Tailoring the Microarchitectures of 3D Printed Bone-like Scaffolds for Tissue Engineering Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Scaffold Design
2.2. Scaffold Fabrication
2.3. Morphological Characterization
2.4. Cell Culture
2.5. Cell Growth Analysis
2.6. Real-Time Quantitative RT-PCR Analysis
2.7. Scanning Electron Microscopy Analysis
2.8. Protein Adsorption
2.9. ELISA Assay
2.10. Statistical Analysis
3. Results and Discussion
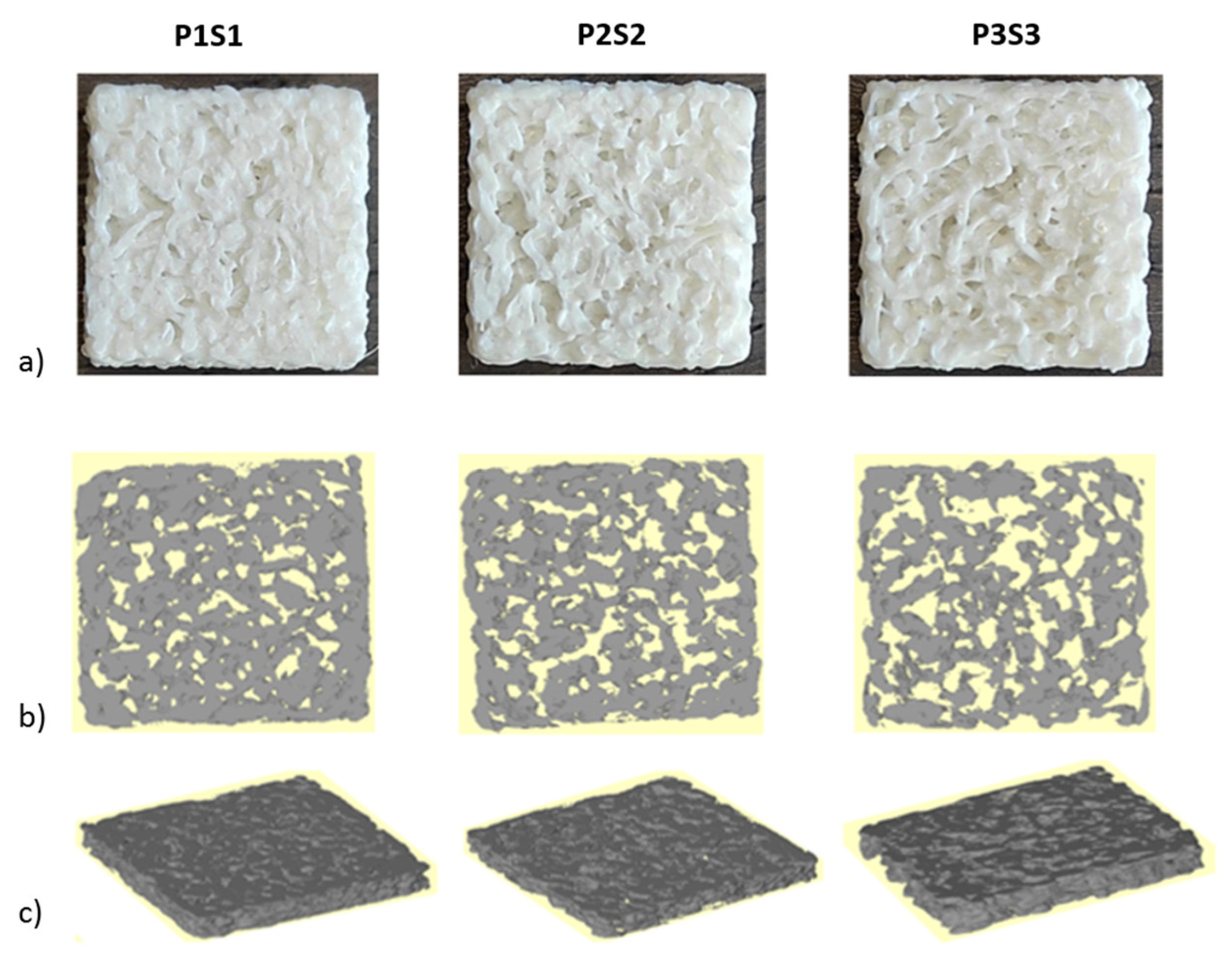
3.1. Scaffold Evaluation
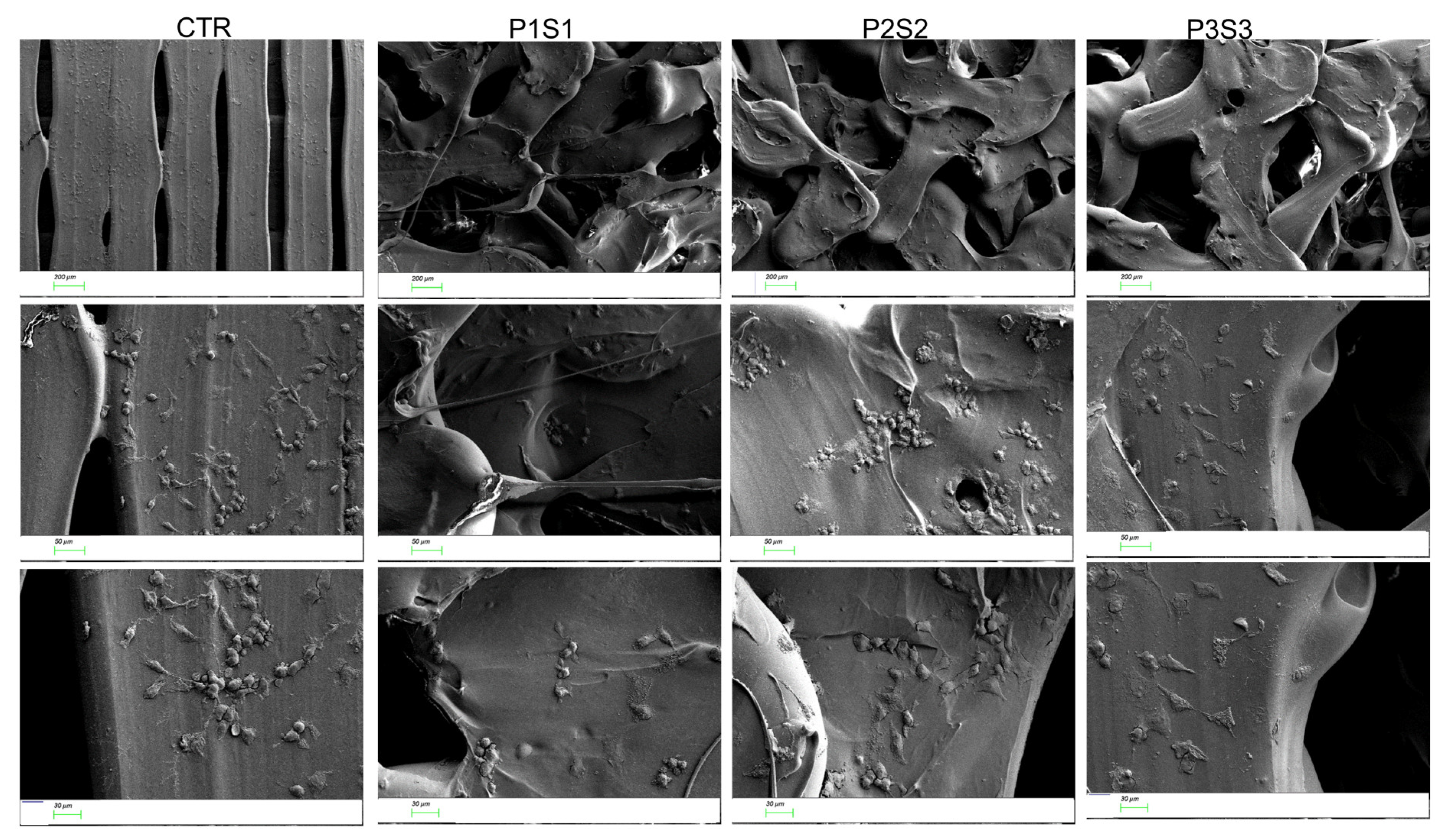
3.2. Adhesion, Growth, and Differentiation of Osteoblast-like Cells on Biomimetic 3D-Printed Scaffolds
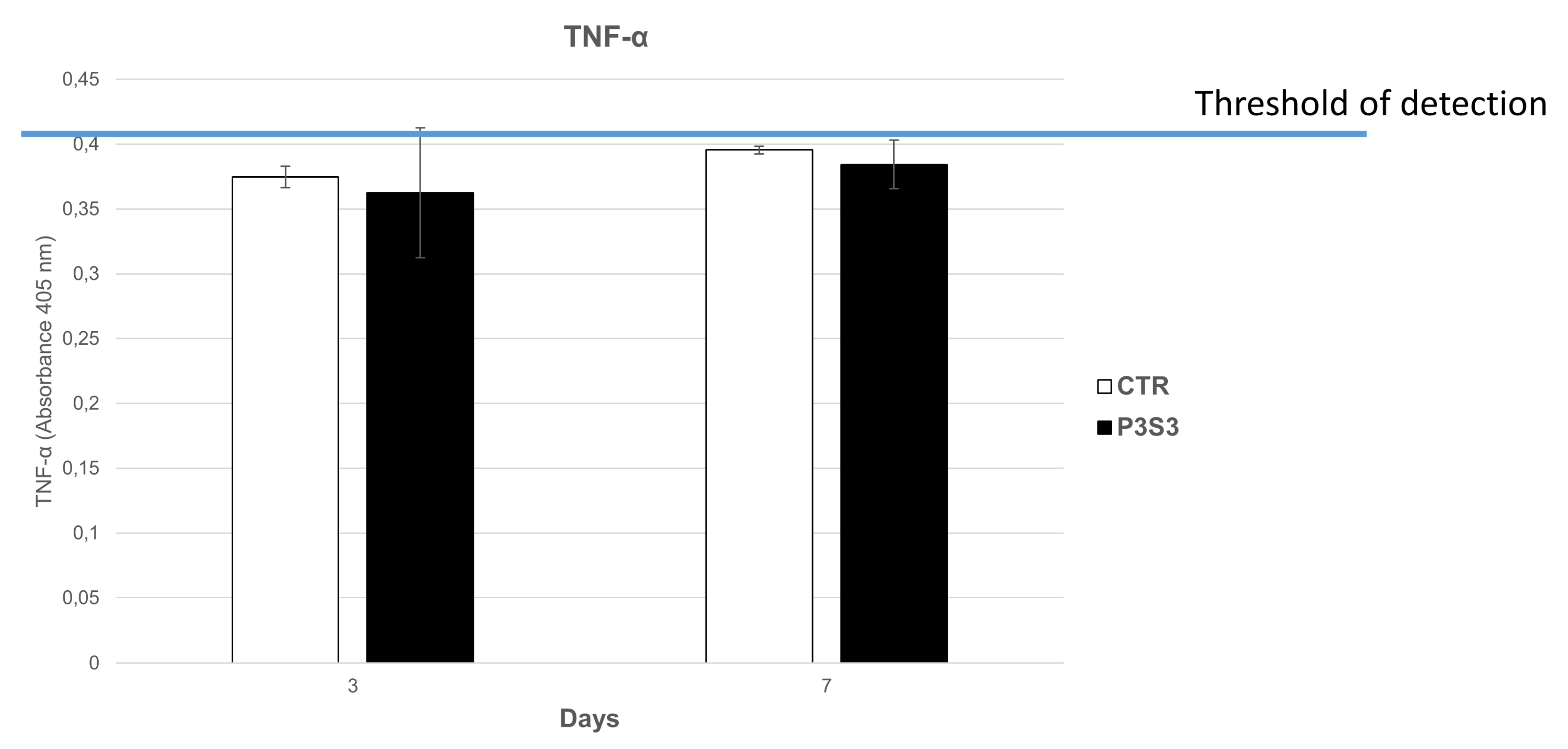
3.3. Protein Adsorption of 3D Printed Scaffolds
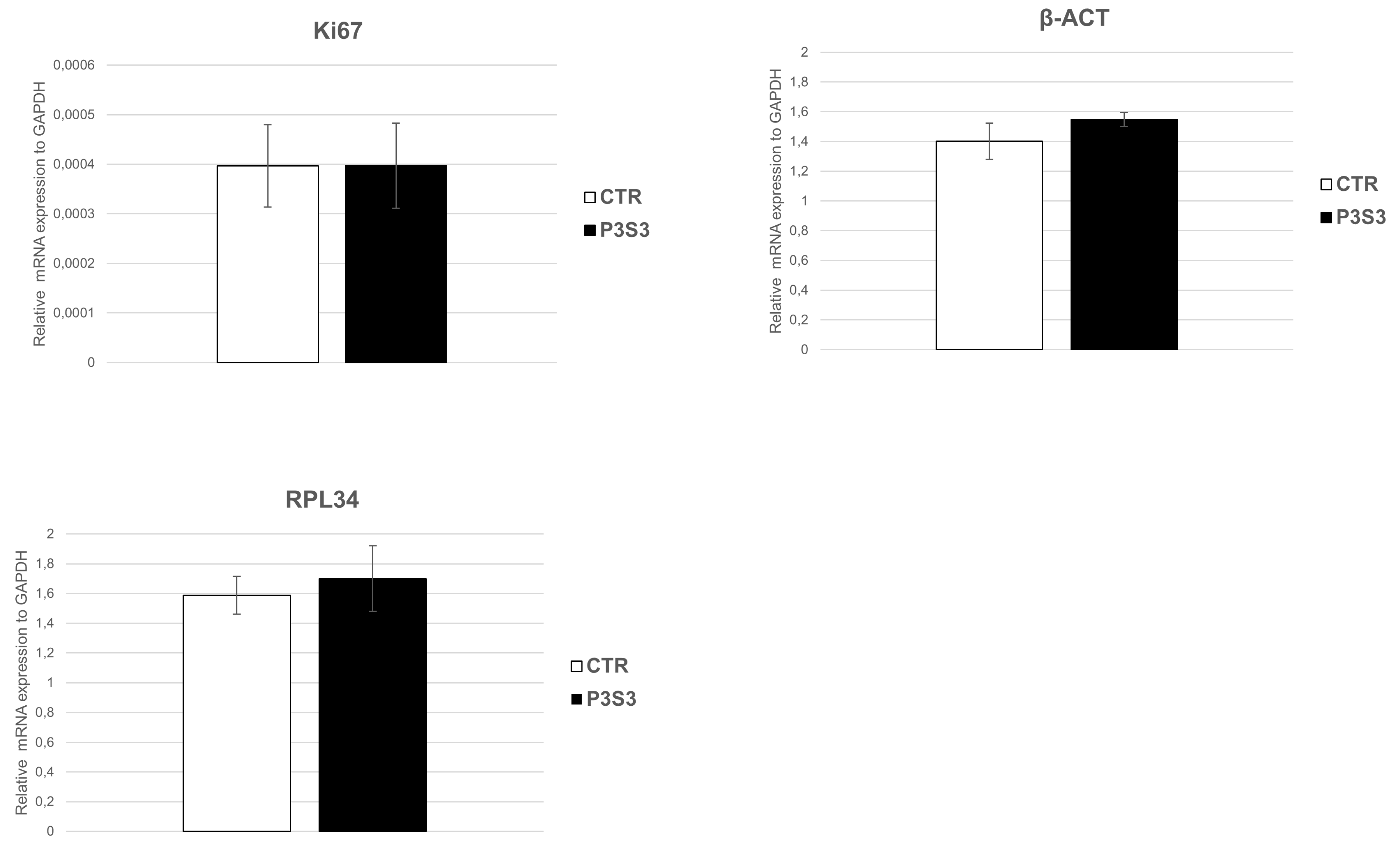
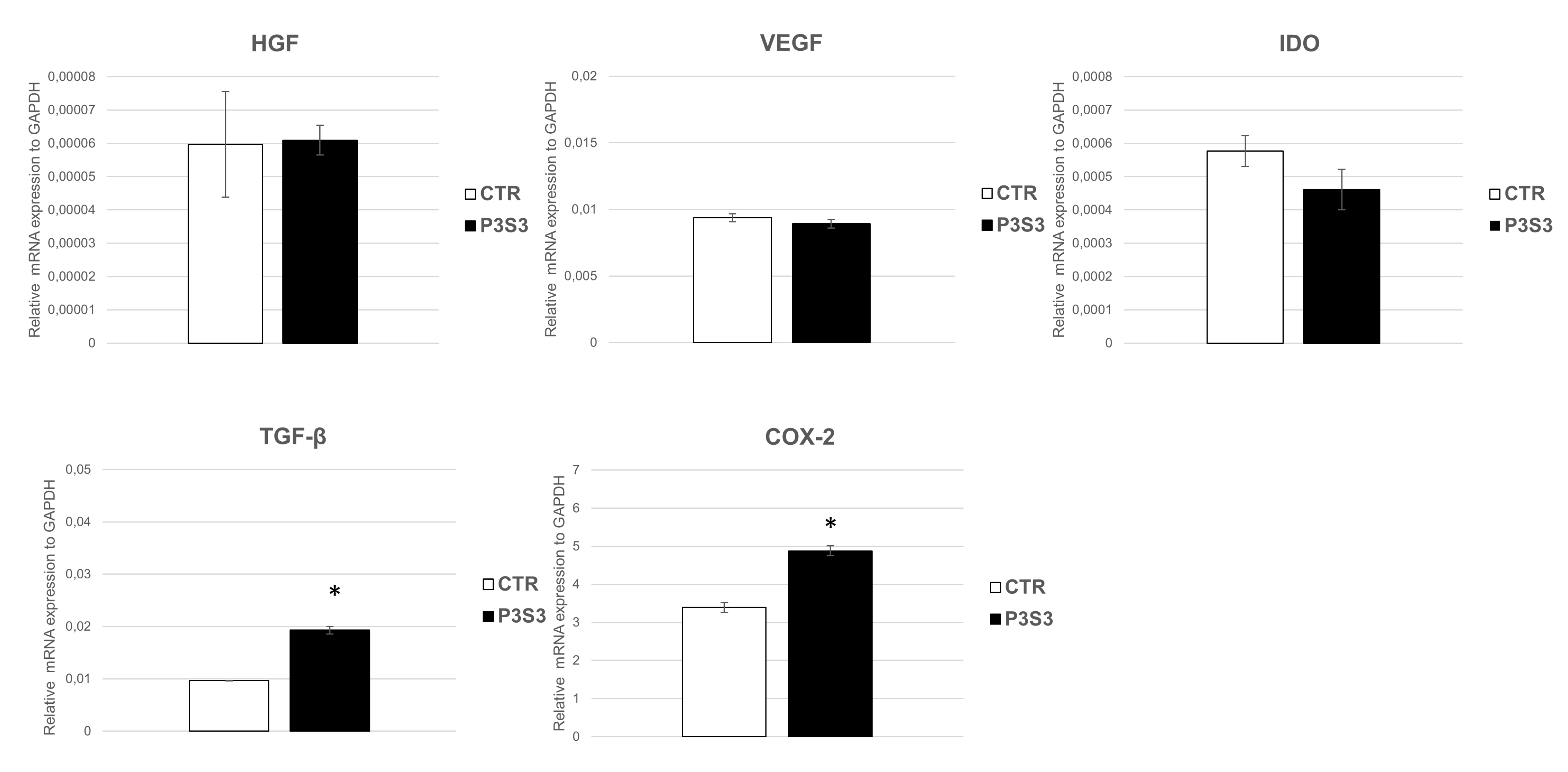
3.4. Human Mesenchymal Stromal Cell Interaction with 3D-Printed Biomimetic Scaffolds (P3S3)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Haleem, A.; Javaid, M. Additive Manufacturing Applications in Industry 4.0: A Review. J. Ind. Integr. Manag. 2019, 4, 1930001. [Google Scholar] [CrossRef]
- Ciocci, M.; Mochi, F.; Carotenuto, F.; Di Giovanni, E.; Prosposito, P.; Francini, R.; De Matteis, F.; Reshetov, I.; Casalboni, M.; Melino, S.; et al. Scaffold-in-Scaffold Potential to Induce Growth and Differentiation of Cardiac Progenitor Cells. Stem Cells Dev. 2017, 26, 1438–1447. [Google Scholar] [CrossRef]
- Baino, F.; Magnaterra, G.; Fiume, E.; Schiavi, A.; Tofan, L.P.; Schwentenwein, M.; Verne, E. Digital light processing stereolithography of hydroxyapatite scaffolds with bone-like architecture, permeability, and mechanical properties. J. Am. Ceram. Soc. 2022, 105, 1648–1657. [Google Scholar] [CrossRef]
- Lee, M.; Dunn, J.C.Y.; Wu, B.M. Scaffold fabrication by indirect three-dimensional printing. Biomaterials 2005, 26, 4281–4289. [Google Scholar] [CrossRef]
- Butscher, A.; Bohner, M.; Hofmann, S.; Gauckler, L.; Muller, R. Structural and material approaches to bone tissue engineering in powder-based three-dimensional printing. Acta Biomater. 2011, 7, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Tirella, A.; Vozzi, F.; De Maria, C.; Vozzi, G.; Sandri, T.; Sassano, D.; Cognolato, L.; Ahluwalia, A. Substrate stiffness influences high resolution printing of living cells with an ink-jet system. J. Biosci. Bioeng. 