoming tensions in designing for socio-emotional and informational needs in online health communities. In Proceedings of the 2017 ACM Conference on Computer Supported Cooperative Work and Social Computing, Portland, OR, USA, 25 February–1 March 2017; pp. 1304–1319. [Google Scholar]
- Nakikj, D.; Mamykina, L. Lost in migration: Information management and community building in an online health community. In Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems, Montreal, QC, Canada, 21–26 April 2018; pp. 1–14. [Google Scholar]
- Chen, Z.H.; Zhang, C.C.; Fan, G.H. Interrelationship between Interpersonal Interaction Intensity and Health Self-Efficacy in People with Diabetes or Prediabetes on Online Diabetes Social Platforms: An In-Depth Survey in China. Int. J. Environ. Res. Public Health 2020, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Zhang, R.T.; Liu, K.C.; Hou, L. Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System. Int. J. Environ. Res. Public Health 2018, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Coden, A.; Savova, G.; Sominsky, I.; Tanenblatt, M.; Masanz, J.; Schuler, K.; Cooper, J.; Guan, W.; de Groen, P.C. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J. Biomed. Inform. 2009, 42, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Yu, H.; Han, W.S. Developing a hybrid dictionary-based bio-entity recognition technique. BMC Med. Inform. Decis. Mak. 2015, 15, 8. [Google Scholar] [CrossRef]
- Khan, A.T.; Cao, X.W.; Liao, B.L.; Francis, A. Bio-inspired Machine Learning for Distributed Confidential Multi-Portfolio Selection Problem. Biomimetics 2022, 7, 20. [Google Scholar] [CrossRef]
- Pletscher-Frankild, S.; Pallejà, A.; Tsafou, K.; Binder, J.X.; Jensen, L.J. DISEASES: Text mining and data integration of disease–gene associations. Methods 2015, 74, 83–89. [Google Scholar] [CrossRef]
- Rais, M.; Lachkar, A.; Lachkar, A.; Ouatik, S.E.A. A comparative study of biomedical named entity recognition methods based machine learning approach. In Proceedings of the 2014 Third IEEE International Colloquium in Information Science and Technology (CIST), Tetouan, Morocco, 20–22 October 2014; pp. 329–334. [Google Scholar]
- Saha, S.K.; Sarkar, S.; Mitra, P. Feature selection techniques for maximum entropy based biomedical named entity recognition. J. Biomed. Inform. 2009, 42, 905–911. [Google Scholar] [CrossRef]
- Francis, A.; Nazeer, K.A. Literature search tool for the extraction of disease-associated genes using frequent itemset mining. In Proceedings of the 2016 International Conference on Computing, Analytics and Security Trends (CAST), Pune, India, 19–21 December 2016; pp. 306–311. [Google Scholar]
- Liang, T.; Shih, P.-K. Empirical textual mining to protein entities recognition from PubMed corpus. In Proceedings of the Natural Language Processing and Information Systems: 10th International Conference on Applications of Natural Language to Information Systems, NLDB 2005, Alicante, Spain, 15–17 June 2005; pp. 56–66. [Google Scholar]
- Cai, X.L.; Dong, S.B.; Hu, J.L. A deep learning model incorporating part of speech and self-matching attention for named entity recognition of Chinese electronic medical records. BMC Med. Inform. Decis. Mak. 2019, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.S.; Chowdhury, S.; Qian, L.J.; Li, X.F.; Guan, Y.; Yang, J.F.; Yu, Q.B. Deep learning for named entity recognition on Chinese electronic medical records: Combining deep transfer learning with multitask bi-directional LSTM RNN. PLoS ONE 2019, 14, e0216046. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Qian, L.; Guan, Y.; Huang, L.; Yu, Q.; Yang, J. A multiclass classification method based on deep learning for named entity recognition in electronic medical records. In Proceedings of the 2016 New York Scientific Data Summit (NYSDS), New York, NY, USA, 14–17 August 2016; pp. 1–10. [Google Scholar]