2011, 112, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, D.M.; Huang, J.H.; Wei, Y.; Xiong, J.Y.; Zhu, W.M.; Duan, L.; Chen, J.L.; Sun, R.; Wang, D.P. Low-temperature deposition manufacturing: A novel and promising rapid prototyping technology for the fabrication of tissue-engineered scaffold. Mat. Sci. Eng. C–Mater. 2017, 70, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Negro, A.; Cherbuin, T.; Lutolf, M.P. 3D Inkjet Printing of Complex, Cell-Laden Hydrogel Structures. Sci. Rep. 2018, 8, 17099. [Google Scholar] [CrossRef]
- Ansari, M.H.B.; Bin Ibrahim, M.H.I. Thermal Characteristic of Waste-Derived Hydroxyapatite (Ha) Reinforced Ultra High Molecular Weight Polyethylene (Uhmwpe) Composites for Fused Deposition Modeling (Fdm) Process. Iop Conf. Ser.–Mat. Sci. 2017, 165, 012014. [Google Scholar]
- Ivanov, E.; Kotsilkova, R.; Xia, H.S.; Chen, Y.H.; Donato, R.K.; Donato, K.; Godoy, A.P.; Di Maio, R.; Silvestre, C.; Cimmino, S.; et al. PLA/Graphene/MWCNT Composites with Improved Electrical and Thermal Properties Suitable for FDM 3D Printing Applications. Appl. Sci. 2019, 9, 1209. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, B.; Fu, F.; You, F.S.; Dong, X.Z.; Dai, M. Resistivity and Its Anisotropy Characterization of 3D-Printed Acrylonitrile Butadiene Styrene Copolymer (ABS)/Carbon Black (CB) Composites. Appl. Sci. 2017, 7, 20. [Google Scholar] [CrossRef]
- Liu, S.D.; Chen, J.M.; Chen, T.; Zeng, Y. Fabrication of trabecular-like beta-tricalcium phosphate biomimetic scaffolds for bone tissue engineering. Ceram. Int. 2021, 47, 13187–13198. [Google Scholar] [CrossRef]
- Baptista, R.; Guedes, M. Morphological and mechanical characterization of 3D printed PLA scaffolds with controlled porosity for trabecular bone tissue replacement. Mater. Sci. Eng. C–Mater. Biol. Appl. 2021, 118, 111528. [Google Scholar] [CrossRef] [PubMed]
- Velioglu, Z.B.; Pulat, D.; Demirbakan, B.; Ozcan, B.; Bayrak, E.; Erisken, C. 3D-printed poly(lactic acid) scaffolds for trabecular bone repair and regeneration: Scaffold and native bone characterization. Connect. Tissue Res. 2019, 60, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Q.; Wang, L.; Song, P.; Pei, X.; Sun, H.; Wu, L.A.; Zhou, C.C.; Wang, K.F.; Fan, Y.J.; Zhang, X.D. 3D printed bone tissue regenerative PLA/HA scaffolds with comprehensive performance optimizations. Mater. Des. 2021, 201, 109490. [Google Scholar] [CrossRef]
- Perez, R.A.; Mestres, G. Role of pore size and morphology in musculo-skeletal tissue regeneration. Mater. Sci. Eng. C–Mater. Biol. Appl. 2016, 61, 922–939. [Google Scholar] [CrossRef]
- Pecci, R.; Baiguera, S.; Ioppolo, P.; Bedini, R.; Del Gaudio, C. 3D printed scaffolds with random microarchitecture for bone tissue engineering applications: Manufacturing and characterization. J. Mech. Behav. Biomed. Mater. 2020, 103, 103583. [Google Scholar] [CrossRef]
- Tsai, M.S.; Hwang, S.M.; Chen, K.D.; Lee, Y.S.; Hsu, L.W.; Chang, Y.J.; Wang, C.N.; Peng, H.H.; Chang, Y.L.; Chao, A.S.; et al. Functional network analysis of the transcriptomes of mesenchymal stem cells derived from amniotic fluid, amniotic membrane, cord blood, and bone marrow. Stem Cells 2007, 25, 2511–2523. [Google Scholar] [CrossRef]
- Amado, L.C.; Saliaris, A.P.; Schuleri, K.H.; St John, M.; Xie, J.S.; Cattaneo, S.; Durand, D.J.; Fitton, T.; Kuang, J.Q.; Stewart, G.; et al. Cardiac repair with intramyocardial injection of allogeneic mesenchymal stem cells after myocardial infarction. Proc. Natl. Acad. Sci. USA 2005, 102, 11474–11479. [Google Scholar] [CrossRef]
- Kim, J.M.; Lee, S.T.; Chu, K.; Jung, K.H.; Song, E.C.; Kim, S.J.; Sinn, D.I.; Kim, J.H.; Park, D.K.; Kang, K.M.; et al. Systemic transplantation of human adipose stem cells attenuated cerebral inflammation and degeneration in a hemorrhagic stroke model. Brain Res. 2007, 1183, 43–50. [Google Scholar] [CrossRef]
- Le Blanc, K.; Rasmusson, I.; Sundberg, B.; Gotherstrom, C.; Hassan, M.; Uzunel, M.; Ringden, O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet 2004, 363, 1439–1441. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Fang, X.H.; Gupta, N.; Serikov, V.; Matthay, M.A. Allogeneic human mesenchymal stem cells for treatment of E. coli endotoxin-induced acute lung injury in the ex vivo perfused human lung. Proc. Natl. Acad. Sci. USA 2009, 106, 16357–16362. [Google Scholar] [CrossRef] [PubMed]
- Gnecchi, M.; Danieli, P.; Malpasso, G.; Ciuffreda, M.C. Paracrine Mechanisms of Mesenchymal Stem Cells in Tissue Repair. Methods Mol. Biol. 2016, 1416, 123–146. [Google Scholar] [PubMed]
- Loukogeorgakis, S.P.; De Coppi, P. Stem cells from amniotic fluid–Potential for regenerative medicine. Best Pract. Clin. Obstet. Gynaecol. 2016, 31, 45–57. [Google Scholar] [CrossRef]
- Wobma, H.M.; Liu, D.; Vunjak-Novakovic, G. Paracrine Effects of Mesenchymal Stromal Cells Cultured in Three-Dimensional Settings on Tissue Repair. Acs Biomater. Sci. Eng. 2018, 4, 1162–1175. [Google Scholar] [CrossRef]
- Bernardo, M.E.; Fibbe, W.E. Mesenchymal stromal cells: Sensors and switchers of inflammation. Cell Stem Cell 2013, 13, 392–402. [Google Scholar] [CrossRef]