- Dreyfus, B.; Chaudhary, A.; Bhardwaj, P.; Shree, V.K. Application of natural language processing techniques to identify off-label drug usage from various online health communities. J. Am. Med. Inf. Assoc. 2021, 28, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Li, X.M.; Zhang, Z. Disease-Pertinent Knowledge Extraction in Online Health Communities Using GRU Based on a Double Attention Mechanism. IEEE Access 2020, 8, 95947–95955. [Google Scholar] [CrossRef]
- Whitfield, C.; Liu, Y.; Anwar, M. Surveillance of COVID-19 Pandemic using Social Media: A Reddit Study in North Carolina. In Proceedings of the 12th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, Gainesville, FL, USA, 1–4 August 2021; Association for Computing Machinery: New York, NY, USA; pp. 1–8. [Google Scholar]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. arXiv 2016, arXiv:1603.01360. [Google Scholar]
- Wang, Z.; Guan, H. Research on named entity recognition of doctor-patient question answering community based on bilstm-crf model. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 1641–1644. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- Zhang, Q.; Wu, M.; Lv, P.; Zhang, M.; Yang, H. Research on named entity recognition of chinese electronic medical records based on multi-head attention mechanism and character-word information fusion. J. Intell. Fuzzy Syst. 2022, 42, 4105–4116. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, J.; Zhao, Q.; Wei, Y.-C.; Jia, Y. Medical named entity recognition of Chinese electronic medical records based on stacked Bidirectional Long Short-Term Memory. In Proceedings of the 2021 IEEE 45th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 12–16 July 2021; pp. 1930–1935. [Google Scholar]
- Fang, A.; Hu, J.H.; Zhao, W.Q.; Feng, M.; Fu, J.; Feng, S.S.; Lou, P.; Ren, H.L.; Chen, X.L. Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records. BMC Med. Inform. Decis. Mak. 2022, 22, 14. [Google Scholar] [CrossRef]
- Han, P.; Li, X.; Wang, X.; Wang, S.; Gao, C.; Chen, W. Exploring the effects of drug, disease, and protein dependencies on biomedical named entity recognition: A comparative analysis. Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef]
- Yang, X.; Bian, J.; Hogan, W.R.; Wu, Y. Clinical concept extraction using transformers. J. Am. Med. Inf. Assoc. 2020, 27, 1935–1942. [Google Scholar] [CrossRef]
- Yang, H.; Gao, H. Toward Sustainable Virtualized Healthcare: Extracting Medical Entities from Chinese Online Health Consultations Using Deep Neural Networks. Sustainability 2018, 10, 3292. [Google Scholar] [CrossRef]
- Yang, L.; Fu, Y.F.; Dai, Y. BIBC: A Chinese Named Entity Recognition Model for Diabetes Research. Appl. Sci. 2021, 11, 14. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Zhou, X.-H. Chinese clinical named entity recognition with variant neural structures based on BERT methods. J. Biomed. Inform. 2020, 107, 103422. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Liu, R.; Li, S.; Tang, J.; Yu, J.; Li, Q.; Xu, W. A BILSTM-CRF method to Chinese electronic medical record named entity recognition. In Proceedings of the 2018 International Conference on Algorithms, Computing and Artificial Intelligence, Sanya, China, 21–23 December 2018; pp. 1–6. [Google Scholar]