- Chung, E.; Son, Y. Crosstalk between Mesenchymal Stem Cells and Macrophages in Tissue Repair. Tissue Eng. Regen. Med. 2014, 11, 431–438. [Google Scholar] [CrossRef]
- Schemitsch, E.H. Size Matters: Defining Critical in Bone Defect Size! J. Orthop. Trauma 2017, 31, S20–S22. [Google Scholar] [CrossRef]
- Liu, X.S.; Zhang, X.H.; Rajapakse, C.S.; Wald, M.J.; Magland, J.; Sekhon, K.K.; Adam, M.F.; Sajda, P.; Wehrli, F.W.; Guo, X.E. Accuracy of High-Resolution In Vivo Micro Magnetic Resonance Imaging for Measurements of Microstructural and Mechanical Properties of Human Distal Tibial Bone. J. Bone Miner. Res. 2010, 25, 2039–2050. [Google Scholar] [CrossRef]
- Karageorgiou, V.; Kaplan, D. Porosity of 3D biomaterial scaffolds and osteogenesis. Biomaterials 2005, 26, 5474–5491. [Google Scholar] [CrossRef]
- Murphy, C.M.; Haugh, M.G.; O’Brien, F.J. The effect of mean pore size on cell attachment, proliferation and migration in collagen-glycosaminoglycan scaffolds for bone tissue engineering. Biomaterials 2010, 31, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Srinivasa, A.; Reddy, J.N.; Dubrowski, A. Topology Optimization of Lightweight Structures with Application to Bone Scaffolds and 3D Printed Shoes for Diabetics. J. Appl. Mech 2022, 89, 041009. [Google Scholar] [CrossRef]
- Cengiz, I.F.; Oliveira, J.M.; Reis, R.L. Micro-CT–A digital 3D microstructural voyage into scaffolds: A systematic review of the reported methods and results. Biomater. Res. 2018, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Ledda, M.; D’Emilia, E.; Lolli, M.G.; Marchese, R.; De Lazzari, C.; Lisi, A. Non-Ionizing Radiation for Cardiac Human Amniotic Mesenchymal Stromal Cell Commitment: A Physical Strategy in Regenerative Medicine. Int. J. Mol. Sci. 2018, 19, 2324. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Adanty, K.; Rabey, K.N.; Doschak, M.R.; Bhagavathula, K.B.; Hogan, J.D.; Romanyk, D.L.; Adeeb, S.; Ouellet, S.; Plaisted, T.A.; Satapathy, S.S.; et al. Cortical and trabecular morphometric properties of the human calvarium. Bone 2021, 148, 115931. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, H.J.; Yun, J.H. Three-dimensional microstructure of human alveolar trabecular bone: A micro-computed tomography study. J. Periodontal Implan. 2017, 47, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Viveen, J.; Perilli, E.; Zahrooni, S.; Jaarsma, R.L.; Doornberg, J.N.; Bain, G.I. Three-dimensional cortical and trabecular bone microstructure of the proximal ulna. Arch. Orthop. Trauma Surg. 2021, 143, 213–223. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzynski, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar]
- Zhou, X.Q.; Zhou, G.; Junka, R.; Chang, N.X.; Anwar, A.; Wang, H.Y.; Yu, X.J. Fabrication of polylactic acid (PLA)-based porous scaffold through the combination of traditional bio-fabrication and 3D printing technology for bone regeneration. Colloid Surface B. 2021, 197, 111420. [Google Scholar] [CrossRef]
- Eckstein, F.; Matsuura, M.; Kuhn, V.; Priemel, M.; Muller, R.; Link, T.M.; Lochmuller, E.M. Sex differences of human trabecular bone microstructure in aging are site-dependent. J. Bone Miner. Res. 2007, 22, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Imamura, E.; Mayahara, M.; Inoue, S.; Miyamoto, M.; Funae, T.; Watanabe, Y.; Matsuki-Fukushima, M.; Nakamura, M. Trabecular structure and composition analysis of human autogenous bone donor sites using micro-computed tomography. J. Oral Biosci. 2021, 63, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Nikodem, A. Correlations between structural and mechanical properties of human trabecular femur bone. Acta Bioeng. Biomech. 2012, 14, 37–46. [Google Scholar] [PubMed]
- Mcquillan, D.J.; Richardson, M.D.; Bateman, J.F. Matrix Deposition by a Calcifying Human Osteogenic-Sarcoma Cell-Line (Saos-2). Bone 1995, 16, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Prideaux, M.; Wijenayaka, A.R.; Kumarasinghe, D.D.; Ormsby, R.T.; Evdokiou, A.; Findlay, D.M.; Atkins, G.J. SaOS2 Osteosarcoma Cells as an In Vitro Model for Studying the Transition of Human Osteoblasts to Osteocytes. Calcified Tissue Int. 2014, 95, 183–193. [Google Scholar] [CrossRef]
- Czekanska, E.M.; Stoddart, M.J.; Richards, R.G.; Hayes, J.S. In Search of an Osteoblast Cell Model for in Vitro Research. Eur. Cells Mater. 2012, 24, 1–17. [Google Scholar] [CrossRef]
- Stein, G.S.; Lian, J.B.; Stein, J.L.; Van Wijnen, A.J.; Montecino, M. Transcriptional control of osteoblast growth and differentiation. Physiol. Rev. 1996, 76, 593–629. [Google Scholar] [CrossRef]
- Beck, G.R.; Zerler, B.; Moran, E. Phosphate is a specific signal for induction of osteopontin gene expression. Proc. Natl. Acad. Sci. USA 2000, 97, 8352–8357. [Google Scholar] [CrossRef]
- Zhang, K.; Fan, Y.B.; Dunne, N.; Li, X.M. Effect of microporosity on scaffolds for bone tissue engineering. Regen. Biomater. 2018, 5, 115–124. [Google Scholar] [CrossRef]
- Depan, D.; Misra, R.D.K. The interplay between nanostructured carbon-grafted chitosan scaffolds and protein adsorption on the cellular response of osteoblasts: Structure-function property relationship. Acta Biomater. 2013, 9, 6084–6094. [Google Scholar] [CrossRef]