- Zhang, Y.; Zhang, Y.; Yin, Y.; Xu, J.; Xing, C.; Chen, H. Chronic disease related entity extraction in online chinese question and answer services. In Proceedings of the Smart Health: International Conference, ICSH 2015, Phoenix, AZ, USA, 17–18 November 2015; pp. 55–67. [Google Scholar]
- Zhang, Q.; Sun, Y.; Zhang, L.; Jiao, Y.; Tian, Y. Named entity recognition method in health preserving field based on BERT. Procedia Comput. Sci. 2021, 183, 212–220. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, P.; Liu, J.; Li, J.; Deng, S. Health-Related Hot Topic Detection in Online Communities Using Text Clustering. PLoS ONE 2013, 8, e56221. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wu, Y.; Liu, J.; Li, J.; Zhang, P. Understanding Health Care Social Media Use From Different Stakeholder Perspectives: A Content Analysis of an Online Health Community. J. Med. Internet Res. 2017, 19, e109. [Google Scholar] [CrossRef]
- Zhang, S.; Grave, E.; Sklar, E.; Elhadad, N. Longitudinal analysis of discussion topics in an online breast cancer community using convolutional neural networks. J. Biomed. Inform. 2017, 69, 1–9. [Google Scholar] [CrossRef]



| Classification | Description | Labeling Case |
|---|---|---|
| Check | Test and examination items, physical examination, review, etc. | A review at the hospital the previous day; a check-up at the hospital. |
| Disease | Disease names, such as hypertension, diabetes, etc. | No diabetes in the family either. |
| Drug | The name of the drug, such as nifedipine, metformin, nifedipine, etc. | The medications being taken are Metformin Hydrochloride and Vildagliptin. |
| Lifestyle | Patient’s lifestyle, e.g., smoking, alcohol consumption, sleep, etc. | Smoking; drinking; staying up late. |
| Mood | Irritable, anxious, worried | So now it is confusing. |
| Social context | Dad (my dad), wife (my wife), medical history, occupation, height, weight, age, gender (pregnancy and gestation), wanting children. | Height and weight: 171 cm, 70 kg. Pregnancy: not pregnant. |
| Symptom | Patient’s subjective description of feelings and signs (skin jaundice), such as dizziness, non-dizziness, nocturia, puffy eyelids, and frequent need to urinate. | Feeling of vertigo when standing suddenly; I urinate frequently and often, but each time the amount of urine is not much, nausea, vomiting, weakness, stomach pain, and breast swelling. |
| Treatment | Chinese medicine treatment, immunotherapy, ventilator, and stent release. | Immunotherapy. |
| Experimental Parameters | Value |
|---|---|
| Sequence_length | 128 |
| Batch_size | Train set 16, test set 16 |
| Pooler_fc_size | 768 |
| Epoch | 50 |
| Learning rate | 3 × 10−5 |
| Optimizer | Adam |
| Input layer dropout | 0.1 |
| Hidden layers dropout | 0.1 |
| Models | F1 | P | R | Acc |
|---|---|---|---|---|
| RBC | 0.807 | 0.786 | 0.829 | 0.812 |
| RC | 0.795 | 0.755 | 0.827 | 0.803 |
| RS | 0.790 | 0.755 | 0.828 | 0.799 |
| BC | 0.751 | 0.743 | 0.759 | 0.754 |
| Model | Index | Check | Disease | Drug | Lifestyle | Mood | Social | Symptoms | Treat |
|---|---|---|---|---|---|---|---|---|---|
| RBC | P | 0.739 | 0.787 | 0.730 | 0.754 | 0.865 | 0.941 | 0.609 | 0.571 |
| R | 0.774 | 0.863 | 0.823 | 0.723 | 0.922 | 0.926 | 0.709 | 0.585 | |
| F1 | 0.756 | 0.823 | 0.774 | 0.738 | 0.892 | 0.933 | 0.655 | 0.578 | |