- Lu, H.H.; El-Amin, S.F.; Scott, K.D.; Laurencin, C.T. Three-dimensional, bioactive, biodegradable, polymer-bioactive glass composite scaffolds with improved mechanical properties support collagen synthesis and mineralization of human osteoblast-like cells in vitro. J. Biomed. Mater. Res. A 2003, 64, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.; Kuralesova, A.I. Osteogenic precursor cells of bone marrow in radiation chimeras. Transplantation 1971, 12, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.; Borgonovo, G.; Pistoia, V.; Raffaghello, L. Immunosuppressive cells and tumour microenvironment: Focus on mesenchymal stem cells and myeloid derived suppressor cells. Histol. Histopathol. 2011, 26, 941–951. [Google Scholar] [PubMed]
- Granero-Molto, F.; Weis, J.A.; Longobardi, L.; Spagnoli, A. Role of mesenchymal stem cells in regenerative medicine: Application to bone and cartilage repair. Expert Opin. Biol. Ther. 2008, 8, 255–268. [Google Scholar] [CrossRef]
- Prockop, D.J. Marrow stromal cells as stem cells for continual renewal of nonhematopoietic tissues and as potential vectors for gene therapy. J. Cell. Biochem. 1998, 30, 284–285. [Google Scholar] [CrossRef]
- Dezawa, M.; Ishikawa, H.; Itokazu, Y.; Yoshihara, T.; Hoshino, M.; Takeda, S.; Ide, C.; Nabeshima, Y. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science 2005, 309, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Salem, H.K.; Thiemermann, C. Mesenchymal Stromal Cells: Current Understanding and Clinical Status. Stem Cells 2010, 28, 585–596. [Google Scholar] [CrossRef]
- Janicki, P.; Schmidmaier, G. What should be the characteristics of the ideal bone graft substitute? Combining scaffolds with growth factors and/or stem cells. Injury 2011, 42, S77–S81. [Google Scholar] [CrossRef]
- Meijer, G.J.; de Bruijn, J.D.; Koole, R.; van Blitterswijk, C.A. Cell based bone tissue engineering in jaw defects. Biomaterials 2008, 29, 3053–3061. [Google Scholar] [CrossRef]
- Mesimaki, K.; Lindroos, B.; Tornwall, J.; Mauno, J.; Lindqvist, C.; Kontio, R.; Miettinen, S.; Suuronen, R. Novel maxillary reconstruction with ectopic bone formation by GMP adipose stem cells. Int. J. Oral. Maxillofac. Surg. 2009, 38, 201–209. [Google Scholar] [CrossRef]
- Inukai, T.; Katagiri, W.; Yoshimi, R.; Osugi, M.; Kawai, T.; Hibi, H.; Ueda, M. Novel application of stem cell-derived factors for periodontal regeneration. Biochem. Biophys. Res. Commun. 2013, 430, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Osugi, M.; Katagiri, W.; Yoshimi, R.; Inukai, T.; Hibi, H.; Ueda, M. Conditioned media from mesenchymal stem cells enhanced bone regeneration in rat calvarial bone defects. Tissue Eng. Part A 2012, 18, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Polacek, M.; Bruun, J.A.; Elvenes, J.; Figenschau, Y.; Martinez, I. The secretory profiles of cultured human articular chondrocytes and mesenchymal stem cells: Implications for autologous cell transplantation strategies. Cell Transplant. 2011, 20, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Kaigler, D.; Krebsbach, P.H.; Polverini, P.J.; Mooney, D.J. Role of vascular endothelial growth factor in bone marrow stromal cell modulation of endothelial cells. Tissue Eng. 2003, 9, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Janssens, K.; ten Dijke, P.; Janssens, S.; Van Hul, W. Transforming growth factor-beta1 to the bone. Endocr. Rev. 2005, 26, 743–774. [Google Scholar] [CrossRef]
- Gao, J.; Symons, A.L.; Bartold, P.M. Expression of transforming growth factor-beta 1 (TGF-β1) in the developing periodontium of rats. J. Dent. Res. 1998, 77, 1708–1716. [Google Scholar] [CrossRef]
- Fu, X.; Liu, G.; Halim, A.; Ju, Y.; Luo, Q.; Song, A.G. Mesenchymal Stem Cell Migration and Tissue Repair. Cells 2019, 8, 784. [Google Scholar] [CrossRef]
- Wu, H.; Gong, J.; Liu, Y. Indoleamine 2, 3-dioxygenase regulation of immune response (Review). Mol. Med. Rep. 2018, 17, 4867–4873. [Google Scholar] [CrossRef]
- Horie, M.; Choi, H.; Lee, R.H.; Reger, R.L.; Ylostalo, J.; Muneta, T.; Sekiya, I.; Prockop, D.J. Intra-articular injection of human mesenchymal stem cells (MSCs) promote rat meniscal regeneration by being activated to express Indian hedgehog that enhances expression of type II collagen. Osteoarthr. Cartilage 2012, 20, 1197–1207. [Google Scholar] [CrossRef]
- Marquez, L.; de Abreu, F.A.; Ferreira, C.L.; Alves, G.D.; Miziara, M.N.; Alves, J.B. Enhanced bone healing of rat tooth sockets after administration of epidermal growth factor (EGF) carried by liposome. Injury 2013, 44, 558–564. [Google Scholar] [CrossRef]



| Target Gene | Primer Sequence | Annealing Temperature (°C) |
|---|---|---|
| β-ACT | 5′-gctcctcctgagcgcaag-3′ 5′catctgctggaaggtggaca-3′ | 60 |
| OPN | 5′-gtgtggtttatggactgagg-3′ 5′-acggggatggccttgtatg-3′ | 60 |
| Ki67 | 5′-tgaacaaaaggcaaagaagac-3′ 5′-gagctttccctattattatggt-3′ | 60 |
| RPL34 | 5′-gaaacatgtcagcagggcc-3′ 5′-tgactctgtgcttgtgcctt-3′ | 60 |