| RC | P | 0.719 | 0.761 | 0.730 | 0.696 | 0.903 | 0.918 | 0.596 | 0.542 |
| R | 0.771 | 0.850 | 0.821 | 0.732 | 0.933 | 0.924 | 0.710 | 0.639 | |
| F1 | 0.744 | 0.803 | 0.773 | 0.713 | 0.917 | 0.921 | 0.647 | 0.586 | |
| RS | P | 0.717 | 0.742 | 0.704 | 0.697 | 0.878 | 0.915 | 0.582 | 0.516 |
| R | 0.772 | 0.866 | 0.824 | 0.726 | 0.956 | 0.923 | 0.690 | 0.624 | |
| F1 | 0.743 | 0.799 | 0.759 | 0.710 | 0.915 | 0.919 | 0.631 | 0.564 | |
| BC | P | 0.682 | 0.722 | 0.697 | 0.702 | 0.857 | 0.918 | 0.533 | 0.559 |
| R | 0.731 | 0.766 | 0.684 | 0.668 | 0.756 | 0.909 | 0.521 | 0.610 | |
| F1 | 0.704 | 0.748 | 0.687 | 0.681 | 0.795 | 0.913 | 0.526 | 0.582 |
| Entity Type | Entity Frequency | Rate | Top 10 Entities | Top 10 Number of Entities | Top 10 Rate |
|---|---|---|---|---|---|
| Check | 1512/1890 | 80% | Blood glucose, fasting blood glucose, fasting, postprandial, glycated hemoglobin, physical examination, high blood glucose, glucose tolerance, review, and postprandial blood glucose. | 3019/4259 | 70.86% |
| Disease | 1513/1890 | 80.05% | Diabetes, hypertension, hyperglycemia, type 2 diabetes, fatty liver, coronary heart disease, cerebral infarction, obesity, hyperlipidemia, and complications of diabetes. | 1790/2447 | 73.15% |
| Drug | 1066/1890 | 56.4% | Insulin, Metformin, Acarbose, Glucose, Dapagliflozin, Glucagon, Glimepiride, Bystolic, Gleevec, and Chinese medicine. | 1306/2504 | 52.16% |
| Life | 490/1890 | 25.92% | Blood sugar control, exercise, diet control, poor sleep, stopping the medication, exercise, not taking medication, losing weight, watching what you eat, and staying up late. | 509/787 | 64.68% |
| Mood | 144/1890 | 7.61% | Worry, doubt, fear, anxiety, hurry, tension, tiredness, anger, uneasiness, and fear. | 132/185 | 71.35% |
| Social | 1695/1890 | 89.68% | Height and weight, greater than six months, pregnant, not pregnant, within six months, within one month, within one week, self, allergy, and father. | 3864/4690 | 82.39% |
| Symptom | 688/1890 | 16.4% | Thirst, bitterness and dryness, dizziness and lightheadedness, excessive urination, weakness, weight loss, nausea, sweating, panic attacks, and frequent urination. | 465/1564 | 29.73% |
| Treat | 421/1890 | 22.08% | Surgery, chemotherapy, radiotherapy, drug therapy, inpatient treatment, weight loss, Chinese medicine, stents, minimally invasive, and immunization. | 429/655 | 65.50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Zhou, Y.; Liao, B.; Xin, Z.; Xie, W.; Hu, C.; Luo, A. Named Entity Recognition of Diabetes Online Health Community Data Using Multiple Machine Learning Models. Bioengineering 2023, 10, 659. https://doi.org/10.3390/bioengineering10060659
Xu Q, Zhou Y, Liao B, Xin Z, Xie W, Hu C, Luo A. Named Entity Recognition of Diabetes Online Health Community Data Using Multiple Machine Learning Models. Bioengineering. 2023; 10(6):659. https://doi.org/10.3390/bioengineering10060659
Chicago/Turabian StyleXu, Qian, Yue Zhou, Bolin Liao, Zirui Xin, Wenzhao Xie, Chao Hu, and Aijing Luo. 2023. "Named Entity Recognition of Diabetes Online Health Community Data Using Multiple Machine Learning Models" Bioengineering 10, no. 6: 659. https://doi.org/10.3390/bioengineering10060659
APA StyleXu, Q., Zhou, Y., Liao, B., Xin, Z., Xie, W., Hu, C., & Luo, A. (2023). Named Entity Recognition of Diabetes Online Health Community Data Using Multiple Machine Learning Models. Bioengineering, 10(6), 659. https://doi.org/10.3390/bioengineering10060659