| RUNX2 | 5′-catcatctctgccccctct-3′ 5′-actcttgcctcgtccactc-3′ | 60 |
| ALP | 5′-caatgagggcaccgtggg-3′ 5′-tcgtggtggtcacaatgcc-3′ | 60 |
| OCL | 5′-cagcgaggtagtgaagag-3′ 5′-gaaagccgatgtggtcagc-3′ | 60 |
| GAPDH | 5′-catcatctctgccccctct-3′ 5′-caaagttgtcatggatgacct-3′ | 60 |
| VEGF | 5′-cttgggtgcattggagcct-3′ 5′-ctgcgctgatagacatccat-3′ | 60 |
| HGF | 5′-caatagcatgtcaagtggag-3′ 5′-ctgtgttcgtgtggtatcat-3′ | 60 |
| TGF β1 | 5′-tcaagttaaaagtggagcagc-3′ 5′-actccggtgacatcaaaaga-3′ | 60 |
| IDO | 5′-tgctaaaggcgctgttggaa-3′ 5′-tacaccagaccgtctgatag-3′ | 60 |
| P1S1 | P2S2 | P3S3 | Human Proximal Ulna [38] | Human Calvarium [36] | Human Hemimandibular and Hemimaxillae [37] | |
|---|---|---|---|---|---|---|
| Percent Bone Volume (%) | 71.99 | 62.78 | 55.26 | 43.70 ± 22.40 | 46.70 | 37.29 ± 17.96 |
| Total Porosity (%) | 28.01 | 37.22 | 44.74 | 56.30 ± 22.40 | 53.30 | 62.71 ± 17.96 |
| Open Porosity (%) | 27.87 | 37.15 | 44.67 | |||
| Trabecular Thickness (mm) | 0.31 | 0.28 | 0.26 | 0.40 ± 0.09 | 0.27 | 0.30 ± 0.08 |
| Trabecular Separation (mm) | 0.19 | 0.24 | 0.28 | 0.63 ± 0.22 | 0.59 | 0.59 ± 0.22 |
| Specific Surface (mm−1) | 10.22 | 12.57 | 14.30 | NA | NA | 12.79 ± 4.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zenobi, E.; Merco, M.; Mochi, F.; Ruspi, J.; Pecci, R.; Marchese, R.; Convertino, A.; Lisi, A.; Del Gaudio, C.; Ledda, M. Tailoring the Microarchitectures of 3D Printed Bone-like Scaffolds for Tissue Engineering Applications. Bioengineering 2023, 10, 567. https://doi.org/10.3390/bioengineering10050567
Zenobi E, Merco M, Mochi F, Ruspi J, Pecci R, Marchese R, Convertino A, Lisi A, Del Gaudio C, Ledda M. Tailoring the Microarchitectures of 3D Printed Bone-like Scaffolds for Tissue Engineering Applications. Bioengineering. 2023; 10(5):567. https://doi.org/10.3390/bioengineering10050567
Chicago/Turabian StyleZenobi, Eleonora, Miriam Merco, Federico Mochi, Jacopo Ruspi, Raffaella Pecci, Rodolfo Marchese, Annalisa Convertino, Antonella Lisi, Costantino Del Gaudio, and Mario Ledda. 2023. "Tailoring the Microarchitectures of 3D Printed Bone-like Scaffolds for Tissue Engineering Applications" Bioengineering 10, no. 5: 567. https://doi.org/10.3390/bioengineering10050567
APA StyleZenobi, E., Merco, M., Mochi, F., Ruspi, J., Pecci, R., Marchese, R., Convertino, A., Lisi, A., Del Gaudio, C., & Ledda, M. (2023). Tailoring the Microarchitectures of 3D Printed Bone-like Scaffolds for Tissue Engineering Applications. Bioengineering, 10(5), 567. https://doi.org/10.3390/bioengineering10050567







